Bugs on Drugs: A Drosophila melanogaster Gut Model to Study In Vivo Antibiotic Tolerance of E. coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultivation of Drosohila melanogaster and Escherichia coli
2.2. Generation of Axenic Flies
2.3. Stability of Antibiotics in 10/10 Fly Food
SpanSlow = (P − Y0) × (100 − PercentFast) × 0.01,
Y = Y0 + SpanFast × (1 − exp(−KFast × X)) + SpanSlow × (1 − exp(−KSlow × X)),
2.4. Development Dynamics on Antibiotics
2.5. Feeding Rate in Adult Flies
2.6. Generating Gnotobiotic Flies
2.7. In Vivo Bacterial Load in Absence and Presence of Antibiotic Treatment
2.8. Microscopy
2.9. Statistics
3. Results
3.1. Effects of Antibiotics on D. melanogaster
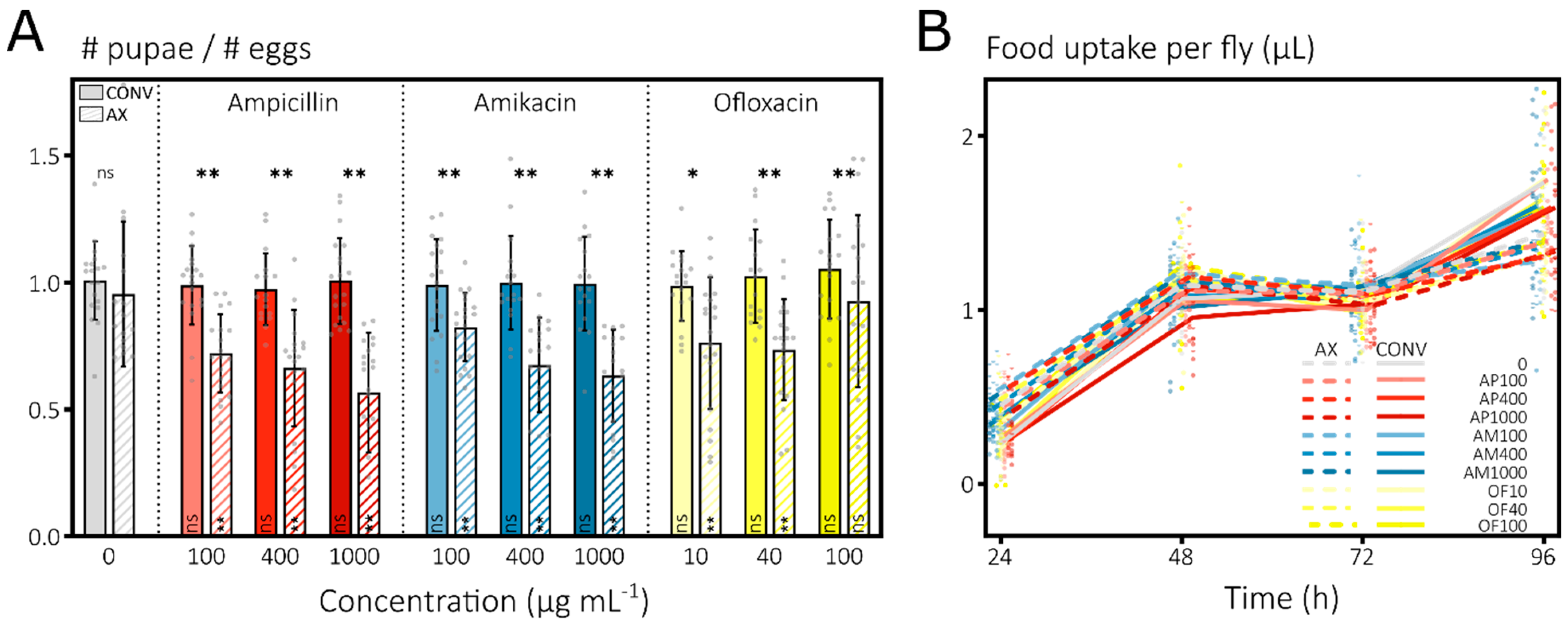
3.1.1. Antibiotics Induce Strong Mortality in Developing D. melanogaster Larvae
3.1.2. Antibiotics Do Not Impact Mortality or Feeding of Adult D. melanogaster Flies
3.2. E. coli Associates with Adult D. melanogaster and Locates Preferentially in the Crop
