Orthohepevirus C: An Expanding Species of Emerging Hepatitis E Virus Variants
Abstract
1. Introduction
2. Detection and Distribution of Orthohepevirus C
2.1. Seroprevalence of Anti-HEV Antibodies
2.2. Detection of Orthohepevirus C Genomes
3. Genomic Characterization of Orthohepevirus C
4. Genetic Variability of Orthohepevirus C
5. Molecular Evolution of Orthohepevirus C
6. Cross-Species Transmission of Orthohepevirus C
7. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Rein, D.B.; Stevens, G.A.; Theaker, J.; Wittenborn, J.S.; Wiersma, S.T. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology 2012, 55, 988–997. [Google Scholar] [CrossRef] [PubMed]
- Bose, P.D.; Das, B.C.; Kumar, A.; Gondal, R.; Kumar, D.; Kar, P. High viral load and deregulation of the progesterone receptor signaling pathway: Association with hepatitis E-related poor pregnancy outcome. J. Hepatol. 2011, 54, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Pavio, N.; Meng, X.J.; Doceul, V. Zoonotic origin of hepatitis E. Curr. Opin. Virol. 2015, 10, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Izopet, J.; Tripon, S.; Bismuth, M.; Hillaire, S.; Dumortier, J.; Radenne, S.; Coilly, A.; Garrigue, V.; D’Alteroche, L.; et al. Ribavirin for chronic hepatitis E virus infection in transplant recipients. N. Engl. J. Med. 2014, 370, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Pischke, S.; Hartl, J.; Pas, S.D.; Lohse, A.W.; Jacobs, B.C.; Van der Eijk, A.A. Hepatitis E virus: Infection beyond the liver? J. Hepatol. 2017, 66, 1082–1095. [Google Scholar] [CrossRef] [PubMed]
- Balayan, M.S.; Andjaparidze, A.G.; Savinskaya, S.S.; Ketiladze, E.S.; Braginsky, D.M.; Savinov, A.P.; Poleschuk, V.F. Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal-oral route. Intervirology 1983, 20, 23–31. [Google Scholar] [CrossRef]
- Tam, A.W.; Smith, M.M.; Guerra, M.E.; Huang, C.C.; Bradley, D.W.; Fry, K.E.; Reyes, G.R. Hepatitis E virus (HEV): Molecular cloning and sequencing of the full-length viral genome. Virology 1991, 185, 120–131. [Google Scholar] [CrossRef]
- Purdy, M.A.; Harrison, T.J.; Jameel, S.; Meng, X.J.; Okamoto, H.; Van der Poel, W.H.M.; Smith, D.B.; Ictv Report, C. ICTV Virus Taxonomy Profile: Hepeviridae. J. Gen. Virol. 2017, 98, 2645–2646. [Google Scholar] [CrossRef]
- Montpellier, C.; Wychowski, C.; Sayed, I.M.; Meunier, J.C.; Saliou, J.M.; Ankavay, M.; Bull, A.; Pillez, A.; Abravanel, F.; Helle, F.; et al. Hepatitis E Virus Lifecycle and Identification of 3 Forms of the ORF2 Capsid Protein. Gastroenterology 2018, 154, e218. [Google Scholar] [CrossRef]
- Rasche, A.; Sander, A.L.; Corman, V.M.; Drexler, J.F. Evolutionary biology of human hepatitis viruses. J. Hepatol. 2019, 70, 501–520. [Google Scholar] [CrossRef]
- Smith, D.B.; Simmonds, P.; International Committee on Taxonomy of Viruses Hepeviridae Study Group; Jameel, S.; Emerson, S.U.; Harrison, T.J.; Meng, X.J.; Okamoto, H.; Van der Poel, W.H.; Purdy, M.A. Consensus proposals for classification of the family Hepeviridae. J. Gen. Virol. 2014, 95, 2223–2232. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Simmonds, P.; Izopet, J.; Oliveira-Filho, E.F.; Ulrich, R.G.; Johne, R.; Koenig, M.; Jameel, S.; Harrison, T.J.; Meng, X.J.; et al. Proposed reference sequences for hepatitis E virus subtypes. J. Gen. Virol. 2016, 97, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Kenney, S.P. The Current Host Range of Hepatitis E Viruses. Viruses 2019, 11, 452. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Yip, C.C.Y.; Wu, S.; Cai, J.; Zhang, A.J.; Leung, K.H.; Chung, T.W.H.; Chan, J.F.W.; Chan, W.M.; Teng, J.L.L.; et al. Rat Hepatitis E Virus as Cause of Persistent Hepatitis after Liver Transplant. Emerg. Infect. Dis. 2018, 24, 2241–2250. [Google Scholar] [CrossRef] [PubMed]
- Andonov, A.; Robbins, M.; Borlang, J.; Cao, J.; Hatchette, T.; Stueck, A.; Deschambault, Y.; Murnaghan, K.; Varga, J.; Johnston, L. Rat Hepatitis E Virus Linked to Severe Acute Hepatitis in an Immunocompetent Patient. J. Infect. Dis. 2019, 220, 951–955. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Yip, C.C.; Wu, S.; Chew, N.F.; Leung, K.H.; Chan, J.F.; Zhao, P.S.; Chan, W.M.; Poon, R.W.; Tsoi, H.W.; et al. Transmission of rat hepatitis E virus infection to humans in Hong Kong: A clinical and epidemiological analysis. Hepatology 2020. [Google Scholar] [CrossRef]
