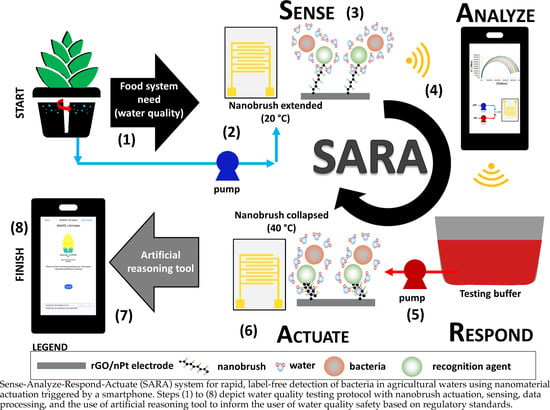
Sense–Analyze–Respond–Actuate (SARA) Paradigm: Proof of Concept System Spanning Nanoscale and Macroscale Actuation for Detection of Escherichia coli in Aqueous Media
Abstract
:1. Introduction
Pathogen Biosensors in Food Systems
2. Materials and Methods
2.1. Materials and Bacteria Cultures
2.2. Image Analysis
2.3. Nanomaterial Deposition and Electrode Biofunctionalization
2.4. Electrochemical Analysis
2.5. Biosensor Performance Testing
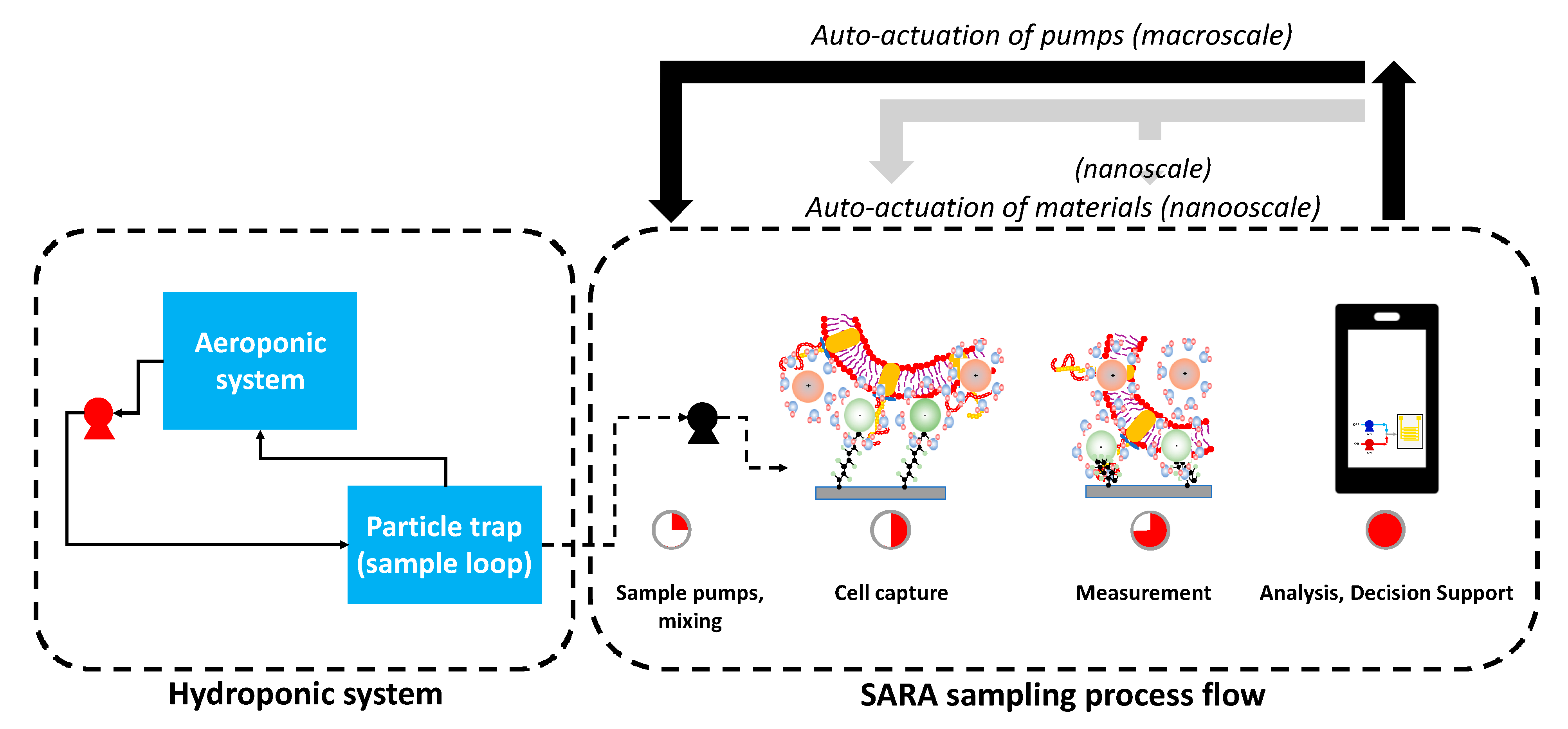
2.6. SARA Paradigm Proof of Concept in Hydroponic System
2.7. Decision Support Application
2.8. Statistical Analysis
3. Results
3.1. Electrochemical Characterization
3.2. Nanobrush Morphology
3.3. Nanobrush Actuation and E. coli Capture
3.4. Nanobrush Sensing in Buffer
3.5. Nanobrush Sensing in Food Samples
| Sensing Platform/Biorecognition Agent | Media | Bacteria | Detection Time (min) | Range (CFU/mL) | LOD (CFU/mL) | Reference |
|---|---|---|---|---|---|---|
| PNIPAAm-ConA | PBS | E. coli K12 | 17 | 3 × 102 to 3 × 105 | 5.0 ± 2.9 | This work |
| PNIPAAm-ConA | PBS | E. coli K12 and Salmonella Enteritidis | 17 | 3 × 102 to 3 × 105 | 2.3 ± 0.8 | This work |
| PNIPAAm-ConA | Vegetable broth | E. coli O157:H7 | 17 | 2.6 × 102 to 2.6 × 106 | 1560.0 ± 202.3 | This work |
| PNIPAAm-Antibody | Vegetable broth | E. coli O157:H7 | 17 | 1.5 × 102 to 1.5 × 106 | 249.3 ± 51.3 | This work |
| Polyanilyne-Antibody | Phosphate Citrate Buffer | E. coli O157:H7 | NR | 102 to 107 | 102 | Chowdhury et al. [80] |
| 11-MUA SAM-ConA * | Carrier buffer | E. coli | <20 | 12 to 1.2 × 106 | 12 | Jantra et al. [81] |
| Hyaluronic acid-Antibody | PBS | E. coli O157:H7 | NR | 10 to 105 | 10 | Joung et al. [78] |
| Hyaluronic acid-Antibody | Whole milk | E. coli O157:H7 | NR | NR | 83 | Joung et al. [78] |