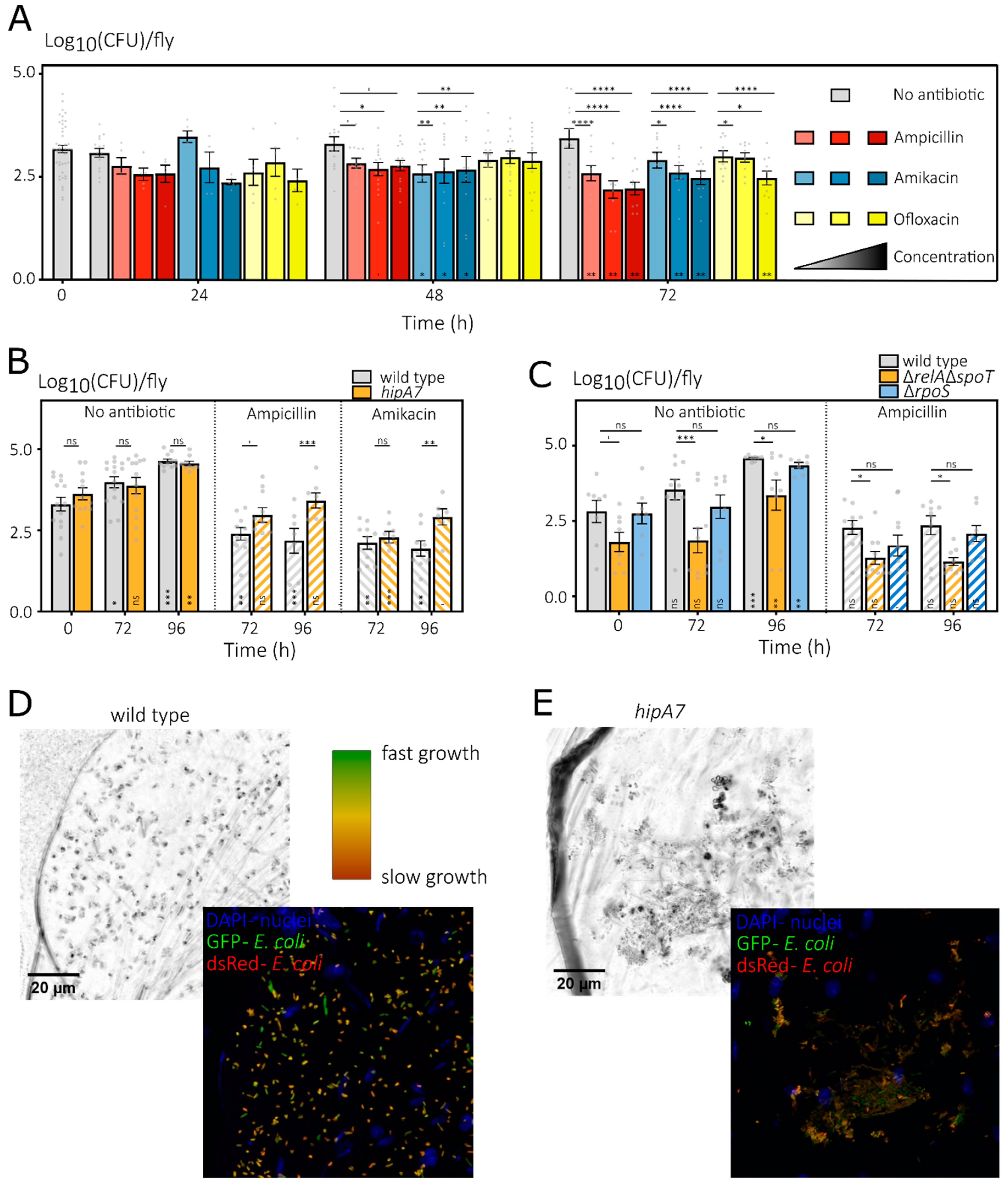
3.2.1. Bacterial Load of E. coli Associated with D. melanogaster Increases over Time
3.2.2. E. coli Is Preferentially Present at the Crop of the Drosophila Digestive Tract
3.3. In Vitro Identified Persistence Mutants Show Mixed Results during In Vivo Antibiotic Treatments
4. Discussion and Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lewis, K. The Science of Antibiotic Discovery. Cell 2020, 181, 29–45. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Rigol, S. A brief history of antibiotics and select advances in their synthesis. J. Antibiot. 2018, 71, 153–184. [Google Scholar] [CrossRef]
- Jones, D.S.; Podolsky, S.H.; Greene, J.A. The Burden of Disease and the Changing Task of Medicine. N. Engl. J. Med. 2012, 366, 2333–2338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laxminarayan, R.; Matsoso, P.; Pant, S.; Brower, C.; Røttingen, J.-A.; Klugman, K.; Davies, S. Access to effective antimicrobials: A worldwide challenge. Lancet 2016, 387, 168–175. [Google Scholar] [CrossRef]
- Karyne, R.; Lechuga, G.C.; Souza, A.L.A.; da Silva Carvalho, J.P.R.; Bôas, M.H.S.V.; De Simone, S.G. Pan-drug resistant Acinetobacter baumannii, but not other strains, are resistant to the bee venom peptide mellitin. Antibiotics 2020, 9, 178. [Google Scholar]
- Karakonstantis, S.; Kritsotakis, E.I.; Gikas, A. Pandrug-resistant gram-negative bacteria: A systematic review of current epidemiology, prognosis and treatment options. J. Antimicrob. Chemother. 2020, 75, 271–282. [Google Scholar] [CrossRef]
- Chen, L.; Todd, R.; Julia, K.; Maroya, W.; Alexander, K. Pan-Resistant New Delhi Metallo-Beta-Lactamase-Producing Klebsiella pneumoniae—Washoe County, Nevada, 2016. US Dep. Health Hum. Serv. Dis. Control Prev. 2017, 66, 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Center for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States. 2019. Available online: https://www.cdc.gov/DrugResistance/Biggest-Threats.html (accessed on 4 January 2022).