- Kabrane-Lazizi, Y.; Fine, J.B.; Elm, J.; Glass, G.E.; Higa, H.; Diwan, A.; Gibbs, C.J., Jr.; Meng, X.J.; Emerson, S.U.; Purcell, R.H. Evidence for widespread infection of wild rats with hepatitis E virus in the United States. Am. J. Trop. Med. Hyg. 1999, 61, 331–335. [Google Scholar] [CrossRef]
- Favorov, M.O.; Kosoy, M.Y.; Tsarev, S.A.; Childs, J.E.; Margolis, H.S. Prevalence of antibody to hepatitis E virus among rodents in the United States. J. Infect. Dis. 2000, 181, 449–455. [Google Scholar] [CrossRef]
- Johne, R.; Heckel, G.; Plenge-Bonig, A.; Kindler, E.; Maresch, C.; Reetz, J.; Schielke, A.; Ulrich, R.G. Novel hepatitis E virus genotype in Norway rats, Germany. Emerg. Infect. Dis. 2010, 16, 1452–1455. [Google Scholar] [CrossRef]
- Johne, R.; Plenge-Bonig, A.; Hess, M.; Ulrich, R.G.; Reetz, J.; Schielke, A. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J. Gen. Virol. 2010, 91, 750–758. [Google Scholar] [CrossRef]
- Easterbrook, J.D.; Kaplan, J.B.; Vanasco, N.B.; Reeves, W.K.; Purcell, R.H.; Kosoy, M.Y.; Glass, G.E.; Watson, J.; Klein, S.L. A survey of zoonotic pathogens carried by Norway rats in Baltimore, Maryland, USA. Epidemiol. Infect. 2007, 135, 1192–1199. [Google Scholar] [CrossRef] [PubMed]
- Hirano, M.; Ding, X.; Li, T.C.; Takeda, N.; Kawabata, H.; Koizumi, N.; Kadosaka, T.; Goto, I.; Masuzawa, T.; Nakamura, M.; et al. Evidence for widespread infection of hepatitis E virus among wild rats in Japan. Hepatol. Res. 2003, 27, 1–5. [Google Scholar] [CrossRef]
- Kanai, Y.; Miyasaka, S.; Uyama, S.; Kawami, S.; Kato-Mori, Y.; Tsujikawa, M.; Yunoki, M.; Nishiyama, S.; Ikuta, K.; Hagiwara, K. Hepatitis E virus in Norway rats (Rattus norvegicus) captured around a pig farm. BMC Res. Notes 2012, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Johne, R.; Dremsek, P.; Kindler, E.; Schielke, A.; Plenge-Bonig, A.; Gregersen, H.; Wessels, U.; Schmidt, K.; Rietschel, W.; Groschup, M.H.; et al. Rat hepatitis E virus: Geographical clustering within Germany and serological detection in wild Norway rats (Rattus norvegicus). Infect. Genet. Evol. 2012, 12, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Guan, D.; Su, J.; Takeda, N.; Wakita, T.; Li, T.C.; Ke, C.W. High prevalence of rat hepatitis E virus in wild rats in China. Vet. Microbiol. 2013, 165, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Li, T.C.; Yoshimatsu, K.; Yasuda, S.P.; Arikawa, J.; Koma, T.; Kataoka, M.; Ami, Y.; Suzaki, Y.; Mai le, T.Q.; Hoa, N.T.; et al. Characterization of self-assembled virus-like particles of rat hepatitis E virus generated by recombinant baculoviruses. J. Gen. Virol. 2011, 92, 2830–2837. [Google Scholar] [CrossRef]
- Koma, T.; Yoshimatsu, K.; Yasuda, S.P.; Li, T.; Amada, T.; Shimizu, K.; Isozumi, R.; Mai, L.T.; Hoa, N.T.; Nguyen, V.; et al. A survey of rodent-borne pathogens carried by wild Rattus spp. in Northern Vietnam. Epidemiol. Infect. 2013, 141, 1876–1884. [Google Scholar] [CrossRef]
- Obana, S.; Shimizu, K.; Yoshimatsu, K.; Hasebe, F.; Hotta, K.; Isozumi, R.; Nguyen, H.T.; Le, M.Q.; Yamashiro, T.; Tsuda, Y.; et al. Epizootiological study of rodent-borne hepatitis E virus HEV-C1 in small mammals in Hanoi, Vietnam. J. Vet. Med. Sci. 2017, 79, 76–81. [Google Scholar] [CrossRef][Green Version]
- Simanavicius, M.; Juskaite, K.; Verbickaite, A.; Jasiulionis, M.; Tamosiunas, P.L.; Petraityte-Burneikiene, R.; Zvirbliene, A.; Ulrich, R.G.; Kucinskaite-Kodze, I. Detection of rat hepatitis E virus, but not human pathogenic hepatitis E virus genotype 1-4 infections in wild rats from Lithuania. Vet. Microbiol. 2018, 221, 129–133. [Google Scholar] [CrossRef]
- Mulyanto; Depamede, S.N.; Sriasih, M.; Takahashi, M.; Nagashima, S.; Jirintai, S.; Nishizawa, T.; Okamoto, H. Frequent detection and characterization of hepatitis E virus variants in wild rats (Rattus rattus) in Indonesia. Arch. Virol. 2013, 158, 87–96. [Google Scholar] [CrossRef]
- Mulyanto; Suparyatmo, J.B.; Andayani, I.G.; Khalid; Takahashi, M.; Ohnishi, H.; Jirintai, S.; Nagashima, S.; Nishizawa, T.; Okamoto, H. Marked genomic heterogeneity of rat hepatitis E virus strains in Indonesia demonstrated on a full-length genome analysis. Virus Res. 2014, 179, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Primadharsini, P.P.; Mulyanto; Wibawa, I.D.N.; Anggoro, J.; Nishizawa, T.; Takahashi, M.; Jirintai, S.; Okamoto, H. The identification and characterization of novel rat hepatitis E virus strains in Bali and Sumbawa, Indonesia. Arch. Virol. 2018, 163, 1345–1349. [Google Scholar] [CrossRef] [PubMed]