| CHIT-MWNTs-SiO2@THI ** | PBS | E. coli O157:H7 | <45 | 4.1 × 102 to 12 × 105 | 250 | Li et al. [82] |
| Bayhydrol 110-Mannose-ConA *** | Water | E. coli O157:H7 | 180 | 6 to 60 × 109 | 60 | Lu et al. [83] |
| Gold electrode array-Antibody | Lettuce Wash Water | E. coli O157:H7 | 10 | 104 to 107 | 104 | Radke & Alocilja [77] |
3.6. SARA: Sense-Analyze-Respond-Actuate
3.7. Decision Support Application
3.8. Decision Support Application—Cybersecurity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Valderrama, W.B.; Dudley, E.G.; Doores, S.; Cutter, C.N. Commercially Available Rapid Methods for Detection of Selected Food-borne Pathogens. Crit. Rev. Food Sci. Nutr. 2016, 56, 1519–1531. [Google Scholar] [CrossRef]
- Vanegas, D.C.; Gomes, C.L.; Cavallaro, N.D.; Giraldo-Escobar, D.; McLamore, E.S. Emerging Biorecognition and Transduction Schemes for Rapid Detection of Pathogenic Bacteria in Food. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1188–1205. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Salazar, J.K. Culture-Independent Rapid Detection Methods for Bacterial Pathogens and Toxins in Food Matrices. Compr. Rev. Food Sci. Food Saf. 2016, 15, 183–205. [Google Scholar] [CrossRef]
- Umesha, S.; Manukumar, H.M. Advanced molecular diagnostic techniques for detection of food-borne pathogens: Current applications and future challenges. Crit. Rev. Food Sci. Nutr. 2018, 58, 84–104. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). National Outbreak Reporting System (NORS). 2020. Available online: https://wwwn.cdc.gov/norsdashboard/ (accessed on 14 October 2020).
- Centers for Disease Control and Prevention (CDC). Estimates of Foodborne Illness in the United States. 2018. Available online: https://www.cdc.gov/foodborneburden/2011-foodborne-estimates.html (accessed on 27 July 2020).
