Comparison of the Transcriptome Response within the Swine Tracheobronchial Lymphnode Following Infection with PRRSV, PCV-2 or IAV-S.
Abstract
:1. Introduction
2. Results
2.1. Clinical Evaluation and Gross Pathology
2.1.1. Serological Analysis
2.1.2. Quantitative PCR for Virus Nucleic Acid and Virus Isolation
2.2. Differentially Expressed Gene (DEG) Analysis During Infection
2.2.1. Venn Diagram Intersection of PRRSV, PCV-2, and IAV-S
2.2.2. Venn Diagram Intersection between PRRSV and PCV-2
2.2.3. Venn Diagram Intersection between PRRSV and IAV-S
2.3. Gene Set Enrichment Analysis and Pathway Analysis
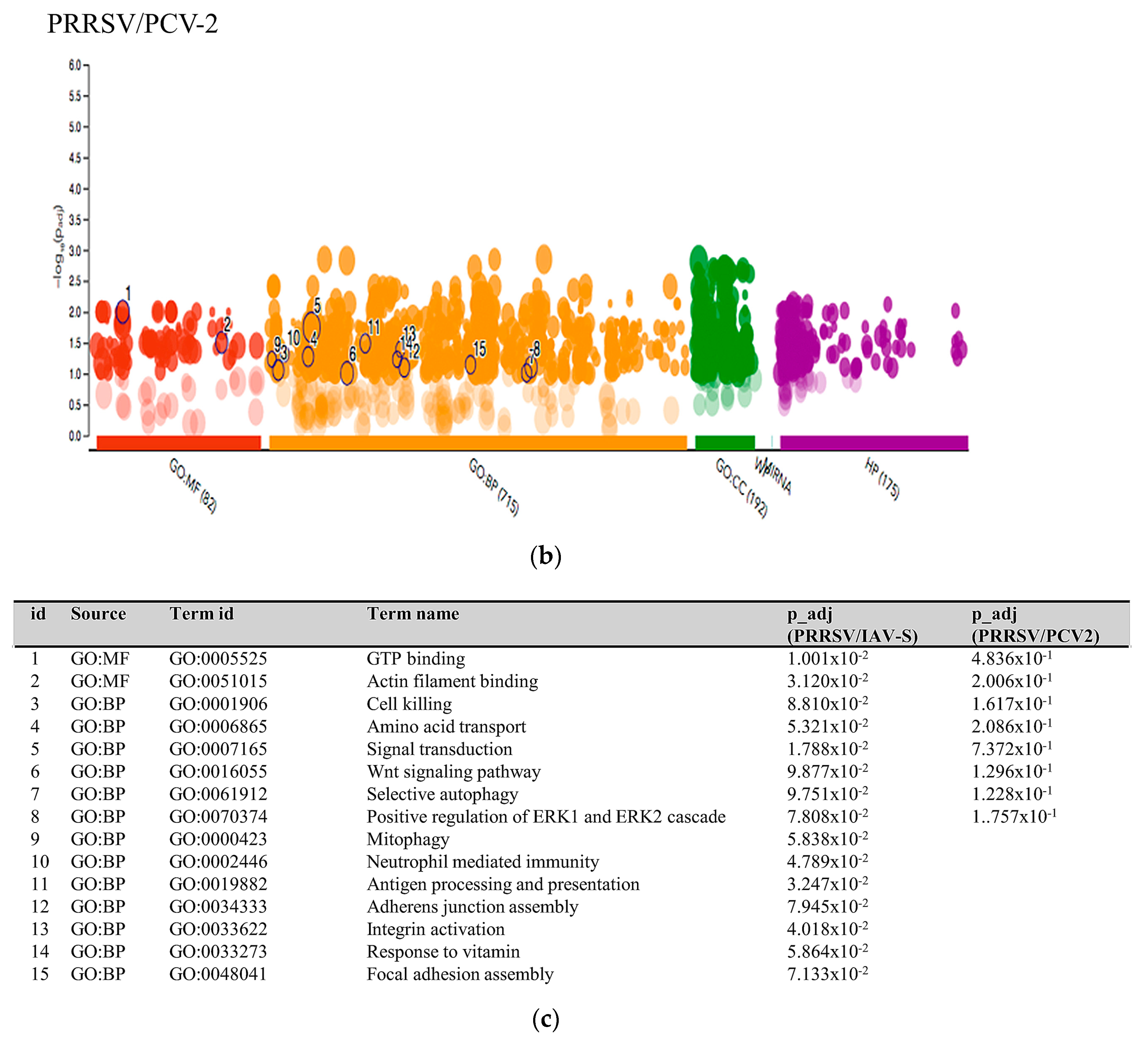
2.3.1. PRRSV and PCV-2 G.O. Analysis
2.3.2. PRRSV and IAV-S G.O. Analysis
2.3.3. Multiquery G.O. Analysis Comparison of PRRSV/PCV-2 vs. PRRSV/IAV-S
3. Discussion
3.1. PRRSV/PCV-2 Intersection
3.2. PRRSV/IAV-S Intersection
4. Materials and Methods
4.1. Ethics Statement
4.2. Virus, Animals and Experimental Design
4.3. Tissue Collection and Total RNA Isolation
4.4. Digital Gene Expression Tag Profiling (DGETP)
4.5. Tag Mapping and Alignment
4.6. Differential Expression Analysis
4.7. Gene Ontology and Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Holtkamp, D.J.; Kliebenstein, J.B.; Neumann, E.; Zimmerman, J.J.; Rotto, H.; Yoder, T.K.; Wang, C.; Yesk, P.E.; Mowrer, C.L.; Haley, C.A. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J. Swine Health Product. 2013, 21, 72. [Google Scholar]
- Nieuwenhuis, N.; Duinhof, T.F.; Van Nes, A. Economic analysis of outbreaks of porcine reproductive and respiratory syndrome virus in nine sow herds. Veter. Rec. 2012, 170, 225. [Google Scholar] [CrossRef] [PubMed]
- Simionatto, S.; Marchioro, S.B.; Maes, M.; Dellagostin, O.A. Mycoplasma hyopneumoniae: From disease to vaccine development. Veter. Microbiol. 2013, 165, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Dobrescu, I.; Levast, B.; Lai, K.; Delgado-Ortega, M.; Walker, S.; Banman, S.; Townsend, H.; Simon, G.; Zhou, Y.; Gerdts, V.; et al. In vitro and ex vivo analyses of co-infections with swine influenza and porcine reproductive and respiratory syndrome viruses. Veter. Microbiol. 2014, 169, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Gale, M.; Tan, S.-L.; Katze, M.G. Translational Control of Viral Gene Expression in Eukaryotes. Microbiol. Mol. Boil. Rev. 2000, 64, 239–280. [Google Scholar] [CrossRef] [Green Version]
- Walsh, D.; Mohr, I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Genet. 2011, 9, 860–875. [Google Scholar] [CrossRef]
- Miller, S.; Krijnse-Locker, J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Genet. 2008, 6, 363–374. [Google Scholar] [CrossRef]
- Guidotti, L.G.; Chisari, F.V. Noncytolytic Control Of Viral Infections By The Innate And Adaptive Immune Response. Annu. Rev. Immunol. 2001, 19, 65–91. [Google Scholar] [CrossRef]
- Barra, N.G.; Gillgrass, A.; Ashkar, A. Effective control of viral infections by the adaptive immune system requires assistance from innate immunity. Expert Rev. Vaccines 2010, 9, 1143–1147. [Google Scholar] [CrossRef]
