Mycobacterium bovis Population Structure in Cattle and Local Badgers: Co-Localisation and Variation by Farm Type
Abstract
:1. Introduction
2. Results
2.1. Summary Statistics
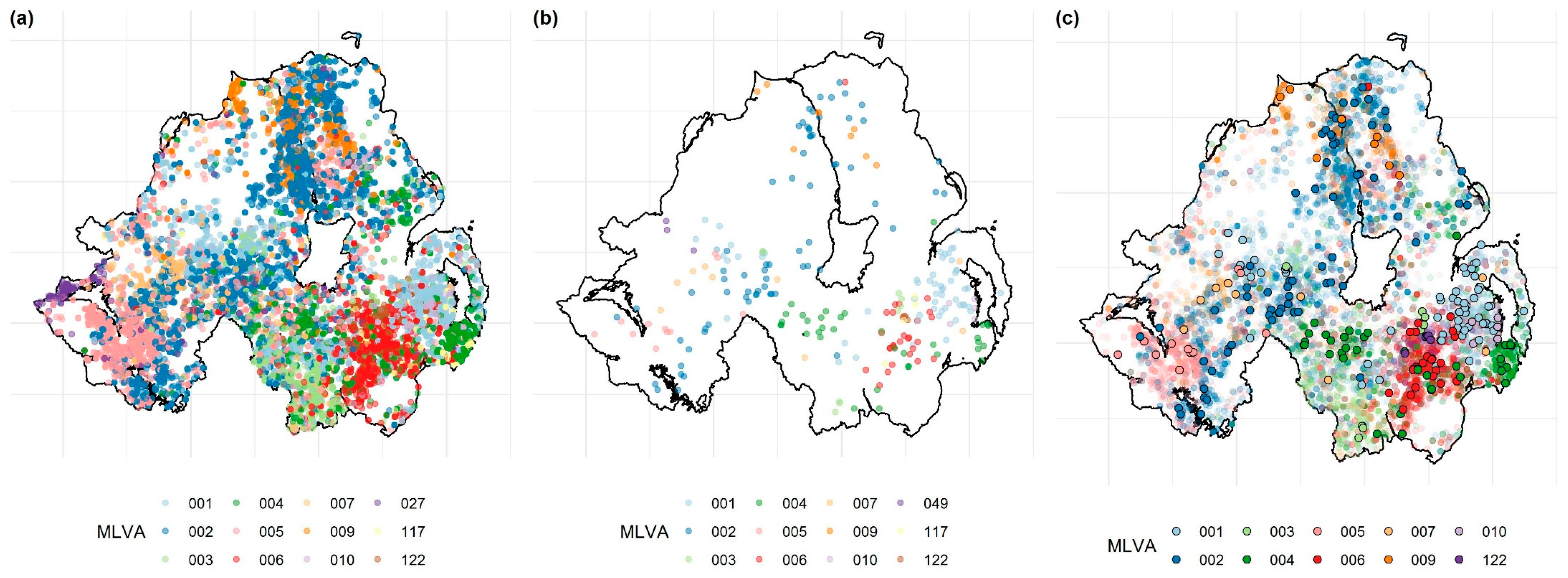
2.2. Assessment of MLVA Clustering within the Cattle Population
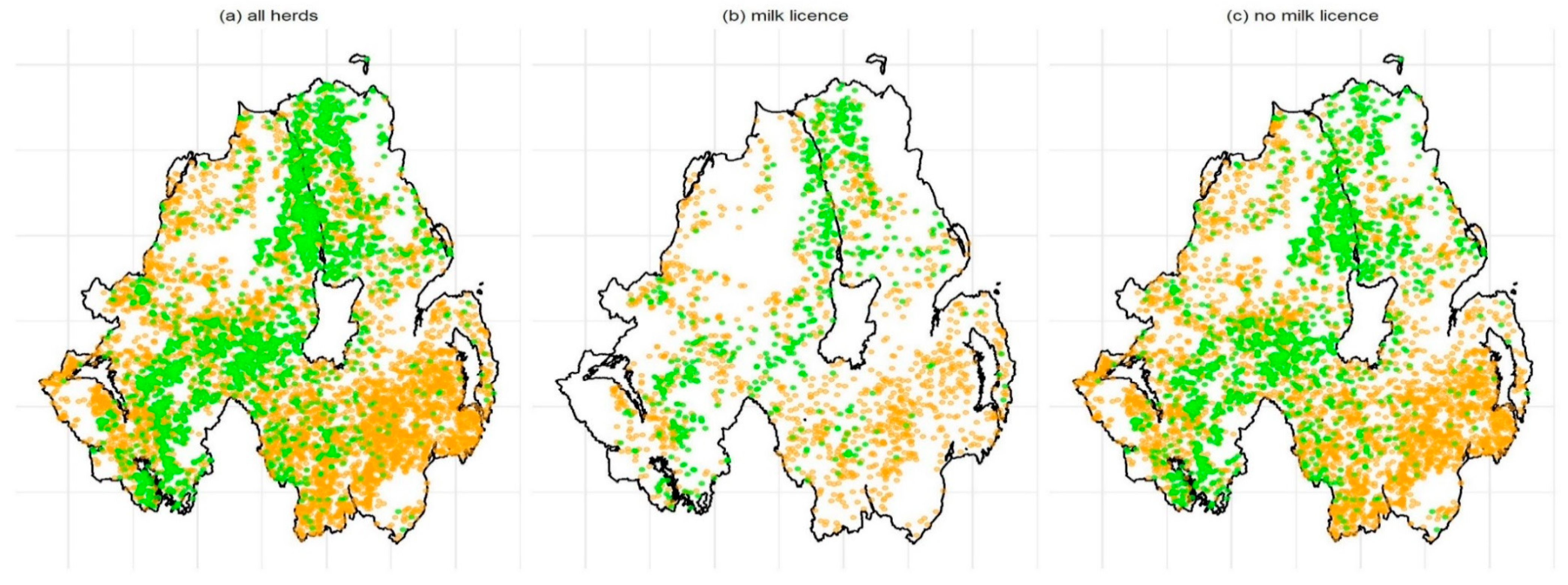
2.3. Intra and Interspecific Nearest Neighbour (NN) Analysis
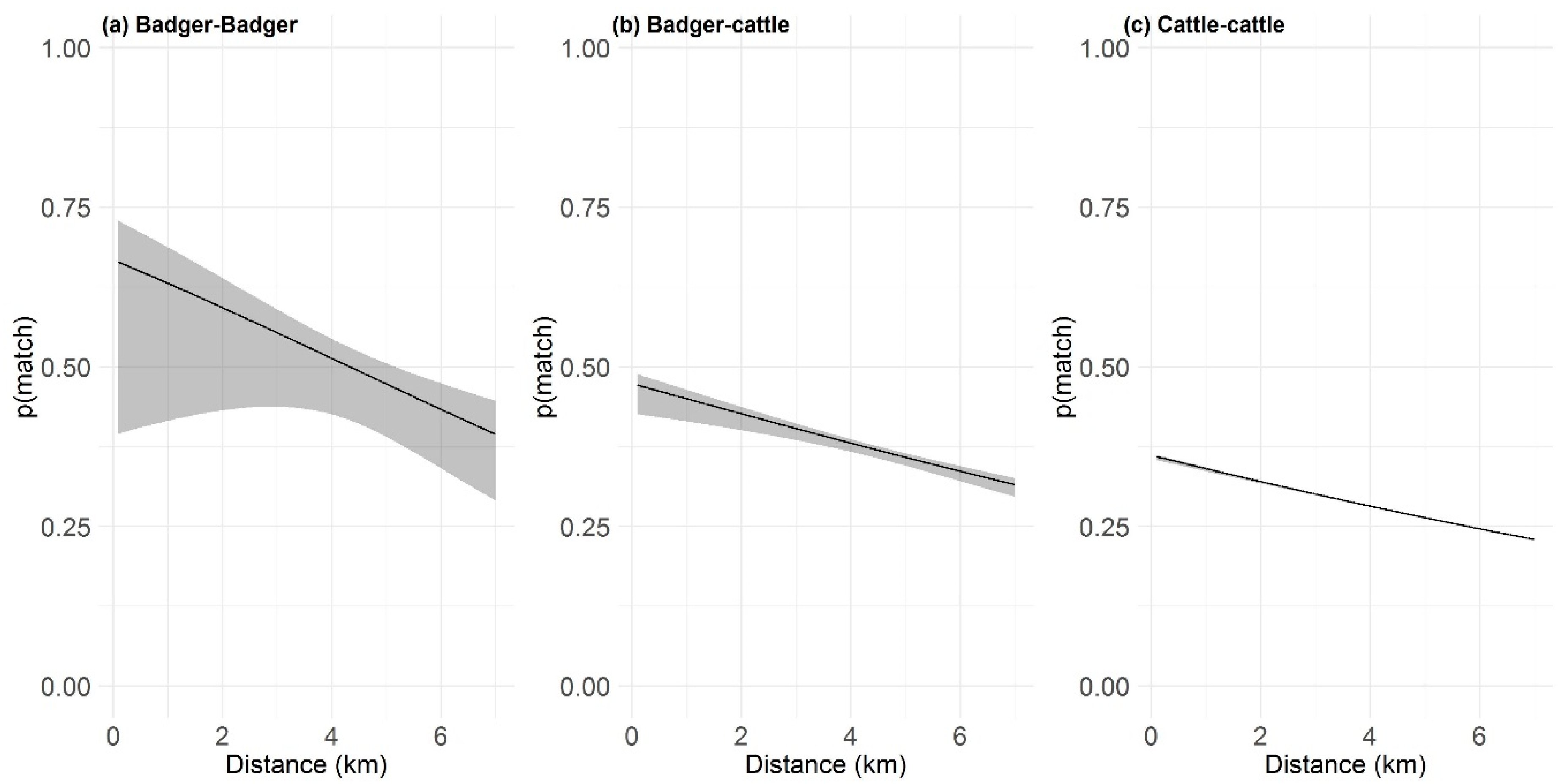
2.4. Distance-Based Similarlty Analysis
2.5. Determinng the Factors Associated with RTA Badgers and Nearby Cattle Herds Sharing MLVA Types
3. Discussion
Limitations
4. Materials and Methods
4.1. Study Area
4.2. Study Data
4.2.1. M. bovis Molecular Typing Data
4.2.2. Cattle Data
4.2.3. Badger Data
4.3. Assessment of MLVA Clustering within the Cattle Population
4.4. Intra- and Interspecific Nearest Neighbour Analysis
4.5. Distance Based Similarlity Analysis
4.6. Determining the Factors Associated with RTA Badgers and Nearby Cattle Herds Sharing MLVA Types
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gortázar, C.; Ferroglio, E.; Höfle, U.; Frölich, K.; Vicente, J. Diseases shared between wildlife and livestock: A European perspective. Eur. J. Wildl. Res. 2007, 53, 241–256. [Google Scholar] [CrossRef]
- De La Rua-Domenech, R.; Goodchild, A.; Vordermeier, H.; Hewinson, R.; Christiansen, K.; Clifton-Hadley, R. Ante mortem diagnosis of tuberculosis in cattle: A review of the tuberculin tests, γ-interferon assay and other ancillary diagnostic techniques. Res. Vet. Sci. 2006, 81, 190–210. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.; Pfeiffer, D.U.; Jackson, R. The epidemiology of Mycobacterium bovis infections. Vet. Microbiol. 1994, 40, 153–177. [Google Scholar] [CrossRef]