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Hede, K. Antibiotic resistance: An infectious arms race. Nature 2014, 509, S2–S3. [Google Scholar] [CrossRef] [Green Version]
- Plackett, B. No money for new drugs. Nat. Outlook 2020, 586, S50–S52. [Google Scholar]
- Waring, M.J.; Arrowsmith, J.; Leach, A.R.; Leeson, P.D.; Mandrell, S.; Owen, R.M.; Pairaudeau, G.; Pennie, W.D.; Pickett, S.D.; Wang, J.; et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 2015, 14, 475–486. [Google Scholar] [CrossRef]
- Tyers, M.; Wright, G.D. Drug combinations: A strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019, 17, 141–155. [Google Scholar] [CrossRef]
- Andersson, D.I.; Balaban, N.Q.; Baquero, F.; Courvalin, P.; Glaser, P.; Gophna, U.; Kishony, R.; Molin, S.; Tønjum, T. Antibiotic resistance: Turning evolutionary principles into clinical reality. FEMS Microbiol. Rev. 2020, 44, 171–188. [Google Scholar] [CrossRef] [PubMed]
- Pa, C.; Pál, C.; Papp, B.; Lázár, V.; Pa, C.; Pál, C.; Papp, B.; Lázár, V. Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 2015, 23, 401–407. [Google Scholar]
- Imamovic, L.; Sommer, M.O.A. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 2013, 5, 204ra132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baym, M.; Stone, L.K.; Kishony, R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science 2016, 351, aad3292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, N.R.; Lobritz, M.A.; Collins, J.J. Microbial persistence and the road to drug resistance. Cell Host Microbe 2013, 13, 632–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Windels, E.M.; Michiels, J.J.E.; Van den Bergh, B.; Fauvart, M.; Michiels, J.J.E. Antibiotics: Combatting tolerance to stop resistance. MBio 2019, 10, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Verstraete, L.; Van den Bergh, B.; Verstraeten, N.A.; Michiels, J.; Baquero, F.; Alvarez-Ortega, C.; Martinez, J.L.; Verstraete, L.; Van den Bergh, B.; Verstraeten, N.A.; et al. Ecology and evolution of antibiotic persistence. Trends Microbiol. 2021, 1, 469–476. [Google Scholar] [CrossRef]
- Van den Bergh, B.; Fauvart, M.; Michiels, J. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol. Rev. 2017, 41, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Gollan, B.; Grabe, G.; Michaux, C.; Helaine, S. Bacterial Persisters and Infection: Past, Present, and Progressing. Annu. Rev. Microbiol. 2019, 73, 359–385. [Google Scholar] [CrossRef] [PubMed]
- Van den Bergh, B.; Michiels, J.J.E.; Wenseleers, T.; Windels, E.M.; Boer, P.V.; Kestemont, D.; De Meester, L.; Verstrepen, K.J.; Verstraeten, N.A.; Fauvart, M.; et al. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat. Microbiol. 2016, 1, 16020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fridman, O.; Goldberg, A.; Ronin, I.; Shoresh, N.; Balaban, N.Q. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 2014, 99, 1–9. [Google Scholar] [CrossRef]
- Khare, A.; Tavazoie, S. Extreme antibiotic persistence via heterogeneity-generating mutations targeting translation. mSystems 2020, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sulaiman, J.E.; Lam, H. Proteomic investigation of tolerant Escherichia coli populations from cyclic antibiotic treatment. J. Proteome Res. 2020, 19, 900–913. [Google Scholar] [CrossRef]
- Mechler, L.; Herbig, A.; Paprotka, K.; Fraunholz, M.; Nieselt, K.; Bertram, R. A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2015, 59, 5366–5376. [Google Scholar] [CrossRef] [Green Version]