- Rai, P.K.; Tripathi, B.D. Microbial contamination in vegetables due to irrigation with partially treated municipal wastewater in a tropical city. Int. J. Environ. Health Res. 2007, 17, 389–395. [Google Scholar] [CrossRef]
- Callejón, R.M.; Rodríguez-Naranjo, M.I.; Ubeda, C.; Hornedo-Ortega, R.; Garcia-Parrilla, M.C.; Troncoso, A.M. Reported Foodborne Outbreaks Due to Fresh Produce in the United States and European Union: Trends and Causes. Foodborne Pathog. Dis. 2015, 12, 32–38. [Google Scholar] [CrossRef]
- Odonkor, S.T.; Ampofo, J.K. Escherichia coli as an indicator of bacteriological quality of water: An overview. Microbiol. Res. 2013, 4, e2. [Google Scholar] [CrossRef]
- Feng, P.; Weagant, S. Chapter 4A: Diarrheagenic Escherichia coli. U.S. Food & Drug Administration (FDA). Bacteriological Analytical Manual. 2011. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-4a-diarrheagenic-escherichia-coli (accessed on 24 June 2020).
- Walker, C.L.F.; Applegate, J.A.; Black, R.E. Haemolytic-uraemic syndrome as a sequela of diarrhoeal disease. J. Health Popul. Nutr. 2012, 30, 257–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasko, D.A.; Rosovitz, M.J.; Myers, G.S.A.; Mongodin, E.F.; Fricke, W.F.; Gajer, P.; Crabtree, J.; Sebaihia, M.; Thomson, N.R.; Chaudhuri, R.; et al. The Pangenome Structure of Escherichia coli: Comparative Genomic Analysis of E. coli Commensal and Pathogenic Isolates. J. Bacteriol. 2008, 190, 6881–6893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centers for Disease Control and Prevention (CDC). Multistate Outbreak of E. coli O157:H7 Infections Linked to Romaine Lettuce. 2018. Available online: https://www.cdc.gov/ecoli/2018/o157h7-04-18/index.html (accessed on 24 July 2020).
- Allende, A.; Datta, A.R.; Smith, W.A.; Adonis, R.; MacKay, A.; Adell, A.D. Implications of new legislation (US FSMA) and guidelines (EC) on the establishment of management systems for agricultural water. Food Microbiol. 2018, 75, 119–125. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration (FDA). FSMA Final Rule on Produce Safety; FDA: Silver Spring, MD, USA, 2020.
- U.S. Food and Drug Administration (FDA). FSMA Rules & Guidance for Industry. 2020. Available online: https://www.fda.gov/food/food-safety-modernization-act-fsma/fsma-rules-guidance-industry (accessed on 24 July 2020).
- Rezaei, A. Food safety: The farmer first health paradigm. One Health 2018, 5, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Ahearn, M.C.; Armbruster, W.; Young, R. Big Data’s Portential to Improve Food Supply Chain Environmental Sustainability and Food Safety. Int. Food Agribus. Manag. Rev. 2016, 19, 155–171. [Google Scholar]
- McLamore, E.S.; Palit Austin Datta, S.; Morgan, V.; Cavallaro, N.; Kiker, G.; Jenkins, D.M.; Rong, Y.; Gomes, C.; Claussen, J.; Vanegas, D.; et al. SNAPS: Sensor Analytics Point Solutions for Detection and Decision Support Systems. Sensors 2019, 19, 4935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armbruster, W.; MacDonell, M.M. Informatics to Support International Food Safety. In Proceedings of the 28th EnviroInfo 2014 Conference, Oldenburg, Germany, 10–12 September 2014. [Google Scholar]
- Marvin, H.J.P.; Janssen, E.M.; Bouzembrak, Y.; Hendriksen, P.J.M.; Staats, M. Big data in food safety: An overview. Crit. Rev. Food Sci. Nutr. 2017, 57, 2286–2295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Food and Drug Administration (FDA). Standards for the Growing, Harvesting, Packing, and Holding of Produce for Human Consumption: What You Need to Know About the FDA Regulation: Guidance for Industry Small Entity Compliance Guide. 2017. Available online: https://www.fda.gov/media/107298/download (accessed on 24 July 2020).
- Rock, C.M.; Brassill, N.; Dery, J.L.; Carr, D.; McLain, J.E.; Bright, K.R.; Gerba, C.P. Review of water quality criteria for water reuse and risk-based implications for irrigated produce under the FDA Food Safety Modernization Act, produce safety rule. Environ. Res. 2019, 172, 616–629. [Google Scholar] [CrossRef]
- Markland, S.M.; Ingram, D.; Kniel, K.E.; Sharma, M. Water for Agriculture: The Convergence of Sustainability and Safety. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef] [Green Version]
- United States Department of Agriculture Food Safety and Inspection Services (USDA FSIS). FSIS Compliance Guideline for Controlling Meat and Poultry Products Pending FSIS Test Results. 2013. Available online: https://www.fsis.usda.gov/wps/wcm/connect/6b7b5a65-9ad3-4f89-927d-078e564aca24/Compliance_Guide_Test_Hold_020113.pdf?MOD=AJPERES (accessed on 24 July 2020).