- Glass, E.J.; Baxter, R.; Leach, R.J.; Jann, O.C. Genes controlling vaccine responses and disease resistance to respiratory viral pathogens in cattle. Vet. Immunol. Immunopathol. 2012, 148, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, A.; Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015, 16, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.C.; Bayles, D.O.; Zanella, E.L.; Lager, K.M. Effects of Pseudorabies Virus Infection on the Tracheobronchial Lymph Node Transcriptome. Bioinform. Biol. Insights. 2015, 9, 25–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, L.C.; Fleming, D.; Arbogast, A.; O Bayles, D.; Guo, B.; Lager, K.M.; Henningson, J.N.; Schlink, S.N.; Yang, H.-C.; Faaberg, K.S.; et al. Analysis of the swine tracheobronchial lymph node transcriptomic response to infection with a Chinese highly pathogenic strain of porcine reproductive and respiratory syndrome virus. BMC Veter- Res. 2012, 8, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.-J. Spread like a wildfire—The omnipresence of porcine circovirus type 2 (PCV2) and its ever-expanding association with diseases in pigs. Virus Res. 2012, 164, 1–3. [Google Scholar] [CrossRef]
- Rajao, D.S.; Vincent, A.L. Swine as a Model for Influenza A Virus Infection and Immunity. ILAR J. 2015, 56, 44–52. [Google Scholar] [CrossRef]
- Talker, S.C.; Stadler, M.; Koinig, H.C.; Mair, K.H.; Rodríguez-Gómez, I.M.; Graage, R.; Zell, R.; Dürrwald, R.; Starick, E.; Harder, T.; et al. Influenza A Virus Infection in Pigs Attracts Multifunctional and Cross-Reactive T Cells to the Lung. J. Virol. 2016, 90, 9364–9382. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.-W.; Jeng, C.-R.; Lin, T.-L.; Liu, J.J.; Chiou, M.-T.; Tsai, Y.-C.; Chia, M.-Y.; Jan, T.-R.; Pang, V.F. Immunopathological effects of porcine circovirus type 2 (PCV2) on swine alveolar macrophages by in vitro inoculation. Veter. Immunol. Immunopathol. 2006, 110, 207–219. [Google Scholar] [CrossRef]
- Segalés, J. Porcine circovirus type 2 (PCV2) infections: Clinical signs, pathology and laboratory diagnosis. Virus Res. 2012, 164, 10–19. [Google Scholar] [CrossRef]
- Borghetti, P.; Morganti, M.; Saleri, R.; Ferrari, L.; De Angelis, E.; Cavalli, V.; Cacchioli, A.; Corradi, A.; Martelli, P. Innate pro-inflammatory and adaptive immune cytokines in PBMC of vaccinated and unvaccinated pigs naturally exposed to porcine circovirus type 2 (PCV2) infection vary with the occurrence of the disease and the viral burden. Veter. Microbiol. 2013, 163, 42–53. [Google Scholar] [CrossRef]
- Harbers, M.; Carninci, P. Tag-based approaches for transcriptome research and genome annotation. Nat. Methods 2005, 2, 495–502. [Google Scholar] [CrossRef]
- Morrissy, S.; Zhao, Y.; Delaney, A.; Asano, J.; Dhalla, N.; Li, I.; McDonald, H.; Pandoh, P.; Prabhu, A.-L.; Tam, A.; et al. Digital Gene Expression by Tag Sequencing on the Illumina Genome Analyzer. Curr. Protoc. Hum. Genet. 2010, 65, 11.11.1–11.11.36. [Google Scholar] [CrossRef] [PubMed]