- Arankalle, V.A.; Joshi, M.V.; Kulkarni, A.M.; Gandhe, S.S.; Chobe, L.P.; Rautmare, S.S.; Mishra, A.C.; Padbidri, V.S. Prevalence of anti-hepatitis E virus antibodies in different Indian animal species. J. Viral. Hepat. 2001, 8, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Vitral, C.L.; Pinto, M.A.; Lewis-Ximenez, L.L.; Khudyakov, Y.E.; dos Santos, D.R.; Gaspar, A.M. Serological evidence of hepatitis E virus infection in different animal species from the Southeast of Brazil. Memorias do Instituto Oswaldo Cruz 2005, 100, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Li, W.; Su, J.; Fang, L.; Takeda, N.; Wakita, T.; Li, T.C.; Ke, C. Asian musk shrew as a reservoir of rat hepatitis E virus, China. Emerg. Infect. Dis. 2013, 19, 1341–1343. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lu, L.; Du, J.; Yang, L.; Ren, X.; Liu, B.; Jiang, J.; Yang, J.; Dong, J.; Sun, L.; et al. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 2018, 6, 178. [Google Scholar] [CrossRef]
- Purcell, R.H.; Engle, R.E.; Rood, M.P.; Kabrane-Lazizi, Y.; Nguyen, H.T.; Govindarajan, S.; St Claire, M.; Emerson, S.U. Hepatitis E virus in rats, Los Angeles, California, USA. Emerg. Infect. Dis. 2011, 17, 2216–2222. [Google Scholar] [CrossRef]
- Wolf, S.; Reetz, J.; Johne, R.; Heiberg, A.C.; Petri, S.; Kanig, H.; Ulrich, R.G. The simultaneous occurrence of human norovirus and hepatitis E virus in a Norway rat (Rattus norvegicus). Arch. Virol. 2013, 158, 1575–1578. [Google Scholar] [CrossRef]
- Widen, F.; Ayral, F.; Artois, M.; Olofson, A.S.; Lin, J. PCR detection and analysis of potentially zoonotic Hepatitis E virus in French rats. Virol. J. 2014, 11, 90. [Google Scholar] [CrossRef]
- Ayral, F.; Artois, J.; Zilber, A.L.; Widen, F.; Pounder, K.C.; Aubert, D.; Bicout, D.J.; Artois, M. The relationship between socioeconomic indices and potentially zoonotic pathogens carried by wild Norway rats: A survey in Rhone, France (2010–2012). Epidemiol. Infect. 2015, 143, 586–599. [Google Scholar] [CrossRef]
- Ryll, R.; Bernstein, S.; Heuser, E.; Schlegel, M.; Dremsek, P.; Zumpe, M.; Wolf, S.; Pepin, M.; Bajomi, D.; Muller, G.; et al. Detection of rat hepatitis E virus in wild Norway rats (Rattus norvegicus) and Black rats (Rattus rattus) from 11 European countries. Vet. Microbiol. 2017, 208, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Spahr, C.; Ryll, R.; Knauf-Witzens, T.; Vahlenkamp, T.W.; Ulrich, R.G.; Johne, R. Serological evidence of hepatitis E virus infection in zoo animals and identification of a rodent-borne strain in a Syrian brown bear. Vet. Microbiol. 2017, 212, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Cai, C.L.; Li, B.; Zhang, W.; Zhu, Y.; Chen, W.H.; Zhuo, F.; Shi, Z.L.; Yang, X.L. Detection and characterization of three zoonotic viruses in wild rodents and shrews from Shenzhen city, China. Virol. Sin. 2017, 32, 290–297. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wen, Y.; Xiong, Y.; Zhang, M.; Cheng, M.; Chen, Q. The prevalence and genomic characteristics of hepatitis E virus in murine rodents and house shrews from several regions in China. BMC Vet. Res. 2018, 14, 414. [Google Scholar] [CrossRef]
- Murphy, E.G.; Williams, N.J.; Jennings, D.; Chantrey, J.; Verin, R.; Grierson, S.; McElhinney, L.M.; Bennett, M. First detection of Hepatitis E virus (Orthohepevirus C) in wild brown rats (Rattus norvegicus) from Great Britain. Zoonoses Public Health 2019, 66, 686–694. [Google Scholar] [CrossRef]
- Lack, J.B.; Volk, K.; Van Den Bussche, R.A. Hepatitis E virus genotype 3 in wild rats, United States. Emerg. Infect. Dis. 2012, 18, 1268–1273. [Google Scholar] [CrossRef]
- De Sabato, L.; Ianiro, G.; Monini, M.; De Lucia, A.; Ostanello, F.; Di Bartolo, I. Detection of hepatitis E virus RNA in rats caught in pig farms from Northern Italy. Zoonoses Public Health 2020, 67, 62–69. [Google Scholar] [CrossRef]