- Pozo, V.F.; Schroeder, T.C. Evaluating the costs of meat and poultry recalls to food firms using stock returns. Food Policy 2016, 59, 66–77. [Google Scholar] [CrossRef]
- Castell-Perez, E.; Gomes, C.; Tahtouh, J.; Moreira, R.; McLamore, E.S.; Knowles, H.S. Food Processing and Waste Within the Nexus Framework. Curr. Sustain. Renew. Energy Rep. 2017, 4, 99–108. [Google Scholar] [CrossRef]
- Keener, L.; Nicholson-Keener, S.M.; Koutchma, T. Harmonization of legislation and regulations to achieve food safety: US and Canada perspective. J. Sci. Food Agric. 2014, 94, 1947–1953. [Google Scholar] [CrossRef]
- Morgan, V.; Casso-Hartmann, L.; Bahamon-Pinzon, D.; McCourt, K.; Hjort, R.G.; Bahramzadeh, S.; Velez-Torres, I.; McLamore, E.; Gomes, C.; Alocilja, E.C.; et al. Sensor-as-a-Service: Convergence of Sensor Analytic Point Solutions (SNAPS) and Pay-A-Penny-Per-Use (PAPPU) Paradigm as a Catalyst for Democratization of Healthcare in Underserved Communities. Diagnostics 2020, 10, 22. [Google Scholar] [CrossRef] [Green Version]
- Vanegas, D.; Gomes, C.; McLamore, E. Biosensors for Indirect Monitoring of Foodborne Bacteria. Biosens. J. 2016, 5, 137–139. [Google Scholar] [CrossRef] [Green Version]
- Duffy, G.; Lynch, O.A.; Cagney, C. Tracking emerging zoonotic pathogens from farm to fork. Meat Sci. 2008, 78, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Wassenaar, T.M. Following an imaginary Campylobacter population from farm to fork and beyond: A bacterial perspective. Lett. Appl. Microbiol. 2011, 53, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Fozouni, P.; Son, S.; Díaz de León Derby, M.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Direct detection of SARS-CoV-2 using CRISPR-Cas13a and a mobile phone. medRxiv 2020. [Google Scholar] [CrossRef]
- Bhunia, A.K.; Taitt, C.R.; Kim, M.S. High Throughput Screening for Food Safety Assessment; Woodhead Publishing: Cambridge, UK, 2015. [Google Scholar] [CrossRef]
- Varshney, M.; Li, Y. Interdigitated array microelectrode based impedance biosensor coupled with magnetic nanoparticle–antibody conjugates for detection of Escherichia coli O157:H7 in food samples. Biosens. Bioelectron. 2007, 22, 2408–2414. [Google Scholar] [CrossRef]
- Daniels, J.S.; Pourmand, N. Label-Free Impedance Biosensors: Opportunities and Challenges. Electroanalysis 2007, 19, 1239–1257. [Google Scholar] [CrossRef]
- Jenkins, D.M.; Lee, B.E.; Jun, S.; Reyes-De-Corcuera, J.; McLamore, E.S. ABE-Stat, a Fully Open-Source and Versatile Wireless Potentiostat Project Including Electrochemical Impedance Spectroscopy. J. Electrochem. Soc. 2019, 166, B3056–B3065. [Google Scholar] [CrossRef]
- Wang, X.; Feng, Y.; Dong, P.; Huang, J. A Mini Review on Carbon Quantum Dots: Preparation, Properties, and Electrocatalytic Application. Front. Chem. 2019, 7, 671. [Google Scholar] [CrossRef]
- Xiao, Z.; Kong, L.B.; Ruan, S.; Li, X.; Yu, S.; Li, X.; Jiang, Y.; Yao, Z.; Ye, S.; Wang, C.; et al. Recent development in nanocarbon materials for gas sensor applications. Sens. Actuators B Chem. 2018, 274, 235–267. [Google Scholar] [CrossRef]
- Taguchi, M.; Schwalb, N.; Rong, Y.; Vanegas, D.C.; Garland, N.; Tan, M.; Yamaguchi, H.; Claussen, J.C.; McLamore, E.S. pulSED: Pulsed sonoelectrodeposition of fractal nanoplatinum for enhancing amperometric biosensor performance. Analyst 2016, 141, 3367–3378. [Google Scholar] [CrossRef]
- Burrs, S.L.; Vanegas, D.C.; Bhargava, M.; Mechulan, N.; Hendershot, P.; Yamaguchi, H.; Gomes, C.; McLamore, E.S. A comparative study of graphene–hydrogel hybrid bionanocomposites for biosensing. Analyst 2015, 140, 1466–1476. [Google Scholar] [CrossRef] [PubMed]
- Vanegas, D.C.; Taguchi, M.; Chaturvedi, P.; Burrs, S.; Tan, M.; Yamaguchi, H.; McLamore, E.S. A comparative study of carbon–platinum hybrid nanostructure architecture for amperometric biosensing. Analyst 2014, 139, 660–667. [Google Scholar] [CrossRef] [PubMed]