- Michiels, J.E.; Van den Bergh, B.; Verstraeten, N.A.; Fauvart, M.; Michiels, J. In vitro emergence of high persistence upon periodic aminoglycoside challenge in the ESKAPE pathogens. Antimicrob. Agents Chemother. 2016, 60, 4630–4637. [Google Scholar] [CrossRef] [Green Version]
- Bakkeren, E.; Huisman, J.S.; Fattinger, S.A.; Hausmann, A.; Furter, M.; Egli, A.; Slack, E.; Sellin, M.E.; Bonhoeffer, S.; Regoes, R.R.; et al. Salmonella persisters promote the spread of antibiotic resistance plasmids in the gut. Nature 2019, 573, 276–280. [Google Scholar] [CrossRef]
- Windels, E.M.; Michiels, J.E.; Fauvart, M.; Wenseleers, T.; Van den Bergh, B.; Michiels, J. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 2019, 13, 1239–1251. [Google Scholar] [CrossRef]
- Levin-Reisman, I.; Ronin, I.; Gefen, O.; Braniss, I.; Shoresh, N.; Balaban, N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science 2017, 355, 826–830. [Google Scholar] [CrossRef]
- Rotem, E.; Loinger, A.; Ronin, I.; Levin-Reisman, I.; Gabay, C.; Shoresh, N.; Biham, O.; Balaban, N.Q. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. USA 2010, 107, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Germain, E.; Castro-Roa, D.; Zenkin, N.; Gerdes, K. Molecular mechanism of bacterial persistence by HipA. Mol. Cell 2013, 52, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Verstraeten, N.A.; Knapen, W.J.W.J.; Kint, C.I.; Liebens, V.; Van den Bergh, B.; Dewachter, L.; Michiels, J.E.; Fu, Q.; David, C.C.; Fierro, A.C.; et al. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell 2015, 59, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Balani, P.; Min, J.; Chinnam, N.B.; Hansen, S.; Vulić, M.; Lewis, K.; Brennan, R.G. HipBA–promoter structures reveal the basis of heritable multidrug tolerance. Nature 2015, 524, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Jurėnas, D.; Chatterjee, S.; Konijnenberg, A.; Sobott, F.; Droogmans, L.; Garcia-Pino, A.; Van Melderen, L. AtaT blocks translation initiation by N-acetylation of the initiator tRNAfMet. Nat. Chem. Biol. 2017, 13, 640–646. [Google Scholar] [CrossRef]
- Wilmaerts, D.; Dewachter, L.; De Loose, P.-J.; Bollen, C.; Verstraeten, N.A.; Michiels, J. HokB monomerization and membrane repolarization control persister awakening. Mol. Cell 2019, 75, 1031–1042.e4. [Google Scholar] [CrossRef] [PubMed]
- Dörr, T.; Vulić, M.; Lewis, K.I.M. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010, 8, e1000317. [Google Scholar] [CrossRef] [Green Version]
- Kaspy, I.; Rotem, E.; Weiss, N.; Ronin, I.; Balaban, N.Q.; Glaser, G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 2013, 4, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radzikowski, J.L.; Vedelaar, S.; Siegel, D.; Ortega, Á.D.; Schmidt, A.; Heinemann, M. Bacterial persistence is an active σS stress response to metabolic flux limitation. Mol. Syst. Biol. 2016, 12, 882. [Google Scholar] [CrossRef]
- Amato, S.M.; Orman, M.A.A.; Brynildsen, M.P. Metabolic control of persister formation in Escherichia coli. Mol. Cell 2013, 50, 475–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, Y.; Li, Y.; Jin, X.; Tian, T.; Ma, Q.; Zhao, Z.; Lin, S.; Chen, Z.; Li, B.; Yao, G.; et al. ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol. Cell 2019, 73, 143–156.e4. [Google Scholar] [CrossRef] [Green Version]
- Conlon, B.P.; Rowe, S.E.; Gandt, A.B.; Nuxoll, A.S.; Donegan, N.P.; Zalis, E.A.; Clair, G.; Adkins, J.N.; Cheung, A.L.; Lewis, K. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 2016, 1, 16051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, Y.; Brown Gandt, A.; Rowe, S.E.; Deisinger, J.P.; Conlon, B.P.; Lewis, K.; Formation, A.P.; Gandt, A.B. ATP-dependent persister formation in Escherichia coli. MBio 2017, 8, e02267-16. [Google Scholar] [CrossRef] [Green Version]