- De Lorgeril, J.; Zenagui, R.; Rosa, R.D.; Piquemal, D.; Bachère, E. Whole Transcriptome Profiling of Successful Immune Response to Vibrio Infections in the Oyster Crassostrea gigas by Digital Gene Expression Analysis. PLoS ONE 2011, 6, 23142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, W.; Olson, E.N.; Moore, S.S.; Basarab, J.A.; Basu, U.; Guan, L.L. Transcriptome analysis of subcutaneous adipose tissues in beef cattle using 3′ digital gene expression-tag profiling1. J. Anim. Sci. 2012, 90, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Lager, K.M.; Gauger, P.C.; Vincent, A.L.; Opriessnig, T.; Kehrli, M.E.; Cheung, A.K. Mortality in pigs given porcine circovirus type 2 subgroup 1 and 2 viruses derived from DNA clones. Veter. Rec. 2007, 161, 428–429. [Google Scholar] [CrossRef] [PubMed]
- Mengeling, W.L.; Lager, K.M.; Vorwald, A.C. Clinical consequences of exposing pregnant gilts to strains of porcine reproductive and respiratory syndrome (PRRS) virus isolated from field cases of “atypical” PRRS. Am. J Vet. Res. 1998, 59, 1540–1544. [Google Scholar] [PubMed]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Gramer, M.R.; Richt, J.A.; Janke, B.H. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes 2009, 39, 176–185. [Google Scholar] [CrossRef]
- UniProt C. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Jenuth, J.P. The NCBI. Publicly available tools and resources on the Web. Methods Mol. Boil. (Clifton, N.J.) 2000, 132, 301–312. [Google Scholar]
- The UniProt C. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Miller, L.C.; Fleming, D.S.; Li, X.; Bayles, D.O.; Blecha, F.; Sang, Y. Comparative analysis of signature genes in PRRSV-infected porcine monocyte-derived cells to different stimuli. PLoS ONE 2017, 12, e0181256. [Google Scholar] [CrossRef]
- Chen, Q.; Kang, J.; Fu, C. The independence of and associations among apoptosis, autophagy, and necrosis. Signal Transduct. Target. Ther. 2018, 3, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, J.J.Y.; Pung, Y.F.; Sze, N.S.-K.; Chin, K.-C. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc. Natl. Acad. Sci. USA 2006, 103, 10735–10740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Brown, G.R.; Hem, V.; Katz, K.S.; Ovetsky, M.; Wallin, C.; Ermolaeva, O.; Tolstoy, I.; Tatusova, T.; Pruitt, K.D.; Maglott, D.R.; et al. Gene: A gene-centered information resource at NCBI. Nucleic Acids Res. 2015, 43, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Joshi, T.; Butchar, J.; Tridandapani, S. Fcγ Receptor Signaling in Phagocytes. Int. J. Hematol. 2006, 84, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qin, Y.; Chen, M. Viral strategies for triggering and manipulating mitophagy. Autophagy 2018, 14, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Wang, J.; Zhou, A.; Khan, F.A.; Hu, L.; Zhang, S. Porcine reproductive and respiratory syndrome virus triggers mitochondrial fission and mitophagy to attenuate apoptosis. Oncotarget 2016, 7, 56002–56012. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Chen, Z.; Zhao, J.; Fang, L.; Fang, R.; Xiao, J.; Chen, X.; Zhou, A.; Zhang, Y.; Ren, L.; et al. Difference in microRNA expression and editing profile of lung tissues from different pig breeds related to immune responses to HP-PRRSV. Sci. Rep. 2015, 5, 9549. [Google Scholar] [CrossRef] [Green Version]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S.; AmiGO, H.; Web Presence Working Group. AmiGO: Online access to ontology and annotation data. Bioinform. 2009, 25, 288–289. [Google Scholar] [CrossRef]