- Mathews, F.; Macdonald, D.W.; Taylor, G.M.; Gelling, M.; Norman, R.; Honess, P.E.; Foster, R.; Gower, C.M.; Varley, S.; Harris, A.; et al. Bovine tuberculosis (Mycobacterium bovis) in British farmland wildlife: The importance to agriculture. Proc. R. Soc. B Boil. Sci. 2005, 273, 357–365. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, J.; Clifton-Hadley, R. Tuberculosis in badgers; a review of the disease and its significance for other animals. Res. Vet. Sci. 2000, 69, 203–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrne, A.; Kenny, K.; Fogarty, U.; O’Keeffe, J.; More, S.J.; McGrath, G.; Teeling, M.; Martin, S.; Dohoo, I. Spatial and temporal analyses of metrics of tuberculosis infection in badgers (Meles meles) from the Republic of Ireland: Trends in apparent prevalence. Prev. Vet. Med. 2015, 122, 345–354. [Google Scholar] [CrossRef]
- Crispell, J.; Benton, C.H.; Balaz, D.; De Maio, N.; Ahkmetova, A.; Allen, A.; Biek, R.; Presho, E.L.; Dale, J.; Hewinson, G.; et al. Combining genomics and epidemiology to analyse bi-directional transmission of Mycobacterium bovis in a multi-host system. eLife 2019, 8, 45833. [Google Scholar] [CrossRef]
- Gortázar, C.; Delahay, R.J.; McDonald, R.A.; Boadella, M.; Wilson, G.J.; Gavier-Widen, D.; Acevedo, P. The status of tuberculosis in European wild mammals. Mammal Rev. 2011, 42, 193–206. [Google Scholar] [CrossRef]
- Böhm, M.; Hutchings, M.; White, P.C.L. Contact networks in a wildlife-livestock host community: Identifying high-risk individuals in the transmission of bovine TB among badgers and cattle. PLoS ONE 2009, 4, e5016. [Google Scholar] [CrossRef]