- Wilmaerts, D.; Bayoumi, M.; Dewachter, L.; Knapen, W.; Mika, J.T.; Hofkens, J.; Dedecker, P.; Maglia, G.; Verstraeten, N.A.; Michiels, J. The persistence-inducing toxin HokB forms dynamic pores that cause ATP leakage. MBio 2018, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Dewachter, L.; Bollen, C.; Wilmaerts, D.; Louwagie, E.; Herpels, P.; Matthay, P.; Khodaparast, L.L.; Khodaparast, L.L.; Rousseau, F.; Schymkowitz, J.; et al. The dynamic transition of persistence towards the VBNC state during stationary phase is driven by protein aggregation. BioRxiv 2021, 12, e0070321. [Google Scholar]
- Goode, O.; Smith, A.; Łapińska, U.; Bamford, R.; Kahveci, Z.; Glover, G.; Attrill, E.; Carr, A.; Metz, J.; Pagliara, S. Heterologous protein expression favors the formation of protein aggregates in persister and viable but nonculturable bacteria. ACS Infect. Dis. 2021, 7, 1848–1858. [Google Scholar] [CrossRef] [PubMed]
- Van den Bergh, B.; Schramke, H.; Michiels, J.E.; Radzikowski, J.L.; Schimpf, J.; Burschel, S.; Loncar, N.; Meiyer, T.; Fauvart, M.; Friedrich, T.; et al. Complex I controls entry into persistence by (p)ppGpp-dependent and -independent pathways through regulation of intracellular acidification. Nat. Commun. 2021; under review. [Google Scholar]
- Goode, O.; Smith, A.; Zarkan, A.; Cama, J.; Invergo, B.M.; Belgami, D.; Caño-Muñiz, S.; Metz, J.; O’Neill, P.; Jeffries, A.; et al. Persister Escherichia coli cells have a lower intracellular pH than susceptible cells but maintain their pH in response to antibiotic treatment. mBio 2021, 12, e0090921. [Google Scholar] [CrossRef]
- Bartek, I.L.; Reichlen, M.J.; Honaker, R.W.; Leistikow, R.L.; Clambey, E.T.; Scobey, M.S.; Hinds, A.B.; Born, S.E.; Covey, C.R.; Schurr, M.J.; et al. Antibiotic bactericidal activity is countered by maintaining pH homeostasis in Mycobacterium smegmatis. mSphere 2016, 1, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Lafleur, M.D.; Qi, Q.; Lewis, K. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob. Agents Chemother. 2010, 54, 39–44. [Google Scholar] [CrossRef] [Green Version]
- Mulcahy, L.R.; Burns, J.L.; Lory, S.; Lewis, K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 2010, 192, 6191–6199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartell, J.A.; Cameron, D.R.; Mojsoska, B.; Haagensen, J.A.J.; Pressler, T.; Sommer, L.M.; Lewis, K.; Molin, S.; Johansen, H.K. Bacterial persisters in long-term infection: Emergence and fitness in a complex host environment. PLoS Pathog. 2020, 16, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Santi, I.; Manfredi, P.; Maffei, E.; Egli, A.; Jenal, U. Evolution of antibiotic tolerance shapes resistance development in chronic Peudomonas aeruginosa infections. MBio 2021, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, P.; Regoes, R.R.; Dolowschiak, T.; Wotzka, S.Y.; Lengefeld, J.; Slack, E.; Grant, A.J.; Ackermann, M.; Hardt, W.-D. Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLoS Biol. 2014, 12, e1001793. [Google Scholar] [CrossRef]
- Eugenin, E.A.; Weiner, D.; Via, L.E.; Blanc, L.; Dartois, V.A.; Boshoff, H.; Barry, C.E.; Sarathy, J.P. Extreme Drug Tolerance of Mycobacterium tuberculosis in Caseum. Antimicrob. Agents Chemother. 2017, 62, 1–11. [Google Scholar]
- Claudi, B.; Spröte, P.; Chirkova, A.; Personnic, N.; Zankl, J.; Schürmann, N.; Schmidt, A.; Bumann, D. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 2014, 158, 722–733. [Google Scholar] [CrossRef] [Green Version]