- Moreth, K.; Iozzo, R.V.; Schaefer, L. Small leucine-rich proteoglycans orchestrate receptor crosstalk during inflammation. Cell Cycle 2012, 11, 2084–2091. [Google Scholar] [CrossRef] [Green Version]
- Nikitovic, D.; Papoutsidakis, A.; Karamanos, N.K.; Tzanakakis, G.N. Lumican affects tumor cell functions, tumor–ECM interactions, angiogenesis and inflammatory response. Matrix Boil. 2014, 35, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.L.; Turner, R.B.; Braciale, T.; Heymann, P.W.; Borish, L. Pathogenesis of rhinovirus infection. Curr. Opin. Virol. 2012, 2, 287–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borregaard, N.; Sørensen, O.E.; Theilgaard-Mönch, K. Neutrophil granules: A library of innate immunity proteins. Trends Immunol. 2007, 28, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Camp, J.V.; Jonsson, C.B. A Role for Neutrophils in Viral Respiratory Disease. Front. Immunol. 2017, 8, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, J.; Wang, L.; Quan, R.; Fu, Y.; Zhang, H.; Feng, W.-H. Induction of interleukin-10 is dependent on p38 mitogen-activated protein kinase pathway in macrophages infected with porcine reproductive and respiratory syndrome virus. Virol. J. 2012, 9, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lager, K.M.; Halbur, P.G. Gross and Microscopic Lesions in Porcine Fetuses Infected with Porcine Reproductive and Respiratory Syndrome Virus. J. Veter- Diagn. Investig. 1996, 8, 275–282. [Google Scholar] [CrossRef] [Green Version]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Janke, B.H.; Webby, R.J.; García-Sastre, A.; Richt, J.A. Efficacy of intranasal administration of a truncated NS1 modified live influenza virus vaccine in swine. Vaccine 2007, 25, 7999–8009. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinform. 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. Feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinform. 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- I Love, M.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Boil 2014, 15, 002832. [Google Scholar] [CrossRef] [Green Version]
- Afgan, E.; Baker, D.; Beek, M.V.D.; Blankenberg, D.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Forner, O.; Marin-Garcia, P.; Arnau, V.; D’Eustachio, P.; Stein, L.; Hermjakob, H. Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinform. 2017, 18, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. g:Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef]







| Group | 3 dpi | 6 dpi | 14 dpi |
|---|---|---|---|
| Sham | 0.34 ± 0.3 | 0.14 ± 0.1 | 0 ± 0 |
| PCV-2 | 0.11 ± 0.1 | 1.36 ± 1.2 | 1.04 ± 0.6 |
| PRRSV | 5.27 ± 1.6 | 8.60 ± 3.4 | 57.1 ± 7.8 |
| IAV-S | 8.96 ± 1.4 | 31.5 ± 2.0 | 1.30 ± 0.5 |
| Tissue | Treatment Group | Virus Tested for | 0 dpi | 1 dpi | 3 dpi | 6 dpi | 14 dpi |
|---|---|---|---|---|---|---|---|