- McLamore, E.S.; Mohanty, S.; Shi, J.; Claussen, J.; Jedlicka, S.S.; Rickus, J.L.; Porterfield, D.M. A self-referencing glutamate biosensor for measuring real time neuronal glutamate flux. J. Neurosci. Methods 2010, 189, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Claussen, J.C.; Hengenius, J.B.; Wickner, M.M.; Fisher, T.S.; Umulis, D.M.; Porterfield, D.M. Effects of Carbon Nanotube-Tethered Nanosphere Density on Amperometric Biosensing: Simulation and Experiment. J. Phys. Chem. C 2011, 115, 20896–20904. [Google Scholar] [CrossRef]
- Claussen, J.C.; Franklin, A.D.; Haque, A.; Porterfield, D.M.; Fisher, T.S. Electrochemical Biosensor of Nanocube-Augmented Carbon Nanotube Networks. ACS Nano 2009, 3, 37–44. [Google Scholar] [CrossRef]
- He, Q.; Das, S.R.; Garland, N.T.; Jing, D.; Hondred, J.A.; Cargill, A.A.; Ding, S.; Karunakaran, C.; Claussen, J.C. Enabling Inkjet Printed Graphene for Ion Selective Electrodes with Postprint Thermal Annealing. ACS Appl. Mater. Interfaces 2017, 9, 12719–12727. [Google Scholar] [CrossRef] [Green Version]
- Taguchi, M.; Ptitsyn, A.; McLamore, E.S.; Claussen, J.C. Nanomaterial-mediated Biosensors for Monitoring Glucose. J. Diabetes Sci. Technol. 2014, 8, 403–411. [Google Scholar] [CrossRef] [Green Version]
- Walcarius, A.; Minteer, S.D.; Wang, J.; Lin, Y.; Merkoçi, A. Nanomaterials for bio-functionalized electrodes: Recent trends. J. Mater. Chem. B 2013, 1, 4878–4908. [Google Scholar] [CrossRef]
- Hills, K.D.; Oliveira, D.A.; Cavallaro, N.D.; Gomes, C.L.; McLamore, E.S. Actuation of chitosan-aptamer nanobrush borders for pathogen sensing. Analyst 2018, 143, 1650–1661. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; ShamsiJazeyi, H.; Pesek, S.L.; Agrawal, A.; Hammouda, B.; Verduzco, R. Thermoresponsive PNIPAAM bottlebrush polymers with tailored side-chain length and end-group structure. Soft Matter 2014, 10, 2008–2015. [Google Scholar] [CrossRef] [Green Version]
- Yin, Z.; Zhang, J.; Jiang, L.-P.; Zhu, J.-J. Thermosensitive Behavior of Poly(N-isopropylacrylamide) and Release of Incorporated Hemoglobin. J. Phys. Chem. C 2009, 113, 16104–16109. [Google Scholar] [CrossRef]
- Leal, D.; De Borggraeve, W.; Encinas, M.V.; Matsuhiro, B.; Müller, R. Preparation and characterization of hydrogels based on homopolymeric fractions of sodium alginate and PNIPAAm. Carbohydr. Polym. 2013, 92, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Meng, S.; Mu, L.; Jin, G.; Zhong, W.; Kong, J. Novel renewable immunosensors based on temperature-sensitive PNIPAAm bioconjugates. Biosens. Bioelectron. 2008, 24, 710–715. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, Y.; Lu, J.; Zhou, J.; Li, J. Temperature-Responsive Polymer/Carbon Nanotube Hybrids: Smart Conductive Nanocomposite Films for Modulating the Bioelectrocatalysis of NADH. Chem. A Eur. J. 2012, 18, 3687–3694. [Google Scholar] [CrossRef] [PubMed]
- Gamella, M.; Campuzano, S.; Parrado, C.; Reviejo, A.J.; Pingarrón, J.M. Microorganisms recognition and quantification by lectin adsorptive affinity impedance. Talanta 2009, 78, 1303–1309. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Orozco, J.; Kagan, D.; Guix, M.; Gao, W.; Sattayasamitsathit, S.; Claussen, J.C.; Merkoçi, A.; Wang, J. Bacterial Isolation by Lectin-Modified Microengines. Nano Lett. 2012, 12, 396–401. [Google Scholar] [CrossRef]
- Vanegas, D.C.; Clark, G.; Cannon, A.E.; Roux, S.; Chaturvedi, P.; McLamore, E.S. A self-referencing biosensor for real-time monitoring of physiological ATP transport in plant systems. Biosens. Bioelectron. 2015, 74, 37–44. [Google Scholar] [CrossRef]