- Onyuok, S.O.; Hu, B.; Li, B.; Fan, Y.; Kering, K.; Ochola, G.O.; Zheng, X.S.; Obanda, V.; Ommeh, S.; Yang, X.L.; et al. Molecular Detection and Genetic Characterization of Novel RNA Viruses in Wild and Synanthropic Rodents and Shrews in Kenya. Front. Microbiol. 2019, 10, 2696. [Google Scholar] [CrossRef]
- Li, T.C.; Ami, Y.; Suzaki, Y.; Yasuda, S.P.; Yoshimatsu, K.; Arikawa, J.; Takeda, N.; Takaji, W. Characterization of full genome of rat hepatitis E virus strain from Vietnam. Emerg. Infect. Dis. 2013, 19, 115–118. [Google Scholar] [CrossRef]
- Wang, B.; Li, W.; Zhou, J.H.; Li, B.; Zhang, W.; Yang, W.H.; Pan, H.; Wang, L.X.; Bock, C.T.; Shi, Z.L.; et al. Chevrier’s Field Mouse (Apodemus chevrieri) and Pere David’s Vole (Eothenomys melanogaster) in China Carry Orthohepeviruses that form Two Putative Novel Genotypes Within the Species Orthohepevirus C. Virol. Sin. 2018, 33, 44–58. [Google Scholar] [CrossRef]
- De Souza, W.M.; Romeiro, M.F.; Sabino-Santos, G., Jr.; Maia, F.G.M.; Fumagalli, M.J.; Modha, S.; Nunes, M.R.T.; Murcia, P.R.; Figueiredo, L.T.M. Novel orthohepeviruses in wild rodents from Sao Paulo State, Brazil. Virology 2018, 519, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Kurucz, K.; Hederics, D.; Bali, D.; Kemenesi, G.; Horvath, G.; Jakab, F. Hepatitis E virus in Common voles (Microtus arvalis) from an urban environment, Hungary: Discovery of a Cricetidae-specific genotype of Orthohepevirus C. Zoonoses Public Health 2019, 66, 259–263. [Google Scholar] [CrossRef]
- Ryll, R.; Heckel, G.; Corman, V.M.; Drexler, J.F.; Ulrich, R.G. Genomic and spatial variability of a European common vole hepevirus. Arch. Virol. 2019, 164, 2671–2682. [Google Scholar] [CrossRef] [PubMed]
- Raj, V.S.; Smits, S.L.; Pas, S.D.; Provacia, L.B.; Moorman-Roest, H.; Osterhaus, A.D.; Haagmans, B.L. Novel hepatitis E virus in ferrets, the Netherlands. Emerg. Infect. Dis. 2012, 18, 1369–1370. [Google Scholar] [CrossRef] [PubMed]
- Li, T.C.; Yang, T.; Ami, Y.; Suzaki, Y.; Shirakura, M.; Kishida, N.; Asanuma, H.; Takeda, N.; Takaji, W. Complete genome of hepatitis E virus from laboratory ferrets. Emerg. Infect. Dis. 2014, 20, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gong, W.; Fu, H.; Li, M.; Zhang, Y.; Luo, Z.; Xu, Q.; Wang, L. Hepatitis E virus detected from Chinese laboratory ferrets and farmed mink. Transbound. Emerg. Dis. 2018, 65, e219–e223. [Google Scholar] [CrossRef] [PubMed]
- Krog, J.S.; Breum, S.O.; Jensen, T.H.; Larsen, L.E. Hepatitis E virus variant in farmed mink, Denmark. Emerg. Infect. Dis. 2013, 19, 2028–2030. [Google Scholar] [CrossRef]
- Bodewes, R.; van der Giessen, J.; Haagmans, B.L.; Osterhaus, A.D.; Smits, S.L. Identification of multiple novel viruses, including a parvovirus and a hepevirus, in feces of red foxes. J. Virol. 2013, 87, 7758–7764. [Google Scholar] [CrossRef]
- Reuter, G.; Boros, A.; Matics, R.; Kapusinszky, B.; Delwart, E.; Pankovics, P. Divergent hepatitis E virus in birds of prey, common kestrel (Falco tinnunculus) and red-footed falcon (F. vespertinus), Hungary. Infect. Genet. Evol. 2016, 43, 343–346. [Google Scholar] [CrossRef]
- Kobayashi, T.; Takahashi, M.; Jirintai, S.; Nishizawa, T.; Nagashima, S.; Nishiyama, T.; Kunita, S.; Hayama, E.; Tanaka, T.; Okamoto, H.; et al. An analysis of two open reading frames (ORF3 and ORF4) of rat hepatitis E virus genome using its infectious cDNA clones with mutations in ORF3 or ORF4. Virus Res. 2018, 249, 16–30. [Google Scholar] [CrossRef]
- Sridhar, S.; Teng, J.L.L.; Chiu, T.H.; Lau, S.K.P.; Woo, P.C.Y. Hepatitis E Virus Genotypes and Evolution: Emergence of Camel Hepatitis E Variants. Int. J. Mol. Sci. 2017, 18, 869. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.H.; Tan, B.H.; Teo, E.C.; Lim, S.G.; Dan, Y.Y.; Wee, A.; Aw, P.P.; Zhu, Y.; Hibberd, M.L.; Tan, C.K.; et al. Chronic Infection With Camelid Hepatitis E Virus in a Liver Transplant Recipient Who Regularly Consumes Camel Meat and Milk. Gastroenterology 2016, 150, e353. [Google Scholar] [CrossRef] [PubMed]
- Li, T.C.; Zhou, X.; Yoshizaki, S.; Ami, Y.; Suzaki, Y.; Nakamura, T.; Takeda, N.; Wakita, T. Production of infectious dromedary camel hepatitis E virus by a reverse genetic system: Potential for zoonotic infection. J. Hepatol. 2016, 65, 1104–1111. [Google Scholar] [CrossRef] [PubMed]