| Serum | Control | PCV-2 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Serum | Control | PRRSV | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Serum | Control | IAV-S | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Serum | PCV-2 | PCV-2 | 0/5 | 4/5 | 0/5 | 0/5 | 0/5 |
| Serum | PRRSV | PRRSV | 0/5 | 5/5 | 5/5 | 5/5 | 5/5 |
| Serum | IAV-S | IAV-S | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| BALF | Control | PCV-2 | 0/5 | 0/5 | 0/5 | 0/5 | |
| BALF | Control | PRRSV | 0/5 | 0/5 | 0/5 | 0/5 | |
| BALF | Control | IAV | 0/5 | 0/5 | 0/5 | 0/5 | |
| BALF | PCV-2 | PCV-2 | 4/5 | 5/5 | 4/5 | 4/5 | |
| BALF | PRRSV | PRRSV | 3/5 | 5/5 | 5/5 | 5/5 | |
| BALF | IAV-S | IAV-S | 0/5 | 1/5 | 1/5 | 0/5 | |
| TBLN | Control | PCV-2 | 0/5 | 0/5 | 0/5 | 0/5 | |
| TBLN | Control | PRRSV | 0/5 | 0/5 | 0/5 | 0/5 | |
| TBLN | Control | IAV-S | 0/5 | 0/5 | 0/5 | 0/5 | |
| TBLN | PCV-2 | PCV-2 | 2/5 | 3/5 | 3/5 | 4/5 | |
| TBLN | PRRSV | PRRSV | 5/5 | 5/5 | 5/5 | 5/5 | |
| TBLN | IAV-S | IAV-S | 0/5 | 1/5 | 0/5 | 0/5 |
| Gene Name | PRRSV (Log2FC) | IAV-S (Log2FC) | PCV-2 (Log2FC) | Biological Function(s) |
|---|---|---|---|---|
| AK2 | 1.21 | 0.81 | 0.51 | Cellular energy homeostasis and hematopoiesis |
| GOLGA2 | −1.51 | −1.58 | −1.00 | Negative regulation of autophagy; cadherin binding |
| GPSM3 | 1.64 | 1.75 | 0.92 | Positive regulation of cytokine production involved in inflammatory response, positive regulation of leukocyte chemotaxis |
| LGALS1 | 1.34 | 1.00 | 0.66 | Apoptotic process, modulates cell-cell and cell-matrix interactions; T cell costimulation; positive regulation of viral entry into host cell; positive regulation of I-kappaB kinase/NF-kappaB signaling |
| LOC100525318 (solute carrier family 23 member 1-like) | 2.17 | 2.53 | 0.86 | N/A |
| LOC110260088 (eukaryotic translation initiation factor 3 subunit C) | 2.94 | 2.13 | 1.12 | N/A |
| PHB2 | 1.41 | 1.33 | 0.73 | Negative regulation of apoptotic process, response to wounding, |
| POLR2E | 1.14 | 0.96 | 0.78 | Viral process |
| PRDX1 | −0.85 | −0.93 | −0.52 | Antioxidant enzyme, cadherin binding; regulation of NIK/NF-kappaB signaling; natural killer cell activation |
| RBM48 | 1.34 | 1.06 | 0.6 | N/A |
| SH3GLB1 | −0.82 | −0.99 | −0.63 | Autophagy; regulation of cytokinesis |
| SLC26A2 | 0.87 | 1.25 | 0.64 | Critical in cartilage for sulfation of proteoglycans and matrix organization, regulation of membrane potential |
| Gene Name | PRRSV (Log2FC) | PCV-2 (Log2FC) | Biological Function(s) |
|---|---|---|---|
| MED15 | 1.90 | 1.03 | Transcriptional coactivator in RNA pol-II transcription, cholesterol-dependent gene regulation |
| POLR2H | 1.70 | 0.72 | Encodes an essential conserved subunit of RNA poly-I, II, and III |
| TMEM50B | 1.26 | 0.79 | Late endosome to vacuole transport via multivesicular body sorting pathway |
| FOLR1 | 1.15 | 0.58 | Regulation of canonical Wnt signaling, TGF-B receptor, and folate binding pathways |
| STK35 | 1.15 | 0.67 | Protein phosphorylation, protein serine/threonine kinase activity |
| RANBP3 | 1.12 | 0.51 | Nuclear export, negative regulator of TGF-Beta signaling |
| TIGAR | 1.08 | 0.65 | Protects cells from reactive oxygen species (ROS) and DNA damage-induced apoptosis; autophagy |