- Vanegas, D.C.; Patiño, L.; Mendez, C.; Oliveira, D.A.D.; Torres, A.M.; Gomes, C.L.; McLamore, E.S. Laser Scribed Graphene Biosensor for Detection of Biogenic Amines in Food Samples Using Locally Sourced Materials. Biosensors 2018, 8, 42. [Google Scholar] [CrossRef] [Green Version]
- Burrs, S.L.; Bhargava, M.; Sidhu, R.; Kiernan-Lewis, J.; Gomes, C.; Claussen, J.C.; McLamore, E.S. A paper based graphene-nanocauliflower hybrid composite for point of care biosensing. Biosens. Bioelectron. 2016, 85, 479–487. [Google Scholar] [CrossRef] [Green Version]
- Soares, R.R.A.; Hjort, R.G.; Pola, C.C.; Parate, K.; Reis, E.L.; Soares, N.F.F.; McLamore, E.S.; Claussen, J.C.; Gomes, C.L. Laser-Induced Graphene Electrochemical Immunosensors for Rapid and Label-Free Monitoring of Salmonella enterica in Chicken Broth. ACS Sens. 2020, 5, 1900–1911. [Google Scholar] [CrossRef]
- Sidhu, R.K.; Cavallaro, N.D.; Pola, C.C.; Danyluk, M.D.; McLamore, E.S.; Gomes, C.L. Planar Interdigitated Aptasensor for Flow-Through Detection of Listeria spp. in Hydroponic Lettuce Growth Media. Sensors 2020, 20, 5773. [Google Scholar] [CrossRef] [PubMed]
- Marhaenanto, B.; Soni, P.; Salakhe, V.M. Development of an internet-based greenhouse control system. Int. Agric. Eng. J. 2013, 22, 72–83. [Google Scholar]
- McLamore, E.S.; Jaroch, D.; Chatni, M.R.; Porterfield, D.M. Self-referencing optrodes for measuring spatially resolved, real-time metabolic oxygen flux in plant systems. Planta 2010, 232, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Dean, D.A.; Ramanathan, T.; Machado, D.; Sundararajan, R. Electrical Impedance Spectroscopy Study of Biological Tissues. J. Electrost. 2008, 66, 165–177. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Nawroth, J.C.; McFall-Ngai, M.J.; Kanso, E. Mixing and transport by ciliary carpets: A numerical study. J. Fluid Mech. 2014, 743, 124–140. [Google Scholar] [CrossRef] [Green Version]
- Tsumori, F.; Saijou, A.; Osada, T.; Miura, H. Development of actuation system for artificial cilia with magnetic elastomer. Jpn. J. Appl. Phys. 2015, 54. [Google Scholar] [CrossRef]
- Hanasoge, S.; Hesketh, P.J.; Alexeev, A. Metachronal motion of artificial magnetic cilia. Soft Matter 2018, 14, 3689–3693. [Google Scholar] [CrossRef] [Green Version]
- Reeke, G.N.; Becker, J.W.; Edelman, G.M. The covalent and three-dimensional structure of concanavalin A. IV. Atomic coordinates, hydrogen bonding, and quaternary structure. J. Biol. Chem. 1975, 250, 1525–1547. [Google Scholar]
- Castillo-Torres, K.Y.; Arnold, D.P.; McLamore, E.S. Rapid isolation of Escherichia coli from water samples using magnetic microdiscs. Sens. Actuators B Chem. 2019, 291, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Danielsen, E.M.; Hansen, G.H. Lipid rafts in epithelial brush borders: Atypical membrane microdomains with specialized functions. Biochim. Biophys. Acta Biomembr. 2003, 1617, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Samuel, G.; Reeves, P. Biosynthesis of O-antigens: Genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 2003, 338, 2503–2519. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Harrison, S.H.; Schoeniger, J.S. Gangliosides as Receptors for Biological Toxins: Development of Sensitive Fluoroimmunoassays Using Ganglioside-Bearing Liposomes. Anal. Chem. 2000, 72, 6019–6024. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.L.; White, A.P.; Snyder, S.D.; Martin, S.; Heiss, C.; Azadi, P.; Surette, M.; Kay, W.W. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J. Bacteriol. 2006, 188, 7722–7730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miura, Y.; Hoshino, Y.; Seto, H. Glycopolymer Nanobiotechnology. Chem. Rev. 2016, 116, 1673–1692. [Google Scholar] [CrossRef]