- Li, T.C.; Bai, H.; Yoshizaki, S.; Ami, Y.; Suzaki, Y.; Doan, Y.H.; Takahashi, K.; Mishiro, S.; Takeda, N.; Wakita, T. Genotype 5 Hepatitis E Virus Produced by a Reverse Genetics System Has the Potential for Zoonotic Infection. Hepatol. Commun. 2019, 3, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Teng, J.L.L.; Lau, S.K.P.; Sridhar, S.; Fu, H.; Gong, W.; Li, M.; Xu, Q.; He, Y.; Zhuang, H.; et al. Transmission of a Novel Genotype of Hepatitis E Virus from Bactrian Camels to Cynomolgus Macaques. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Cossaboom, C.M.; Cordoba, L.; Sanford, B.J.; Pineyro, P.; Kenney, S.P.; Dryman, B.A.; Wang, Y.; Meng, X.J. Cross-species infection of pigs with a novel rabbit, but not rat, strain of hepatitis E virus isolated in the United States. J. Gen. Virol. 2012, 93, 1687–1695. [Google Scholar] [CrossRef]
- Liu, P.; Bu, Q.N.; Wang, L.; Han, J.; Du, R.J.; Lei, Y.X.; Ouyang, Y.Q.; Li, J.; Zhu, Y.H.; Lu, F.M.; et al. Transmission of hepatitis E virus from rabbits to cynomolgus macaques. Emerg. Infect. Dis. 2013, 19, 559–565. [Google Scholar] [CrossRef]
- Meerburg, B.G.; Singleton, G.R.; Kijlstra, A. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 2009, 35, 221–270. [Google Scholar] [CrossRef]
- Maneerat, Y.; Clayson, E.T.; Myint, K.S.; Young, G.D.; Innis, B.L. Experimental infection of the laboratory rat with the hepatitis E virus. J. Med. Virol. 1996, 48, 121–128. [Google Scholar] [CrossRef]
- Huang, F.; Zhang, W.; Gong, G.; Yuan, C.; Yan, Y.; Yang, S.; Cui, L.; Zhu, J.; Yang, Z.; Hua, X. Experimental infection of Balb/c nude mice with Hepatitis E virus. BMC Infect. Dis. 2009, 9, 93. [Google Scholar] [CrossRef]
- Schlosser, J.; Dahnert, L.; Dremsek, P.; Tauscher, K.; Fast, C.; Ziegler, U.; Groner, A.; Ulrich, R.G.; Groschup, M.H.; Eiden, M. Different Outcomes of Experimental Hepatitis E Virus Infection in Diverse Mouse Strains, Wistar Rats, and Rabbits. Viruses 2018, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Shukla, P.; Nguyen, H.T.; Torian, U.; Engle, R.E.; Faulk, K.; Dalton, H.R.; Bendall, R.P.; Keane, F.E.; Purcell, R.H.; Emerson, S.U. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc. Natl. Acad. Sci. USA 2011, 108, 2438–2443. [Google Scholar] [CrossRef] [PubMed]
- Li, T.C.; Yoshizaki, S.; Ami, Y.; Suzaki, Y.; Yasuda, S.P.; Yoshimatsu, K.; Arikawa, J.; Takeda, N.; Wakita, T. Susceptibility of laboratory rats against genotypes 1, 3, 4, and rat hepatitis E viruses. Vet. Microbiol. 2013, 163, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Dremsek, P.; Wenzel, J.J.; Johne, R.; Ziller, M.; Hofmann, J.; Groschup, M.H.; Werdermann, S.; Mohn, U.; Dorn, S.; Motz, M.; et al. Seroprevalence study in forestry workers from eastern Germany using novel genotype 3- and rat hepatitis E virus-specific immunoglobulin G ELISAs. Med. Microbiol. Immunol. 2012, 201, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Hamaguchi, S.; Ngo, C.C.; Li, T.C.; Ando, S.; Yoshimatsu, K.; Yasuda, S.P.; Koma, T.; Isozumi, R.; Tsuda, Y.; et al. Serological evidence of infection with rodent-borne hepatitis E virus HEV-C1 or antigenically related virus in humans. J. Vet. Med. Sci. 2016, 78, 1677–1681. [Google Scholar] [CrossRef] [PubMed]
- Jirintai, S.; Suparyatmo, J.B.; Takahashi, M.; Kobayashi, T.; Nagashima, S.; Nishizawa, T.; Okamoto, H. Rat hepatitis E virus derived from wild rats (Rattus rattus) propagates efficiently in human hepatoma cell lines. Virus Res. 2014, 185, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Li, T.C.; Yang, T.; Yoshizaki, S.; Ami, Y.; Suzaki, Y.; Ishii, K.; Kishida, N.; Shirakura, M.; Asanuma, H.; Takeda, N.; et al. Ferret hepatitis E virus infection induces acute hepatitis and persistent infection in ferrets. Vet. Microbiol. 2016, 183, 30–36. [Google Scholar] [CrossRef]

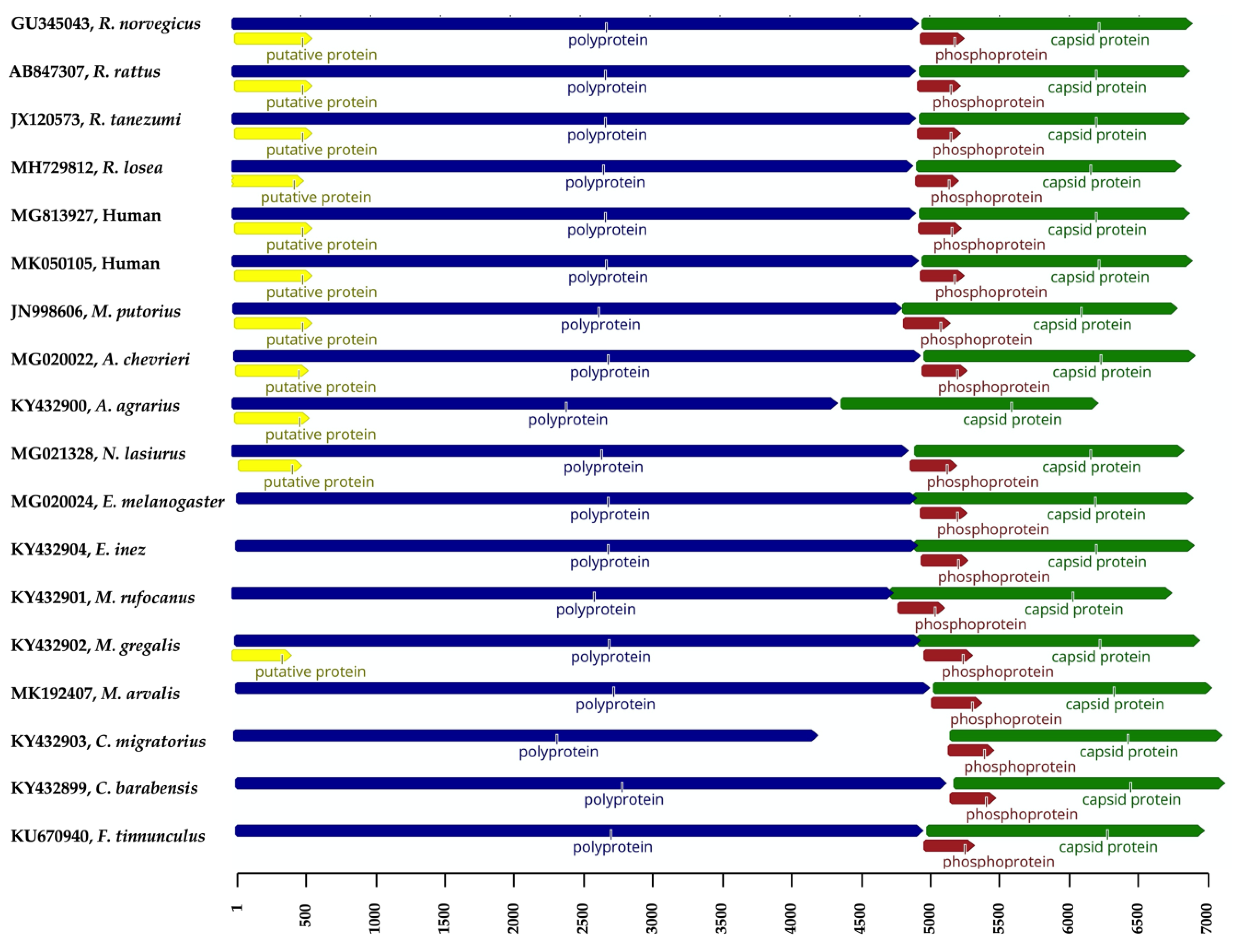
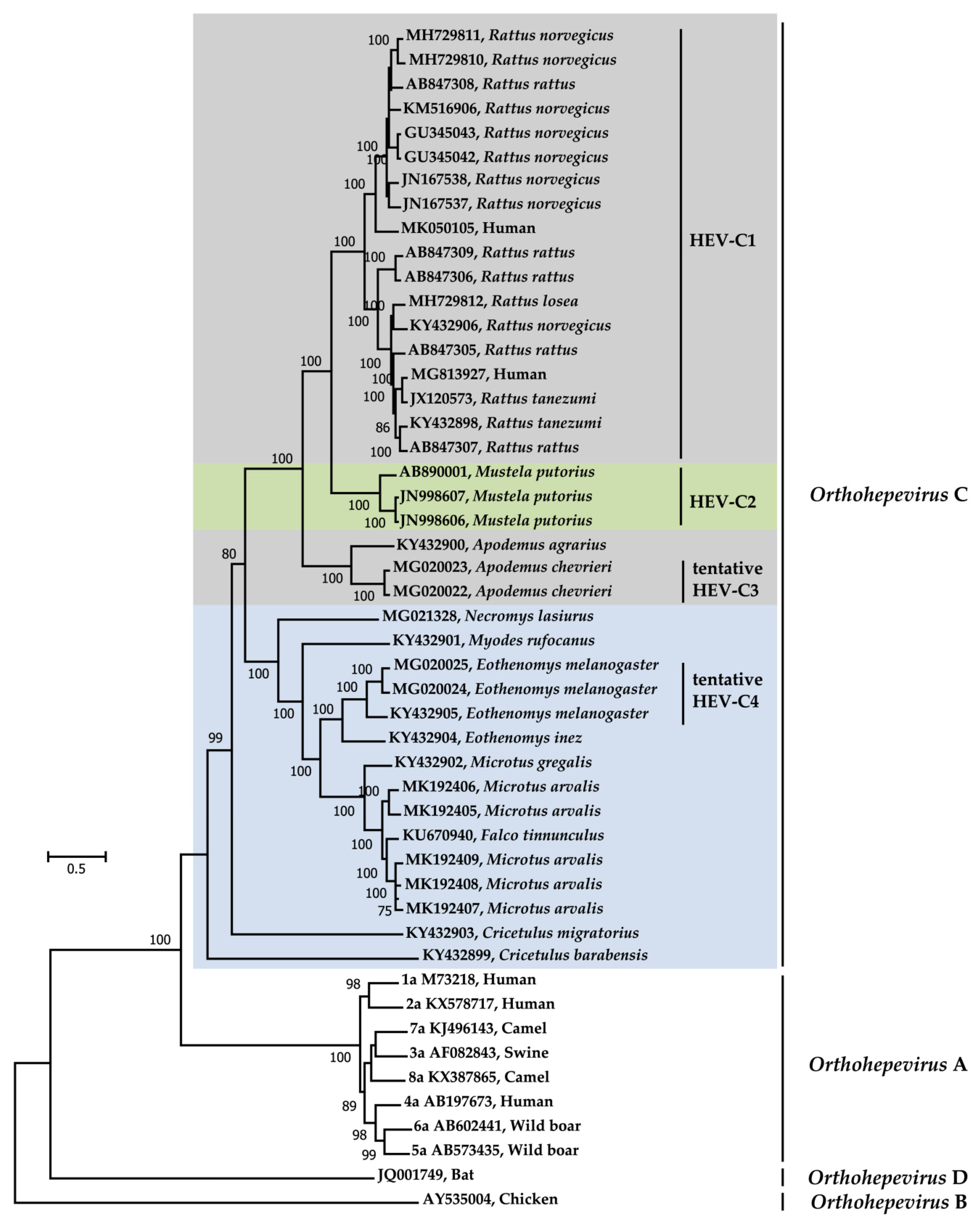
| Order | Family | Species (Scientific Name) | Common Name 1 | No. Positive/No. Tested (%) 3 | Sampling Site (s) (Year) | Assay | Reference |
|---|---|---|---|---|---|---|---|
| Rodentia | Muridae | Rattus norvegicus | Brown rat/Norway rat | 83/108 (76.9) | The USA (1986–1997) | In-house anti-HEV ELISA | [17] |
| 135/197 (68.5) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | ||||
| 144/196 (73.5) | The USA (2005–2006) | In-house anti-HEV ELISA | [21] | ||||
| 114/362 (31.5) | Japan (2000–2002) | In-house anti-HEV ELISA | [22] | ||||