| WDR1 | 0.99 | 0.64 | protein-protein interactions |
| RRP9 | 0.93 | 0.54 | Component of the nucleolar small nuclear ribonucleoprotein particles (snoRNP), ribosome synthesis |
| GDI1 | 0.86 | 0.41 | Regulates the GDP-GTP exchange reaction, involved cellular trafficking in organelles |
| PPFIA3 | 0.83 | 0.49 | Focal adhesion disassembly |
| RPL27A | −0.59 | −0.43 | Structural constituent of ribosome, cytoplasmic translation |
| CCDC107 | −0.87 | −0.50 | N/A |
| RBBP4 | −0.92 | −0.47 | Helps form co-repressor complexes involved in transcriptional silencing, histone binding |
| TMEM128 | −1.06 | −0.60 | Transmembrane protein |
| LCP1 | −1.08 | −0.55 | Actin and Integrin binding; T-cell activation, Extracellular matrix disassembly |
| SPG21 | −1.10 | −0.62 | Binds to CD4 for repression of T-cell activation, stimulatory activity of CD4, antigen receptor-mediated signaling |
| RAB4A | −1.17 | −0.47 | Membrane trafficking regulation, endosome sorting and recycling |
| APBB2 | −1.21 | −0.67 | Extracellular matrix organization, positive regulation of apoptotic process |
| TOPBP1 | −1.29 | −0.69 | N/A |
| RAB11A | −1.34 | −0.70 | Regulation of membrane delivery during cytokinesis, exocytosis |
| NCKAP1 | −1.45 | −0.52 | Rac GTPase binding, apoptotic process, viral process |
| NSA2 | −1.51 | −0.55 | Involved in cell cycle regulation and proliferation, RNA binding |
| TCF12 | −1.52 | −0.54 | Immune response, transcriptional activator, SMAD binding |
| MYOF | −1.62 | −0.74 | Plasma membrane regeneration and repair, phospholipid binding, cellular response to heat |
| PITHD1 | −1.75 | −0.95 | N/A |
| CDH5 (CD144) | −1.76 | −0.73 | Regulation of complement-dependent cytotoxicity, homophilic cell adhesion via plasma membrane adhesion molecules; negative regulation of inflammatory response |
| CPZ | −1.76 | −0.73 | Wnt signaling pathway, metallocarboxypeptidase activity |
| RAB18 | −1.76 | −1.15 | Membrane trafficking, immune functions |
| FERMT2 | −1.91 | −0.68 | Focal adhesion, integrin activation, cell-cell communication |
| SPART | −2.00 | −0.64 | Ubiquitin protein ligase binding, negative regulation of BMP signaling pathway |
| COL5A2 | −2.03 | −0.59 | Collagen formation, SMAD binding, extracellular matrix organization |
| SDC2 | −2.24 | −0.77 | Cell binding, cell signaling, and cytoskeletal organization, leukocyte migration |
| DAAM1 | −2.38 | −0.76 | Rho GTPase binding |
| LUM | −2.38 | −1.00 | Extracellular matrix structural constituent, collagen binding, damage associated molecular pattern signaler (DAMPs) |
| NTRK2 | −2.82 | −1.37 | MAP kinase activity, cellular response to amino acid stimulus |
| GORAB | −3.61 | −1.12 | Protein binding |
| Gene Name | PRRSV (Log2FC) | IAV-S (Log2FC) | Biological Function(s) |
|---|---|---|---|
| ABHD17B | −1.09 | −1.09 | Palmitoyl-(protein) hydrolase activity, regulation of dendritic spine maintenance |
| ACKR4 | −2.19 | −1.97 | Receptor for C-C type chemokines; bind dendritic cell- and T cell-activated chemokines; immune response, chemotaxis |