- Campbell, E.L.; Byrne, A.; Menzies, F.D.; McBride, K.R.; McCormick, C.M.; Scantlebury, M.; Reid, N. Interspecific visitation of cattle and badgers to fomites: A transmission risk for bovine tuberculosis? Ecol. Evol. 2019, 9, 8479–8489. [Google Scholar] [CrossRef]
- Barasona, J.; Vicente, J.; Díez-Delgado, I.; Aznar, J.; Gortázar, C.; Torres, M. Environmental presence of mycobacterium tuberculosis complex in aggregation points at the wildlife/livestock interface. Transbound. Emerg. Dis. 2016, 64, 1148–1158. [Google Scholar] [CrossRef] [PubMed]
- NIAO. The control of Bovine Tuberculosis in Northern Ireland; Northern Ireland Audit Office: Belfast, Northern Ireland, 2018.
- Allen, R.A.; Skuce, R.A.; Byrne, A.W. Bovine tuberculosis in Britain and Ireland a perfect storm? The confluence of potential ecological and epidemiological impediments to controlling a chronic infectious disease. Front. Vet. Sci. 2018, 5, 109. [Google Scholar] [CrossRef] [PubMed]
- Reid, N.; Etherington, T.R.; Wilson, G.J.; Montgomery, W.I.; McDonald, R.A. Monitoring and population estimation of the European badger Meles meles in Northern Ireland. Wildl. Biol. 2012, 18, 46–57. [Google Scholar] [CrossRef] [Green Version]
- Abernethy, D.A.; Walton, E.; Menzies, F.; Courtier, E.; Robinson, P. Mycobacterium bovis surveillance in European badgers (Meles meles) killed by vehicles in Northern Ireland: An epidemiological evaluation. Épidémiologie et Santé Animale 2011, 216–218. Available online: https://pdfs.semanticscholar.org/7313/a7149c10fed40e4432c8fb9d8c0653445e25.pdf (accessed on 30 June 2020).