- Adams, K.N.; Takaki, K.; Connolly, L.E.; Wiedenhoft, H.; Winglee, K.; Humbert, O.; Edelstein, P.H.; Cosma, C.L.; Ramakrishnan, L. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 2011, 145, 39–53. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, Z.; Klinkenberg, L.G.; Pinn, M.L.; Fraig, M.M.; Peloquin, C.A.; Bishai, W.R.; Nuermberger, E.L.; Grosset, J.H.; Karakousis, P.C. Biphasic kill curve of isoniazid reveals the presence of drug-tolerant, not drug-resistant, Mycobacterium tuberculosis in the guinea pig. J. Infect. Dis. 2009, 200, 1136–1143. [Google Scholar] [CrossRef] [Green Version]
- Conlon, B.P.; Nakayasu, E.S.; Fleck, L.E.; LaFleur, M.D.; Isabella, V.M.; Coleman, K.; Leonard, S.N.; Smith, R.D.; Adkins, J.N. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 2013, 503, 365–370. [Google Scholar] [CrossRef] [Green Version]
- Allison, K.R.; Brynildsen, M.P.; Collins, J.J. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 2011, 473, 216–220. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Sambandan, D.; Halder, R.; Wang, J.; Batt, S.M.; Weinrick, B.; Ahmad, I.; Yang, P.; Zhang, Y.; Kim, J.; et al. Identification of a small molecule with activity against drug-resistant and persistent tuberculosis. Proc. Natl. Acad. Sci. USA 2013, 110, E2510–E2517. [Google Scholar] [CrossRef] [Green Version]
- Lebeaux, D.; Chauhan, A.; Letoffe, S.; Fischer, F.; De Reuse, H.; Beloin, C.; Ghigo, J.-M.M.; Létoffé, S.; Fischer, F.; De Reuse, H.; et al. pH-mediated potentiation of aminoglycosides kills bacterial persisters and eradicates in vivo biofilms. J. Infect. Dis. 2014, 210, 1357–1366. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, D.; Joshi-Datar, A.; Lepine, F.; Bauerle, E.; Olakanmi, O.; Beer, K.D.; Mckay, G.; Siehnel, R.; Schafhauser, J.; Wang, Y.; et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 2011, 334, 982–986. [Google Scholar] [CrossRef] [Green Version]
- Fisher, R.A.; Thurston, T.L.; Saliba, A.; Blommestein, I.; Vogel, J.; Helaine, S. Salmonella persisters undermine host immune defenses during antibiotic treatment. Science 2018, 1160, 1156–1160. [Google Scholar]
- Tiwari, P.; Arora, G.; Singh, M.; Kidwai, S.; Narayan, O.P.; Singh, R. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 2015, 6, 6059. [Google Scholar] [CrossRef] [Green Version]
- Dhar, N.; McKinney, J.D. Mycobacterium tuberculosis persistence mutants identified by screening in isoniazid-treated mice. Proc. Natl. Acad. Sci. USA 2010, 107, 12275–12280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helaine, S.; Cheverton, A.M.; Watson, K.G.; Faure, L.M.; Matthews, S.A.; Holden, D.W. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 2014, 343, 204–208. [Google Scholar] [CrossRef]
- Arnoldini, M.; Vizcarra, I.A.; Peña-Miller, R.; Stocker, N.; Diard, M.; Vogel, V.; Beardmore, R.E.; Hardt, W.-D.; Ackermann, M. Bistable expression of virulence genes in Salmonella leads to the formation of an antibiotic-tolerant subpopulation. PLoS Biol. 2014, 12, e1001928. [Google Scholar] [CrossRef] [PubMed]
- Manina, G.; Dhar, N.; McKinney, J.D. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 2015, 17, 32–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vega, N.M.; Allison, K.R.; Samuels, A.N.; Klempner, M.S.; Collins, J.J. Salmonella Typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. USA 2013, 110, 14420–14425. [Google Scholar] [CrossRef] [Green Version]