| ACLY | −2.59 | −2.46 | Catalyzes the formation of acetyl-CoA and oxaloacetate, neutrophil degranulation, positive regulation of cellular metabolic process |
| AIP | 2.17 | 2.14 | Encoded protein can bind specifically to and inhibit the activity of hepatitis B virus; interleukin-12-mediated signaling pathway |
| CCL11 | 2.07 | 1.37 | Antimicrobial chemokine involved in immunoregulatory and inflammatory processes; Monocyte, neutrophil, and lymphocyte chemotaxis, chronic inflammatory response |
| CD5 | 0.72 | 1.54 | Type-I transmembrane glycoprotein found on the surface of thymocytes, T lymphocytes and a subset of B lymphocytes; may be involved in T cell proliferation. T cell costimulation, apoptotic signaling pathway, scavenger receptor activity |
| CLRN1 | −3.08 | −2.81 | May be important in development and homeostasis of the inner ear and retina, actin filament organization |
| CMPK1 | −0.69 | −1.54 | Encodes one of the enzymes required for cellular nucleic acid biosynthesis, nucleoside monophosphate kinase activity |
| CTNNAL1 | −1.12 | −0.98 | Cell adhesion |
| EIF3K | 1.45 | 1.24 | Contributes to translation initiation factor activity, regulation of translational initiation |
| EMP3 | 1.00 | 1.04 | Involved in cell proliferation, cell-cell interactions and function as a tumor suppressor, negative regulation of cell proliferation |
| ETV6 | 1.38 | 1.60 | Transcription factor, negative regulation of transcription by RNA polymerase II, |
| FASN | −1.50 | −1.36 | Oxidation-reduction process, cellular response to interleukin-4, cadherin binding |
| GAPDH | 1.35 | 0.99 | Regulation of macroautophagy, antimicrobial humoral immune response mediated by antimicrobial peptide, positive regulation of cytokine secretion |
| GINS2 | 2.79 | 2.54 | Double-strand break repair via break-induced replication, double-strand break repair via break-induced replication |
| GRHPR | 2.55 | 2.93 | Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; role in metabolism |
| HERC5 | 2.88 | 2.48 | Defense response to virus, negative regulation of type I interferon production, ISG15-protein conjugation, ubiquitin-protein transferase activity |
| ILK | 1.08 | 1.25 | Regulates integrin-mediated signal transduction, MAPK cascade, cell-matrix adhesion, negative regulation of cardiac muscle cell apoptotic process |
| KDELC2 | 1.62 | 1.57 | Protein O-linked glycosylation via serine |
| COX4I1 | 0.77 | 0.73 | Mitochondrial electron transport, cytochrome c to oxygen |
| LRFN1 | −2.60 | −2.15 | Integral component of membrane |
| LRRC8D | 0.83 | 1.26 | Protein binding, cell volume homeostasis |
| MAPK14 | 2.33 | 2.00 | MAP kinase activity, activated by various environmental stresses and proinflammatory cytokines, roles of in stress related transcription and cell cycle regulation, as well as in genotoxic stress response |
| MIER1 | 0.92 | 1.62 | Transcriptional regulator |
| MINDY3 | 1.21 | 1.47 | Deubiquitinase, apoptotic process, Lys48-specific deubiquitinase activity |
| NCSTN | 0.89 | 1.23 | T cell proliferation, neutrophil degranulation |
| NFIX | 1.30 | 1.24 | Transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific DNA binding |