- Courcier, E.A.; Menzies, F.D.; Strain, S.A.J.; Skuce, R.A.; Robinson, P.A.; Patterson, I.A.P.; McBride, K.R.; McCormick, C.M.; Walton, E.; McDowell, S.W.J.; et al. Monitoring Mycobacterium bovis in Eurasian badgers (Meles meles) killed by vehicles in Northern Ireland between 1998 and 2011. Vet. Rec. 2017, 182, 259. [Google Scholar] [CrossRef]
- Kamerbeek, J.; Schouls, L.; Kolk, A.; Van Agterveld, M.; Van Soolingen, D.; Kuijper, S.; Bunschoten, A.; Molhuizen, H.; Shaw, R.; Goyal, M.; et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997, 35, 907–914. [Google Scholar] [CrossRef] [Green Version]
- Skuce, R.; Mallon, T.R.; McBride, S.H.; Couzens, C.; Gordon, A.W.; McDowell, S.; Bsc; McCormick, C.M.; Hnc; Pgdip(Comp), H.B.; et al. Mycobacterium bovis genotypes in Northern Ireland: Herd-level surveillance (2003 to 2008). Vet. Rec. 2010, 167, 684–689. [Google Scholar] [CrossRef]
- Skuce, R.; Breadon, E.; Allen, A.; Milne, G.; McCormick, C.; Hughes, C.; Rutherford, D.; Smith, G.; Thompson, S.; Graham, J.; et al. Longitudinal dynamics of herd-level Mycobacterium bovis MLVA type surveillance in cattle in Northern Ireland 2003–2016. Infect. Genet. Evol. 2020, 79, 104131. [Google Scholar] [CrossRef]
- Trewby, H. The genetic and Spatial Epidemiology of Bovine Tuberculosis in the UK: From Molecular Typing to Bacterial Whole Genome Sequencing 2016; University of Glasgow: Glasgow, UK, 2016. [Google Scholar]
- Trewby, H.; Wright, D.; Breadon, E.L.; Lycett, S.J.; Mallon, T.R.; McCormick, C.; Johnson, P.; Orton, R.J.; Allen, A.R.; Galbraith, J.; et al. Use of bacterial whole-genome sequencing to investigate local persistence and spread in bovine tuberculosis. Epidemics 2016, 14, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Fielding, H.R.; McKinley, T.J.; Silk, M.J.; Delahay, R.J.; McDonald, R.A. Contact chains of cattle farms in Great Britain. R. Soc. Open Sci. 2019, 6, 180719. [Google Scholar] [CrossRef] [Green Version]
- Doyle, L.; Courcier, E.; Gordon, A.; O’Hagan, M.; Menzies, F. Bovine tuberculosis in Northern Ireland: Risk factors associated with duration and recurrence of chronic herd breakdowns. Prev. Vet. Med. 2016, 131, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Menzies, D.F.; Neill, S.D. Cattle-to-cattle transmission of bovine tuberculosis. Vet. J. 2000, 160, 92–106. [Google Scholar] [CrossRef]
- Milne, G.; Graham, J.; Allen, A.; McCormick, C.; Presho, E.; Skuce, R.; Byrne, A. Variation in Mycobacterium bovis genetic richness suggests that inwards cattle movements are a more important source of infection in beef herds than in dairy herds. BMC Microbiol. 2019, 19, 154. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, M.; Matthews, D.; Laird, C.; McDowell, S. Herd-level risk factors for bovine tuberculosis and adoption of related biosecurity measures in Northern Ireland: A case-control study. Vet. J. 2016, 213, 26–32. [Google Scholar] [CrossRef] [Green Version]
- Goodchild, A.V.; Watkins, G.H.; Sayers, A.R.; Jones, J.R.; Clifton-Hadley, R.S. Geographical association between the genotype of bovine tuberculosis in found dead badgers and in cattle herds. Vet. Rec. 2012, 170, 259. [Google Scholar] [CrossRef]
- Woodroffe, R.; Donnelly, C.A.; Johnston, W.T.; Bourne, F.J.; Cheeseman, C.L.; Clifton-Hadley, R.S.; Cox, D.R.; Gettinby, G.; Hewinson, R.G.; Le Fevre, A.M.; et al. Spatial association of Mycobacterium bovis infection in cattle and badgers Meles meles. J. Appl. Ecol. 2005, 42, 852–862. [Google Scholar] [CrossRef]
- Olea-Popelka, F.; Flynn, O.; Costello, E.; McGrath, G.; Collins, J.; O’Keeffe, J.; Kelton, D.; Berke, O.; Martin, S. Spatial relationship between Mycobacterium bovis strains in cattle and badgers in four areas in Ireland. Prev. Vet. Med. 2005, 71, 57–70. [Google Scholar] [CrossRef]
- Rodríguez, L.D.; Romero, B.; Bezos, J.; De Juan, L.; Álvarez, J.; Castellanos, E.; Moya, N.; Lozano, F.; Gonzalez, S.; Sáez-Llorente, J.L.; et al. High spoligotype diversity within a Mycobacterium bovis population: Clues to understanding the demography of the pathogen in Europe. Vet. Microbiol. 2010, 141, 89–95. [Google Scholar] [CrossRef]