- McLellan, J.S.; Chen, M.; Joyce, M.G.; Sastry, M.; Stewart-Jones, G.B.E.; Yang, Y.; Zhang, B.; Chen, L.; Srivatsan, S.; Zheng, A.; et al. Structure-Based Design of a Fusion Glycoprotein Vaccine for Respiratory Syncytial Virus. Science 2013, 342, 592–598. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, C.-L.; Goldsmith, J.A.; Schaub, J.M.; DiVenere, A.M.; Kuo, H.-C.; Javanmardi, K.; Le, K.C.; Wrapp, D.; Lee, A.G.; Liu, Y.; et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020, 369, 1501–1505. [Google Scholar] [CrossRef]
- Radke, S.M.; Alocilja, E.C. A high density microelectrode array biosensor for detection of E. coli O157:H7. Biosens. Bioelectron. 2005, 20, 1662–1667. [Google Scholar] [CrossRef]
- Joung, C.-K.; Kim, H.-N.; Lim, M.-C.; Jeon, T.-J.; Kim, H.-Y.; Kim, Y.-R. A nanoporous membrane-based impedimetric immunosensor for label-free detection of pathogenic bacteria in whole milk. Biosens. Bioelectron. 2013, 44, 210–215. [Google Scholar] [CrossRef]
- Chowdhury, A.; Raut, S.A.; Narman, H.S. DA-DRLS: Drift adaptive deep reinforcement learning based scheduling for IoT resource management. J. Netw. Comput. Appl. 2019, 138, 51–65. [Google Scholar] [CrossRef]
- Chowdhury, A.D.; De, A.; Chaudhuri, C.R.; Bandyopadhyay, K.; Sen, P. Label free polyaniline based impedimetric biosensor for detection of E. coli O157:H7 Bacteria. Sens. Actuators B Chem. 2012, 171, 916–923. [Google Scholar] [CrossRef]
- Jantra, J.; Kanatharana, P.; Asawatreratanakul, P.; Hedström, M.; Mattiasson, B.; Thavarungkul, P. Real-time label-free affinity biosensors for enumeration of total bacteria based on immobilized concanavalin A. J. Environ. Sci. Health Part A 2011, 46, 1450–1460. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cheng, P.; Gong, J.; Fang, L.; Deng, J.; Liang, W.; Zheng, J. Amperometric immunosensor for the detection of Escherichia coli O157:H7 in food specimens. Anal. Biochem. 2012, 421, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Lin, H.; Ge, S.; Luo, S.; Cai, Q.; Grimes, C.A. Wireless, Remote-Query, and High Sensitivity Escherichia coli O157:H7 Biosensor Based on the Recognition Action of Concanavalin A. Anal. Chem. 2009, 81, 5846–5850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aloul, F.; Zahidi, S.; El-Hajj, W. Two factor authentication using mobile phones. In Proceedings of the 2009 IEEE/ACS International Conference on Computer Systems and Applications, Rabat, Morocco, 10–13 May 2009; pp. 641–644. [Google Scholar]
- Fielding, R.T. Architectural Styles and the Design of Network-Based Software Architectures. Ph.D. Thesis, University of California, Irvine, CA, USA, 2000. [Google Scholar]
- Surbatovich, M.; Aljuraidan, J.; Bauer, L.; Das, A.; Jia, L. Some Recipes Can Do More Than Spoil Your Appetite: Analyzing the Security and Privacy Risks of IFTTT Recipes. In Proceedings of the 26th International Conference on World Wide Web, Perth, Australia, 3–7 May 2017; pp. 1501–1510. [Google Scholar]
- TSN. Time-Sensitive Networking (TSN) Task Group. Available online: https://1.ieee802.org/tsn/ (accessed on 24 October 2020).
- NIST. NIST Cybersecurity for IoT Program. 2020. Available online: https://www.nist.gov/programs-projects/nist-cybersecurity-iot-program (accessed on 23 August 2020).
- Datta, S.P.A. Emergence of Digital Twins: Is This the March of Reason? 2016. Available online: https://dspace.mit.edu/handle/1721.1/104429 (accessed on 20 October 2020).
- CISA. UDP-Based Amplification Attacks. Available online: https://us-cert.cisa.gov/ncas/alerts/TA14-017A (accessed on 25 October 2020).
- Galeano-Brajones, J.; Carmona-Murillo, J.; Valenzuela-Valdés, J.F.; Luna-Valero, F. Detection and Mitigation of DoS and DDoS Attacks in IoT-Based Stateful SDN: An Experimental Approach. Sensors 2020, 20, 816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NIST. Mitigating IoT-Based DDoS. Available online: https://www.nccoe.nist.gov/projects/building-blocks/mitigating-iot-based-ddos (accessed on 25 October 2020).