| 16/56 (28.6) | Japan (N.A.) | Viragent HEV-Ab kit | [23] | ||||
| 36/147 (24.5) | Germany (2007–2010) | In-house anti-rat HEV ELISA | [24] | ||||
| 64/230 (27.8) | China (2011–2012) | In-house anti-rat HEV ELISA | [25] | ||||
| 25/123 (20.3) | Vietnam (2011) | In-house anti-rat HEV ELISA | [26] | ||||
| 21/94 (22.3) | Vietnam (2011) | In-house anti-rat HEV ELISA | [27] | ||||
| 48/389 (12.3) | Vietnam (2012–2013) | In-house anti-rat HEV ELISA | [28] | ||||
| 34/109 (31.2) 4 | Lithuania (2014–2017) | In-house anti-rat HEV ELISA | [29] | ||||
| Rattus rattus | Black rat | 102/113 (90.2) | The USA (1986–1997) | In-house anti-HEV ELISA | [17] | ||
| 31/81 (38.3) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | ||||
| 21/116 (18.1) | Indonesia (2011–2012) | In-house anti-HEV ELISA | [30] | ||||
| 137/369 (37.1) | Indonesia (2012) | In-house anti-rat HEV ELISA | [31] | ||||
| 10/242 (4.1) | Indonesia (2014–2016) | In-house anti-rat HEV ELISA | [32] | ||||
| 12/90 (13.3) | Japan (2000–2002) | In-house anti-HEV ELISA | [22] | ||||
| Rattus tanezumi | Oriental house rat | 4/16 (25.0) | Vietnam (2011) | In-house anti-rat HEV ELISA | [26] | ||
| 2/6 (33.3) | Vietnam (2011) | In-house anti-rat HEV ELISA | [27] | ||||
| 9/46 (19.6) | Vietnam (2012–2013) | In-house anti-rat HEV ELISA | [28] | ||||
| Rattus rattoides losea | Losea rat | 26/121 (21.5) | China (2011–2012) | In-house anti-rat HEV ELISA | [25] | ||
| Rattus flavipectus | Yellow-breasted rat | 34/171 (19.9) | China (2011–2012) | In-house anti-rat HEV ELISA | [25] | ||
| Rattus rattus hainanus | N.A. 2 | 2/17 (11.8) | China (2011–2012) | In-house anti-rat HEV ELISA | [25] | ||
| Bandicota indica | Greater bandicoot rat | 40/174 (23.0) | China (2011–2012) | In-house anti-rat HEV ELISA | [25] | ||
| Rattus rattus rufescens | Indian black rat | 9/57 (15.8) | India (1985) | In-house anti-HEV ELISA | [33] | ||
| Rattus rattus andamanesis | Sikkim rat | 2/55 (3.6) | India (1990) | In-house anti-HEV ELISA | [33] | ||
| Bandicota bengalensis | Lesser bandicoot rat | 12/22 (54.5) | India (1985) | In-house anti-HEV ELISA | [33] | ||
| Rattus exulans | Polynesian rat | 15/18 (83.3) | The USA (1986–1997) | In-house anti-HEV ELISA | [17] | ||
| Mus musculus | House rat | 2/14 (14.3) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | ||
| Cricetidae | Clethrionomys gapperi | Southern red-backed vole | 4/6 (66.7) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | |
| Neotoma albigula | White-throated woodrat | 13/22 (59.1) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | ||
| Neotoma mexicana | Mexican woodrat | 48/84 (57.1) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | ||
| Neotoma micropus | Southern plains woodrat | 1/8 (12.5) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | ||
| Oryzomys palustris | Marsh rice rat | 10/41 (24.4) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | ||
| Peromyscus boylei | Brush mice | 2/24 (8.3) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | ||
| Peromyscus eremicus | Cactus mouse | 3/7 (42.9) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | ||
| Peromyscus leucopus | White-footed mouse | 5/50 (10.0) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | ||
| Peromyscus maniculatus | North American deermouse | 11/91 (11.0) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | ||
| Sigmodon hispidus | Hispid cotton rat | 37/113 (32.7) | The USA (1994–1998) | In-house anti-HEV ELISAs | [18] | ||
| Soricomorpha | Soricidae | Suncus murinus | House shrew | 27/260 (10.4) | China (2011–2012) | In-house anti-rat HEV ELISA | [35] |
| Order | Family | Species (Scientific Name) | Common Name 1 | Sampling Site (s) (Year) | Virus Genotype | Genomic Sequence | Reference |
|---|---|---|---|---|---|---|---|
| Rodentia | Muridae | Rattus norvegicus | Brown rat/Norway rat | The USA (2003), Germany (2007–2010), Germany (2009–2016), France (2011–2012), Denmark (2012), China (2011–2012; 2013–2016; 2014–2017), Lithuania (2014–2017), 11 European countries (2005–2016) 2, England (2014–2016), Vietnam (2011; 2012–2013), Museum collections 3 | HEV-C1 | Partial, Nearly complete 5, Complete | [19,20,24,25,28,29,36,37,38,39,40,41,42,43,44,45,46] |