- Romero, B.; Aranaz, A.; Sandoval, A.; Álvarez, J.; De Juan, L.; Bezos, J.; Sánchez, C.; Galka, M.; Fernández, P.; Mateos, A.; et al. Persistence and molecular evolution of Mycobacterium bovis population from cattle and wildlife in Doñana National Park revealed by genotype variation. Vet. Microbiol. 2008, 132, 87–95. [Google Scholar] [CrossRef]
- De La Cruz, M.L.; Pérez, A.; Bezos, J.; Pages, E.; Casal, C.; Carpintero, J.; Romero, B.; Dominguez, L.; Barker, C.M.; Díaz, R.; et al. Spatial dynamics of bovine tuberculosis in the autonomous community of Madrid, Spain (2010–2012). PLoS ONE 2014, 9, e115632. [Google Scholar] [CrossRef] [Green Version]
- Green, D.; Kiss, I.Z.; Mitchell, A.P.; Kao, R. Estimates for local and movement-based transmission of bovine tuberculosis in British cattle. Proc. R. Soc. B Boil. Sci. 2008, 275, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- White, P.W.; Martin, S.W.; De Jong, M.C.; O’Keeffe, J.; More, S.J.; Frankena, K. The importance of ‘neighbourhood’ in the persistence of bovine tuberculosis in Irish cattle herds. Prev. Vet. Med. 2013, 110, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Guta, S.; Casal, J.; Napp, S.; Saez, J.L.; Garcia-Saenz, A.; De Val, B.P.; Allepuz, A. Epidemiological investigation of bovine tuberculosis herd breakdowns in Spain 2009/2011. PLoS ONE 2014, 9, e104383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrne, A.W.; White, P.W.; McGrath, G.; James, O.; Martin, S.W. Risk of tuberculosis cattle herd breakdowns in Ireland: Effects of badger culling effort, density and historic large-scale interventions. Vet. Res. 2014, 45, 109. [Google Scholar] [CrossRef]
- Denny, G.O.; Wilesmith, J.W. Bovine tuberculosis in Northern Ireland: A case-control study of herd risk factors. Vet. Rec. 1999, 144, 305–310. [Google Scholar] [CrossRef]
- Vernon, M.C. Demographics of cattle movements in the UK. BMC Vet. Res. 2011, 7, 31. [Google Scholar] [CrossRef] [Green Version]
- Mekonnen, G.A.; Ameni, G.; Wood, J.; The ETHICOBOTS consortium; Berg, S.; Conlan, A. Network analysis of dairy cattle movement and associations with bovine tuberculosis spread and control in emerging dairy belts of Ethiopia. BMC Vet. Res. 2019, 15, 262. [Google Scholar] [CrossRef]
- Picasso, C.; Alvarez, J.; VanderWaal, K.L.; Fernandez, F.; Gil, A.; Wells, S.J.; Perez, A. Epidemiological investigation of bovine tuberculosis outbreaks in Uruguay (2011–2013). Prev. Vet. Med. 2017, 138, 156–161. [Google Scholar] [CrossRef] [Green Version]
- Gopal, R.; Goodchild, A.; Hewinson, G.; Domenech, R.D.L.R.; Clifton-Hadley, R. Introduction of bovine tuberculosis to north-east England by bought-in cattle. Vet. Rec. 2006, 159, 265–271. [Google Scholar] [CrossRef]
- Adkin, A.; Brouwer, A.; Downs, S.; Kelly, L. Assessing the impact of a cattle risk-based trading scheme on the movement of bovine tuberculosis infected animals in England and Wales. Prev. Vet. Med. 2016, 123, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Palisson, A.; Courcoul, A.; Durand, B. Role of Cattle Movements in Bovine Tuberculosis Spread in France between 2005 and 2014. PLoS ONE 2016, 11, e0152578. [Google Scholar] [CrossRef]
- Brown, E.; Marshall, A.H.; Mitchell, H.J.; Byrne, A. Cattle movements in Northern Ireland form a robust network: Implications for disease management. Prev. Vet. Med. 2019, 170, 104740. [Google Scholar] [CrossRef]
- Trewby, H.; Wright, D.M.; Skuce, R.A.; McCormick, C.; Mallon, T.R.; Presho, E.L.; Kao, R.R.; Haydon, D.T.; Biek, R. Relative abundance of Mycobacterium bovis molecular types in cattle: A simulation study of potential epidemiological drivers. BMC Vet. Res. 2017, 13, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DAERA. The Test and Vaccinate or Remove (TVR) Wildlife Intervention Research Project. Year 4 Report 2017; DAERA: Belfast, UK, 2017.