| NUP188 | 1.50 | 1.65 | Viral process, structural constituent of nuclear pore |
| OIP5 | 1.12 | 1.56 | CENP-A containing nucleosome assembly, protein binding |
| PLA2G2D | 1.40 | 1.19 | Gene may be involved in inflammation and immune response, inflammatory response, negative regulation of T cell proliferation, heparan sulfate proteoglycan binding |
| PRPF3 | 1.01 | 0.83 | Pre-mRNA processing factor, mRNA splicing, via spliceosome |
| PRPF8 | −3.25 | −2.43 | Cellular response to lipopolysaccharide, cellular response to tumor necrosis factor, mRNA splicing, via spliceosome |
| PTDSS1 | 1.52 | 1.57 | Transferase activity, phosphatidylserine biosynthetic process |
| RAB7A | 1.07 | 1.26 | Phagosome-lysosome fusion, neutrophil degranulation, antigen processing and presentation of exogenous peptide antigen via MHC class II (ALL HUMAN) |
| RBM26 | −1.37 | −1.22 | Regulation of mRNA processing |
| RFC2 | 1.11 | 1.32 | Binding ATP and may help promote cell survival; DNA damage response, detection of DNA damage |
| RPF1 | −0.71 | −1.15 | rRNA processing |
| SMARCA5 | −1.16 | −1.67 | Regulation of transcription by RNA polymerase II, cellular response to leukemia inhibitory factor |
| SNX8 | 2.25 | 2.90 | Intracellular protein transport, phosphatidylinositol binding |
| SORBS2 | −3.49 | −3.29 | Actin filament organization, cytoskeletal adaptor activity |
| SZRD1 | 1.42 | 1.15 | N/A |
| TSEN54 | 1.08 | 1.00 | Removal of introns from precursor tRNAs, tRNA splicing, via endonucleolytic cleavage and ligation |
| TXNDC5 | 3.30 | 1.75 | Role may be to protect hypoxic cells from apoptosis, cell redox homeostasis, negative regulation of apoptotic process, neutrophil degranulation |
| UBE2V1 | 1.29 | 1.12 | Positive regulation of NF-kappaB transcription factor activity, activation of MAPK activity, positive regulation of I-kappaB kinase/NF-kappaB signaling |
| UTRN | −2.66 | −2.57 | Positive regulation of cell-matrix adhesion, integrin binding |
| WDFY2 | 1.13 | 0.82 | Positive regulation of fat cell differentiation, positive regulation of protein phosphorylation |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, L.C.; Fleming, D.S.; Lager, K.M. Comparison of the Transcriptome Response within the Swine Tracheobronchial Lymphnode Following Infection with PRRSV, PCV-2 or IAV-S. Pathogens 2020, 9, 99. https://doi.org/10.3390/pathogens9020099
Miller LC, Fleming DS, Lager KM. Comparison of the Transcriptome Response within the Swine Tracheobronchial Lymphnode Following Infection with PRRSV, PCV-2 or IAV-S. Pathogens. 2020; 9(2):99. https://doi.org/10.3390/pathogens9020099
Chicago/Turabian StyleMiller, Laura C., Damarius S. Fleming, and Kelly M. Lager. 2020. "Comparison of the Transcriptome Response within the Swine Tracheobronchial Lymphnode Following Infection with PRRSV, PCV-2 or IAV-S." Pathogens 9, no. 2: 99. https://doi.org/10.3390/pathogens9020099
APA StyleMiller, L. C., Fleming, D. S., & Lager, K. M. (2020). Comparison of the Transcriptome Response within the Swine Tracheobronchial Lymphnode Following Infection with PRRSV, PCV-2 or IAV-S. Pathogens, 9(2), 99. https://doi.org/10.3390/pathogens9020099