| Rattus rattus | Black rat | Indonesia (2011–2012; 2014–2016), Lithuania (2014–2017), Italy (2015), 11 European countries (2005–2016), Kenya (2016); Museum collections | HEV-C1 | Partial, Complete | [29,30,32,41,47,48] | ||
| Rattus tanezumi | Oriental house rat | Vietnam (2011; 2012–2013), China (2013–2016; 2014–2017) | HEV-C1 | Partial, Complete | [26,28,36,44,49] | ||
| Rattus losea | Losea rat | China (2011–2012; 2014–2017) | HEV-C1 | Partial, Nearly complete | [25,44] | ||
| Rattus flavipectus | Yellow-breasted rat | China (2011–2012) | HEV-C1 | Partial | [25,43] | ||
| Bandicota indica | Greater bandicoot rat | China (2011–2012) | HEV-C1 | Partial | [25] | ||
| Apodemus chevrieri | Chevrier’s field mouse | China (2013–2015) | N.A. 4 | Partial, Complete | [50] | ||
| Apodemus agrarius | Striped field mouse | China (2013–2016) | N.A. | Complete | [36] | ||
| Cricetidae | Eothenomys melanogaster | Père David’s vole | China (2013–2015) | N.A. | Partial, Complete | [36,50] | |
| Eothenomys inez | Inez’s red-backed vole | China (2013–2016) | N.A. | Complete | [36] | ||
| Myodes rufocanus | Grey red-backed vole | China (2013–2016) | N.A. | Nearly complete | [36] | ||
| Microtus gregalis | Narrow-headed vole | China (2013–2016) | N.A. | Complete | [36] | ||
| Cricetulus migratorius | Gray dwarf Hamster | China (2013–2016) | N.A. | Complete | [36] | ||
| Cricetulus barabensis | Striped dwarf hamster | China (2013–2016) | N.A. | Complete | [36] | ||
| Microtus arvalis | Common vole | Hungary (2016), Germany and the Czech Republic (2009–2013) | N.A. | Partial, Complete | [52,53] | ||
| Myodes glareolus | Bank vole | Germany (2009–2013) | N.A. | Partial | [53] | ||
| Necromys lasiurus | Hairy-tailed bolo mouse | Brazil (2008–2013) | N.A. | Nearly complete | [51] | ||
| Calomys tener | Delicate vesper mouse | Brazil (2008–2013) | N.A. | Partial | [51] | ||
| Soricomorpha | Soricidae | Suncus murinus | House shrew/Asian musk shrew | China (2011–2012; 2013–2016; 2014–2017) | HEV-C1 | Partial | [35,43,44] |
| Crocidura olivieri | Olivier’s shrew | Kenya (2016) | N.A. | Partial | [48] | ||
| Carnivora | Ursidae | Ursus arctos syriacus | Syrian brown bear | Germany (2011–2016) | HEV-C1 | Partial | [42] |
| Mustelidae | Mustela putorius | Western polecat/ferret | The Netherlands (2010), the USA (2013), Japan (2009–2013), China (2016), | HEV-C2 | Partial, Complete | [54,55,56] | |
| Neovison vison | American mink | Denmark (2008–2011), China (2016), | HEV-C2 | Partial | [56,57] | ||
| Falconiformes | Falconidae | Falco tinnunculus | Common kestrel | Hungary (2014) | N.A. | Partial, Complete | [59] |
| Falco vespertinus | Red-footed falcon | Hungary (2014) | N.A. | Partial, | [59] | ||
| Primates | Hominidae | Homo sapiens | Human | Hong Kong (2017–2019), Uganda (2017) | HEV-C1 | Complete | [14,15,16] |
| Orthohepevirus C (No. of Variants) 1 | % Identity | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HEV-C1 (20) 2 | HEV-C2 (10) | RdAcHEV (2) | RdAaHEV (1) | RdEmHEV (3) | RdEiHEV (1) | RdMrHEV (1) 3 | RdMgHEV (1) | RdMaHEV (6) 4 | RdNlHEV (1) | RdCmHEV (1) | RdCbHEV (1) | |
| Complete genome (nt) | ||||||||||||
| HEV-C1 | 65.3–67.7 | 62.6–65.0 | 56.5–59.1 | 53.2–55.4 | 54.3–55.4 | 53.1–55.2 | 53.8–55.5 | 52.9–55.3 | 54.8–56.0 | 49.9–51.3 | 52.6–54.1 | |
| HEV-C2 | 63.5–65.9/69.5–74.0 | 58.5–58.6 | 62.1–62.8 | 54.6–55.6 | 54.7–54.9 | 53.3–53.7 | 54.8–55.0 | 53.1–54.4 | 54.9–55.1 | 53.4–53.6 | 51.1–51.5 | |
| RdAcHEV | 61.0–63.9/64.4–67.8 | 61.0–61.6/67.8–68.5 | 66.2–66.4 | 54.3–55.0 | 54.5–54.7 | 53.6–53.8 | 54.5–54.9 | 53.0–54.1 | 54.5–54.6 | 52.7 | 50.4–51.0 | |
| RdAaHEV | 54.8–56.5/57.1–61.1 | 56.3–56.6/61.9–62.1 | 63.2–63.5/72.9–73.6 | 51.8–51.9 | 52.3 | 51.0 | 51.6 | 49.7–50.8 | 52.0 | 50.5 | 48.2 | |
| RdEmHEV | 51.4–53.4/49.6–53.5 | 53.1–53.7/52.5–54.3 | 51.9–52.9/51.3–53.2 | 49.1–49.5/47.6–48.6 | 72.2–72.6 | 62.1–63.0 | 66.9–67.3 | 65.7–67.2 | 59.5–59.9 | 54.5–54.6 | 52.2–52.5 | |
| RdEiHEV | 52.7–53.7/50.0–52.7 | 53.1–53.2/53.3–53.6 | 52.6–52.7/51.8–52.1 | 49.6/48.4 | 70.3–70.9/80.2–82.4 | 62.6 | 67.1 | 64.9–65.9 | 60.1 | 55.0 | 52.7 | |
| RdMrHEV | 51.7–53.3/51.6–54.2 | 51.4–52.9/52.8–53.1 | 51.8–52.0/52.3–52.7 | 48.3/47.9 | 60.6–62.0/67.8–69.1 | 61.0/66.8 | 61.4 | 62.0–62.4 | 58.3 | 54.0 | 52.2 | |
| RdMgHEV | 52.3–53.8/50.4–52.9 | 53.0–53.1/52.9–53.8 | 52.5–53.0/51.8–52.1 | 49.3/48.2 | 66.2–67.0/73.0–74.4 | 66.4/73.1 | 60.1/66.0 | 76.0–77.3 | 59.3 | 54.0 | 52.8 | |