- Srinidhi, N.N.; Dilip Kumar, S.M.; Venugopal, K.R. Network optimizations in the Internet of Things: A review. Eng. Sci. Technol. Int. J. 2019, 22, 1–21. [Google Scholar] [CrossRef]
- Lee, E.A. Computing needs time. Commun. ACM 2009, 52, 70–79. [Google Scholar] [CrossRef]
- Humphreys, T.E.; Ledvina, B.M.; Psiaki, M.L.; O’Hanlon, B.W.; Kintner, P.M. Assessing the Spoofing Threat: Development of a Portable GPS Civilian Spoofer. In Proceedings of the ION GNSS Conference, Savannah, GA, USA, 16–19 September 2008. [Google Scholar]
- King, C.; Chu, J.; Mellinger, A.O. Emerging Technology Domains Risk Survey; Carnegie Mellon University (CMU): Pittsburgh, PA, USA, 2015. [Google Scholar]
- Wei, X.; Sikdar, B. Impact of GPS Time Spoofing Attacks on Cyber Physical Systems. In Proceedings of the 2019 IEEE International Conference on Industrial Technology (ICIT), Melbourne, Australia, 13–15 February 2019; pp. 1155–1160. [Google Scholar]
- Zhao, B.; Chen, Q. Location Spoofing in a Location-Based Game: A Case Study of Pokémon Go. In Proceedings of the International Cartographic Conference, Durban, South Africa, 10–16 August 2003; pp. 21–32. [Google Scholar]
- National Institute of Standards and Technology. Cybersecurity Profile for the Responsible Use of Positioning Navigation and Timing (PNT) Sevices; NIST: Gaithersburg, MD, USA, 2020.
- Psiaki, M.L.; Humphreys, T.E. Protecting GPS from Spoofers is Critical to the Future of Navigation. Available online: https://spectrum.ieee.org/telecom/security/protecting-gps-from-spoofers-is-critical-to-the-future-of-navigation (accessed on 25 October 2020).
- Eldosouky, A.; Ferdowsi, A.; Saad, W. Drones in Distress: A Game-Theoretic Countermeasure for Protecting UAVs against GPS Spoofing. IEEE Internet Things J. 2020, 7, 2840–2854. [Google Scholar] [CrossRef] [Green Version]
- Mallikarjun, B.C.; Kiranmayi, K.J.; Lavanya, N.; Prateeksha, K.H.; Sushmitha, J. Intruder Detection System—A LoRa Based Approach. In Proceedings of the 2020 5th International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 10–12 June 2020; pp. 255–258. [Google Scholar]
- Microsoft. Azure Active Directory Authentication Documentation. Available online: https://docs.microsoft.com/en-us/azure/active-directory/authentication/ (accessed on 24 October 2020).
- Das, M.L.; Samdaria, N. On the security of SSL/TLS-enabled applications. Appl. Comput. Inform. 2014, 10, 68–81. [Google Scholar] [CrossRef] [Green Version]
- MIT. Certificates. Available online: https://ist.mit.edu/certificates (accessed on 25 October 2020).
- Sarma, S.; Brock, D.; Engels, D. Radio frequency identification and the electronic product code. IEEE Micro 2001, 21, 50–54. [Google Scholar] [CrossRef]
- Sarma, S.E.; Weis, S.A.; Engels, D.W. RFID Systems and Security and Privacy Implications; Springer: Berlin/Heidelberg, Germany, 2002; pp. 454–469. [Google Scholar]
- Shabandri, B.; Maheshwari, P. Enhancing IoT Security and Privacy Using Distributed Ledgers with IOTA and the Tangle. In Proceedings of the 2019 6th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 7–8 March 2019; pp. 1069–1075. [Google Scholar]
- Djaja, F. Send IoT Data to the IOTA Tangle with SAP HANA XSA and Analytics Cloud. Available online: https://blogs.sap.com/2019/10/08/send-iot-data-to-the-iota-tangle-with-sap-hana-xsa-and-analytics-cloud/ (accessed on 24 October 2020).







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giacobassi, C.A.; Oliveira, D.A.; Pola, C.C.; Xiang, D.; Tang, Y.; Datta, S.P.A.; McLamore, E.S.; Gomes, C.L. Sense–Analyze–Respond–Actuate (SARA) Paradigm: Proof of Concept System Spanning Nanoscale and Macroscale Actuation for Detection of Escherichia coli in Aqueous Media. Actuators 2021, 10, 2. https://doi.org/10.3390/act10010002
Giacobassi CA, Oliveira DA, Pola CC, Xiang D, Tang Y, Datta SPA, McLamore ES, Gomes CL. Sense–Analyze–Respond–Actuate (SARA) Paradigm: Proof of Concept System Spanning Nanoscale and Macroscale Actuation for Detection of Escherichia coli in Aqueous Media. Actuators. 2021; 10(1):2. https://doi.org/10.3390/act10010002
Chicago/Turabian StyleGiacobassi, Cassie A., Daniela A. Oliveira, Cicero C. Pola, Dong Xiang, Yifan Tang, Shoumen Palit Austin Datta, Eric S. McLamore, and Carmen L. Gomes. 2021. "Sense–Analyze–Respond–Actuate (SARA) Paradigm: Proof of Concept System Spanning Nanoscale and Macroscale Actuation for Detection of Escherichia coli in Aqueous Media" Actuators 10, no. 1: 2. https://doi.org/10.3390/act10010002