- Lahuerta-Marin, A.; Milne, G.; McNair, J.; Skuce, R.; McBride, S.; Menzies, F.; McDowell, S.; Byrne, A.; Handel, I.; Bronsvoort, B.D.C. Bayesian latent class estimation of sensitivity and specificity parameters of diagnostic tests for bovine tuberculosis in chronically infected herds in Northern Ireland. Vet. J. 2018, 238, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Byrne, A.; Graham, J.; Brown, C.; Donaghy, A.; Guelbenzu, M.; McNair, J.; Skuce, R.A.; Allen, A.R.; McDowell, S.W. Bovine tuberculosis visible lesions in cattle culled during herd breakdowns: The effects of individual characteristics, trade movement and co-infection. BMC Vet. Res. 2017, 13, 1–14. [Google Scholar] [CrossRef]
- DAERA. Final results of the June agricultural census 2019. In 2020, Statistics and Analytical Services Branch, Policy, Economics and Statistics Division Belfast; DAERA: Belfast, UK, 2020. [Google Scholar]
- Skuce, R.A.; McDowell, S.; Mallon, T.R.; Luke, B.; Breadon, E.L.; Lagan, P.L.; McCormick, C.M.; McBride, S.H.; Pollock, J.M. Discrimination of isolates of Mycobacterium bovis in Northern Ireland on the basis of variable numbers of tandem repeats (VNTRs). Vet. Rec. 2005, 157, 501–504. [Google Scholar] [CrossRef] [PubMed]
- Houston, R. A computerised database system for bovine traceability. Rev. Sci. Tech. Off. Int. Epizoot. 2001, 20, 652–661. [Google Scholar] [CrossRef]
- Abernethy, D.A.; Denny, G.; Pfeiffer, D.U.; Wrigley, K. Survey for mycobacterium bovis infection in road-traffic-accident badgers in Northern Ireland. In Proceedings of the 10th International Symposium on Veterinary Epidemiology and Economics, Vina del Mar, Chile, 17–21 November 2003. [Google Scholar]
- Byrne, A.; Quinn, J.L.; O’Keeffe, J.; Green, S.; Sleeman, D.P.; Martin, S.W.; Davenport, J. Large-scale movements in European badgers: Has the tail of the movement kernel been underestimated? J. Anim. Ecol. 2014, 83, 991–1001. [Google Scholar] [CrossRef] [Green Version]
- Zuur, A.F.; Ieno, E.N.; Walker, N.; Saveliev, A.A.; Smith, G.M. Mixed Effects Models and Extensions in Ecology with R; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Bivand, R.; Keitt, T.; Rowlingson, B.; Pebesma, E.D.Z.E.R. rgdal: Bindings for the Geospatial Data Abstraction Library. 2010. Available online: https://www.researchgate.net/publication/247474582_rgdal_Bindings_for_the_Geospatial_Data_Abstraction_Library (accessed on 30 June 2020).
- Bivand, R.; Rundel, C. rgeos: Interface to Geometry Engine—Open Source (’GEOS’). 2017. Available online: https://rdrr.io/cran/rgeos/ (accessed on 30 June 2020).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Wickham, H.; Francois, R.; Henry, L.; Müller, K. dplyr: A Grammar of Data Manipulation. 2017. Available online: https://dplyr.tidyverse.org/reference/dplyr-package.html (accessed on 30 June 2020).

| MLVA | Dataset | Total Infected Herds | 25th Percentile (km) | 50th Percentile (km) | 75th Percentile (km) | 100th Percentile (km) |