| RdMaHEV | 50.9–53.9/49.6–54.0 | 51.1–52.5/51.7–54.0 | 50.8–52.0/51.2–52.7 | 47.4–48.7/49.4–50.6 | 65.1–67.1/70.9–74.5 | 64.0–65.5/69.9–72.4 | 60.5–61.4/64.6–62.2 | 74.0–75.9/86.6–89.6 | 58.9–59.5 | 53.4–54.2 | 51.3–52.1 | |
| RdNlHEV | 53.5–54.9/53.4–55.8 | 54.6/55.4–55.5 | 53.5–53.7/53.7–53.9 | 50.4/50.5 | 59.4–59.8/60.9–62.3 | 59.9/61.4 | 57.5/61.5 | 58.5/60.9 | 57.4–58.7/59.6–61.0 | 53.4 | 51.4 | |
| RdCmHEV | 48.0–49.5/47.5–50.8 | 48.1–48.9/49.7–50.4 | 47.7/49.2–49.4 | 44.6/45.0 | 49.3–49.9/50.1–51.8 | 49.6/50.7 | 49.7/49.9 | 49.6/50.0 | 47.6–49.1/48.4–50.3 | 49.6/50.1 | 51.1 | |
| RdCbHEV | 48.3–49.7/46.5–48.8 | 49.6–49.9/49.4–50.2 | 48.6–49.1/48.6–48.8 | 45.7/45.2 | 50.6–50.8/48.4–49.6 | 49.9/48.9 | 50.5/48.3 | 51.5/49.1 | 49.4–50.5/46.7–48.0 | 49.9/48.6 | 46.1/45.4 | |
| ORF1 (nt/aa) | ||||||||||||
| ORF2 (nt/aa) | ||||||||||||
| HEV-C1 | 66.5–72.8/76.4–81.6 | 63.2–69.7/70.7–74.9 | 59.6–66.1/68.2–73.3 | 57.0–61.9/60.7–63.0 | 56.8–61.3/59.7–62.0 | 56.5–61.1/60.2–63.6 | 57.2–61.7/61.2–64.3 | 56.6–61.0/58.7–63.2 | 56.6–60.6/63.0–65.7 | 56.6–60.4/60.1–61.8 | 53.1–57.1/56.4–59.4 | |
| HEV-C2 | 59.0–63.6/37.0–45.4 | 64.5–65.4/70.4–71.0 | 62.9–63.4/69.8–70.2 | 58.5–60.4/58.9–59.6 | 58.1–58.6/58.0–59.3 | 57.7/60.9–61.8 | 59.6–60.9/60.6–61.5 | 57.7–59.2/57.7–59.9 | 58.9–59.4/61.3–62.5 | 57.9–58.3/59.3–59.8 | 56.7/56.8–58.1 | |
| RdAcHEV | 59.2–62.6/39.6–50.0 | 54.6–56.4/32.1–35.7 | 70.1–70.4/85.6–85.8 | 60.4–61.5/62.3–62.9 | 59.6–60.3/61.6–61.8 | 59.1–59.6/61.2–61.8 | 60.3–60.8/63.2–63.5 | 58.1–60.0/60.1–61.6 | 59.4–59.5/61.8–62.3 | 59.3/59.2 | 55.1–56.3/57.4–58.0 | |
| RdAaHEV | N.A. 5 | N.A. | N.A. | 58.0–58.4/60.4–60.7 | 58.7/60.4 | 57.4/59.0 | 57.4/60.2 | 55.1–56.4/57.9–58.7 | 57.6/60.4 | 57.1/57.8 | 53.8/55.6 | |
| RdEmHEV | 40.1–44.8/20.0–27.8 | 41.3–46.7/21.0–26.9 | 40.9–44.3/22.2–28.2 | N.A. | 76.5–77.5/89.0–89.2 | 65.4–66.2/70.6–70.9 | 68.9–70.0/76.3–76.6 | 66.6–68.3/72.1–73.6 | 62.6–63.8/68.4–69.0 | 58.0–59.1/60.3–60.6 | 57.3–58.4/60.2–61.0 | |
| RdEiHEV | 42.2–47.5/20.7–29.3 | 40.7–43.6/20.3–26.3 | 42.6–42.9/23.7–25.4 | N.A. | 80.4–81.8/62.8–67.3 | 66.7/70.9 | 69.6/77.1 | 67.8–68.6/72.7–73.9 | 63.9/69.3 | 60.7/61.4 | 59.5/61.7 | |
| RdMrHEV | 37.1–39.4/19.5–24.6 | 37.8–38.1/19.7–20.5 | 35.4–37.1/22.7 | N.A. | 52.0–53.4/30.8–32.5 | 56.2/39.0 | 66.1/69.7 | 65.1–66.2/67.4–68.6 | 63.2/68.5 | 59.0/60.6 | 57.8/60.9 | |
| RdMgHEV | 38.6–41.6/19.5–24.4 | 39.1–41.0/19.7–22.8 | 36.9–38.2/20.8–21.6 | N.A. | 59.4–60.3/43.7–44.5 | 61.1/44.2 | 57.1/36.3 | 78.9–80.2/90.8–92.1 | 64.1/67.1 | 59.9/62 | 57.3/61.4 | |
| RdMaHEV | 36.3–40.2/10.2–18.9 | 36.5–38.9/10.9–17.8 | 37.8–39.5/10.1–15.5 | N.A. | 55.6–59.7/25.8–31.5 | 56.9–61.1/26.2–31.7 | 58.0–61.8/21.1–26.8 | 81.9–82.2/36.3–41.9 | 61.9–62.9/67.3–67.7 | 57.1–58.4/59.7–60.3 | 56.3–57.0/58.7–59.6 | |
| RdNlHEV | 32.9–35.0/14.9–24.0 | 31.7–32.0/14.4–15.2 | 32.9–34.0/17.2–18.0 | N.A. | 40.8–41.9/24.6–27.9 | 44.6/27.6 | 37.9/24.2 | 41.4/30.3 | 43.5–45.9/20.3–23.4 | 59.8/64.1 | 57.5/61.2 | |
| RdCmHEV | 38.2–43.1/17.8–27.1 | 37.6–38.1/17.7–22.6 | 40.3–41.7/23.3–25.0 | N.A. | 40.5–41.3/20.2–21.0 | 44.5/22.6 | 39.6/22.2 | 43.3/21.7 | 40.2–42.3/14.9–19.5 | 34.2/18.9 | 58.5/63.5 | |
| RdCbHEV | 36.6–39.4/20.5–25.0 | 36.9–38.0/17.2–21.6 | 37.6–37.9/21.7–23.5 | N.A. | 33.9–35.0/16.0–20.2 | 35.8/21.8 | 33.2/15.6 | 32.3/19.7 | 31.2–32.2/15.9–21.4 | 33.0/15.9 | 39.9/18.9 | |
| ORF3 (nt/aa) | ||||||||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Harms, D.; Yang, X.-L.; Bock, C.-T. Orthohepevirus C: An Expanding Species of Emerging Hepatitis E Virus Variants. Pathogens 2020, 9, 154. https://doi.org/10.3390/pathogens9030154
Wang B, Harms D, Yang X-L, Bock C-T. Orthohepevirus C: An Expanding Species of Emerging Hepatitis E Virus Variants. Pathogens. 2020; 9(3):154. https://doi.org/10.3390/pathogens9030154
Chicago/Turabian StyleWang, Bo, Dominik Harms, Xing-Lou Yang, and C.-Thomas Bock. 2020. "Orthohepevirus C: An Expanding Species of Emerging Hepatitis E Virus Variants" Pathogens 9, no. 3: 154. https://doi.org/10.3390/pathogens9030154
APA StyleWang, B., Harms, D., Yang, X.-L., & Bock, C.-T. (2020). Orthohepevirus C: An Expanding Species of Emerging Hepatitis E Virus Variants. Pathogens, 9(3), 154. https://doi.org/10.3390/pathogens9030154