|---|---|---|---|---|---|---|
| 001 | All | 1141 | 28.7 km (19.3%) | 52.7 km (35.5%) | 77.6 km (52.2%) | 148.5 km |
| 001 | Dairy | 347 | 29.2 km (19.2%) | 52.8 km (34.7%) | 76.4 km (50.1%) | 152.4 km |
| 001 | Non Dairy | 794 | 27.8 km (16.9%) | 52.5 km (31.9%) | 78.2 km (47.5%) | 164.5 km |
| 002 | All | 1885 | 30.9 km (18.5%) | 52 km (31.3%) | 76.5 km (46.0%) | 166.4 km |
| 002 | Dairy | 545 | 28.3 km (18.9%) | 54.1 km (36.1%) | 80.9 km (54.0%) | 149.9 km |
| 002 | Non Dairy | 1340 | 31.1 km (18.7%) | 50.8 km (30.6%) | 74.5 km (44.8%) | 166.4 km |
| 003 | All | 460 | 14.9 km (10.0%) | 29 km (19.4%) | 51.2 km (34.2%) | 149.6 km |
| 003 | Dairy | 118 | 17.4 km (13.6%) | 32.2 km (25.1%) | 51.5 km (40.1%) | 128.4 km |
| 003 | Non Dairy | 342 | 13.5 km (9.1%) | 27 km (18.1%) | 50.7 km (33.9%) | 149.6 km |
| 004 | All | 736 | 26.1 km (16.7%) | 45.6 km (29.1%) | 64.2 km (41.0%) | 156.5 km |
| 004 | Dairy | 204 | 27.6 km (19.7%) | 48.1 km (34.3%) | 68.3 km (48.7%) | 140.5 km |
| 004 | Non Dairy | 532 | 25.4 km (16.3%) | 44.4 km (28.6%) | 62.7 km (40.4%) | 155.3 km |
| 005 | All | 1001 | 23.8 km (14.3%) | 51.3 km (30.7%) | 79.3 km (47.4%) | 167.3 km |
| 005 | Dairy | 266 | 17.9 km (12.0%) | 41.2 km (27.4%) | 77.6 km (51.7%) | 150.1 km |
| 005 | Non Dairy | 735 | 26.1 km (15.6%) | 53.4 km (31.9%) | 79.8 km (47.7%) | 167.3 km |
| 006 | All | 810 | 13.9 km (9.5%) | 23.6 km (16.1%) | 40.9 km (27.9%) | 146.4 km |
| 006 | Dairy | 241 | 12.9 km (8.8%) | 23.2 km (15.9%) | 44.7 km (30.5%) | 146.4 km |
| 006 | Non Dairy | 569 | 14 km (10.3%) | 23.5 km (17.3%) | 39.6 km (29.2%) | 135.8 km |
| 007 | All | 388 | 19.8 km (13.8%) | 39.6 km (27.6%) | 65.6 km (45.6%) | 143.8 km |
| 007 | Dairy | 120 | 14.3 km (11.0%) | 27.6 km (21.3%) | 57 km (44.1%) | 129.4 km |
| 007 | Non Dairy | 268 | 22.7 km (15.8%) | 43.7 km (30.4%) | 67.9 km (47.2%) | 143.8 km |
| 009 | All | 355 | 17.1 km (10.5%) | 27.1 km (16.7%) | 44.9 km (27.7%) | 162.0 km |
| 009 | Dairy | 140 | 12 km (9.4%) | 20.4 km (15.9%) | 30.6 km (23.9%) | 128.1 km |
| 009 | Non Dairy | 215 | 19.3 km (11.9%) | 31.7 km (19.5%) | 52.5 km (32.4%) | 162.0 km |
| 122 | All | 157 | 8.5 km (5.8%) | 16.3 km (11.1%) | 32.1 km (21.7%) | 147.5 km |
| 122 | Dairy | 60 | 6.6 km (8.5%) | 11.9 km (15.2%) | 22.7 km (29.1%) | 77.9 km |
| 122 | Non Dairy | 97 | 10.4 km (7.1%) | 19 km (12.9%) | 36.7 km (24.9%) | 147.5 km |
| 010 | All | 148 | 14.2 km (10.2%) | 39.7 km (28.6%) | 59.6 km (42.9%) | 138.9 km |
| 010 | Dairy | 46 | 15 km (12.0%) | 43.7 km (35%) | 59.9 km (48.1%) | 124.7 km |
| 010 | Non Dairy | 102 | 12.9 km (9.7%) | 37.7 km (28.3%) | 59.3 km (44.4%) | 133.5 km |
| 027 | All | 157 | 18.7 km (11.4%) | 46.9 km (28.6%) | 88.5 km (54.1%) | 163.8 km |
| 027 | Dairy | 26 | 27.8 km (20.3%) | 61.2 km (44.8%) | 84.1 km (61.6%) | 136.4 km |
| 027 | Non Dairy | 131 | 13.8 km (9.2%) | 38.9 km (25.9%) | 84.1 km (56.1%) | 150.0 km |
| 117 | All | 116 | 15 km (10.0%) | 26.2 km (17.5%) | 38.8 km (25.9%) | 149.7 km |
| 117 | Dairy | 37 | 12.3 km (9.0%) | 23.7 km (17.3%) | 37.4 km (27.3%) | 136.8 km |
| 117 | Non Dairy | 79 | 14.6 km (9.8%) | 25.2 km (17.0%) | 39.5 km (26.6%) | 148.5 km |
| NN Distance for Hosts Which Share an MLVA Type | NN Distance for Hosts Which Do Not Share an MLVA Type | ||||||
|---|---|---|---|---|---|---|---|
| Median (km) | Q1 (km) | Q4 (km) | Median (km) | Q1 (km) | Q4 (km) | Difference in Medians (km) | |
| Badger-Badger | 2.44 | 1.22 | 4.04 | 3.33 | 1.94 | 5.22 | 0.89 |
| Badger-Cattle (all herds) | 0.82 | 0.50 | 1.64 | 1.49 | 0.83 | 2.25 | 0.67 |
| Badger-Cattle (non-dairy) | 1.14 | 0.61 | 1.94 | 2.55 | 1.62 | 3.74 | 1.41 |
| Badger-Cattle (dairy) | 1.70 | 0.85 | 2.73 | 2.88 | 1.85 | 4.03 | 1.18 |
| Cattle-Cattle (all herds) | 1.12 | 0.61 | 2.32 | 0.82 | 0.51 | 1.27 | 0.29 |
| Cattle-Cattle (dairy) | 1.60 | 0.85 | 2.95 | 1.39 | 0.82 | 2.02 | 0.21 |
| Cattle-Cattle (non-dairy) | 1.28 | 0.67 | 2.66 | 0.93 | 0.58 | 1.50 | 0.35 |
| Variable | Match n = 9471 (61.4%) | No match n = 5964 (38.6%) | Est. | Std. Error | z Value | OR | 95% CI Lower | 95% CI Upper |
|---|---|---|---|---|---|---|---|---|
| Intercept | - | - | 0.19 | 0.19 | 0.99 | 1.21 | 0.83 | 1.75 |
| Number of MLVA types (per type) | Med: 1 IQR: 1–1; Max: 10 | Med: 1; IQR: 1–1; Max: 5 | 0.85 | 0.04 | 20.28 | 2.34 | 2.16 | 2.54 |
| Distance (per km) | Med: 4.4 km IQR: 2.9–5.7 km | Med: 4.9 km IQR: 3.3–6 km | −0.12 | 0.01 | −11.61 | 0.89 | 0.87 | 0.91 |
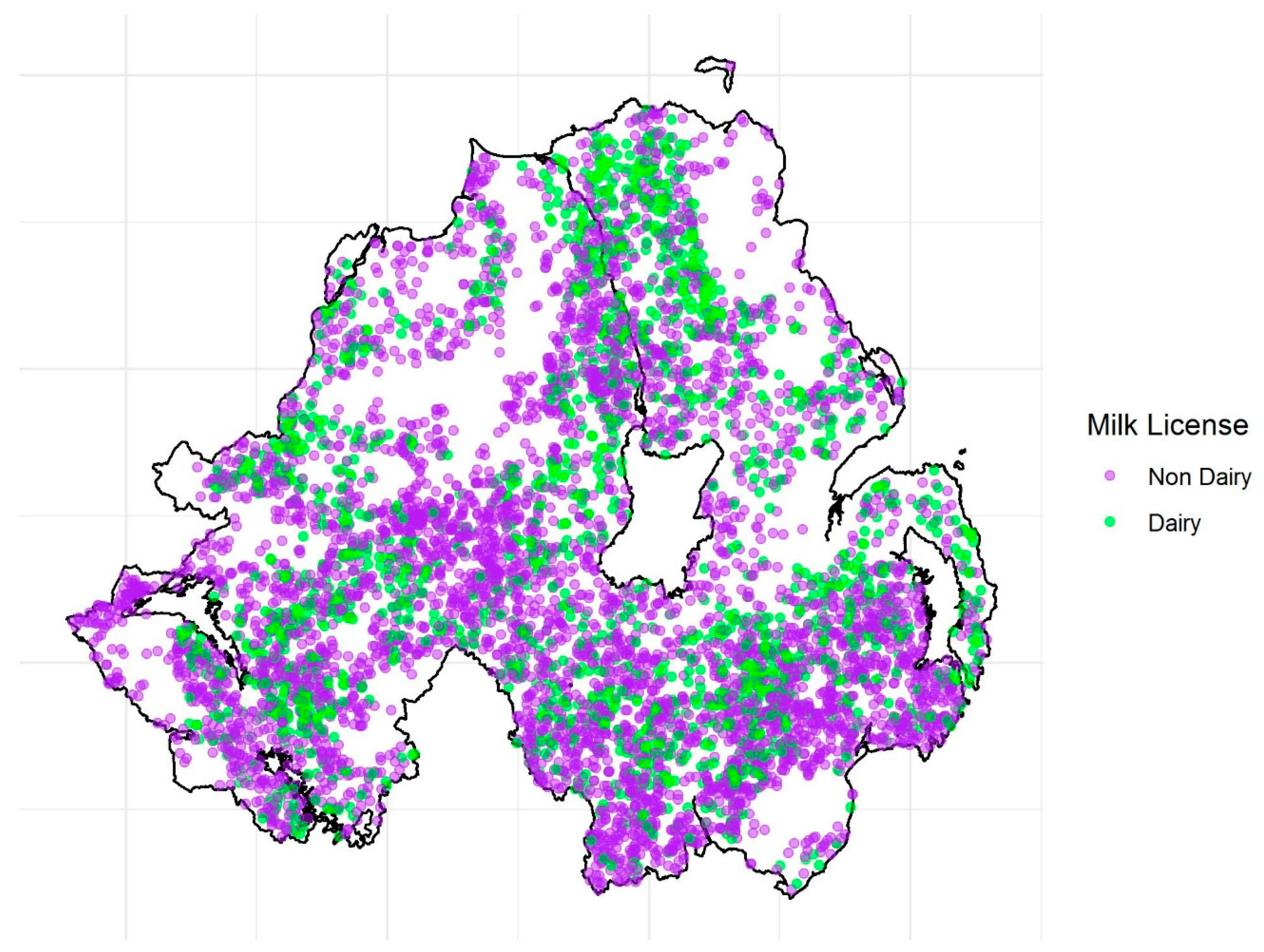
| Milk license (absent) | 6083 (64.2%) | 4261 (71.5%) | −0.28 | 0.04 | −7.42 | 0.75 | 0.70 | 0.81 |
| Inwards cattle movements (per 10 animals) | Med: 4 IQR: 2–9 | Med: 5 IQR: 2–11 | −0.02 | 0.001 | −11.00 | 0.99 | 0.98 | 0.99 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Milne, G.; Allen, A.; Graham, J.; Kirke, R.; McCormick, C.; Presho, E.; Skuce, R.; Byrne, A.W. Mycobacterium bovis Population Structure in Cattle and Local Badgers: Co-Localisation and Variation by Farm Type. Pathogens 2020, 9, 592. https://doi.org/10.3390/pathogens9070592
Milne G, Allen A, Graham J, Kirke R, McCormick C, Presho E, Skuce R, Byrne AW. Mycobacterium bovis Population Structure in Cattle and Local Badgers: Co-Localisation and Variation by Farm Type. Pathogens. 2020; 9(7):592. https://doi.org/10.3390/pathogens9070592
Chicago/Turabian StyleMilne, Georgina, Adrian Allen, Jordon Graham, Raymond Kirke, Carl McCormick, Eleanor Presho, Robin Skuce, and Andrew W. Byrne. 2020. "Mycobacterium bovis Population Structure in Cattle and Local Badgers: Co-Localisation and Variation by Farm Type" Pathogens 9, no. 7: 592. https://doi.org/10.3390/pathogens9070592
APA StyleMilne, G., Allen, A., Graham, J., Kirke, R., McCormick, C., Presho, E., Skuce, R., & Byrne, A. W. (2020). Mycobacterium bovis Population Structure in Cattle and Local Badgers: Co-Localisation and Variation by Farm Type. Pathogens, 9(7), 592. https://doi.org/10.3390/pathogens9070592