- Douglas, A.E. Which experimental systems should we use for human microbiome science? PLoS Biol. 2018, 16, e2005245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glavis-Bloom, J.; Muhammed, M.; Mylonakis, E. Of Model Hosts and Man: Using Caenorhabditis elegans, Drosophila melanogaster and Galleria mellonella as Model Hosts for Infectious Disease Research. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2012; Volume 710, pp. 11–17. ISBN 9781441956378. [Google Scholar]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Irazoqui, J.E.; Urbach, J.M.; Ausubel, F.M. Evolution of host innate defence: Insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 2010, 10, 47–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, A.E. The Drosophila model for microbiome research. Lab Anim. 2018, 47, 157–164. [Google Scholar] [CrossRef]
- Tsai, C.J.Y.; Loh, J.M.S.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Baruah, A.; Tomioka, M.; Iino, Y.; Kalita, M.C.; Khan, M. Caenorhabditis elegans: A model to understand host–microbe interactions. Cell. Mol. Life Sci. 2020, 77, 1229–1249. [Google Scholar] [CrossRef]
- Pereira, M.F.; Rossi, C.C.; Da Silva, G.C.; Rosa, J.N.; Bazzolli, D.M.S. Galleria mellonella as an infection model: An in-depth look at why it works and practical considerations for successful application. Pathog. Dis. 2020, 78, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Buchon, N.; Broderick, N.A.; Lemaitre, B. Gut homeostasis in a microbial world: Insights from Drosophila melanogaster. Nat. Rev. Microbiol. 2013, 11, 615–626. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Kou, S.H.; Xie, R.; VanNieuwenhze, M.S.; Qu, J.; Peng, B.; Zheng, J. Non-walled spherical Acinetobacter baumannii is an important type of persister upon β-lactam antibiotic treatment. Emerg. Microbes Infect. 2020, 9, 1149–1159. [Google Scholar] [CrossRef]
- Rosenberg, A.; Ene, I.V.; Bibi, M.; Zakin, S.; Segal, E.S.; Ziv, N.; Dahan, A.M.; Colombo, A.L.; Bennett, R.J.; Berman, J. Antifungal tolerance is a subpopulation effect distinct from resistance and is associated with persistent candidemia. Nat. Commun. 2018, 9, 2470. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.; Conery, A.L.; Rajamuthiah, R.; Fuchs, B.B.; Ausubel, F.M.; Mylonakis, E. Identification of an antimicrobial agent effective against methicillin-resistant Staphylococcus aureus persisters using a fluorescence-based screening strategy. PLoS ONE 2015, 10, e0127640. [Google Scholar]
- Cruz-Muñiz, M.Y.; López-Jacome, L.E.; Hernández-Durán, M.; Franco-Cendejas, R.; Licona-Limón, P.; Ramos-Balderas, J.L.; Martinéz-Vázquez, M.; Belmont-Díaz, J.A.; Wood, T.K.; García-Contreras, R. Repurposing the anticancer drug mitomycin C for the treatment of persistent Acinetobacter baumannii infections. Int. J. Antimicrob. Agents 2017, 49, 88–92. [Google Scholar] [CrossRef]
- Buchon, N.; Silverman, N.; Cherry, S. Immunity in Drosophila melanogaster-from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 2014, 14, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Tafin, U.F.; Majic, I.; Zalila, C.B.; Betrisey, B.; Corvec, S.; Zimmerli, W.; Trampuz, A. Gentamicin improves the activities of daptomycin and vancomycin against Enterococcus faecalis in vitro and in an experimental foreign-body infection model. Antimicrob. Agents Chemother. 2011, 55, 4821–4827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearl Mizrahi, S.; Gabay, C.; Kishony, R.; Oppenheim, A.; Balaban, N.Q.; Pearl, S.; Gabay, C.; Kishony, R.; Oppenheim, A.; Balaban, N.Q. Nongenetic individuality in the host-phage interaction. PLoS Biol. 2008, 6, e120. [Google Scholar]
- Taniguchi, Y.; Choi, P.P.J.; Li, G.-W.; Chen, H.; Babu, M.; Hearn, J.; Emili, A.; Xie, X.S. Quantifying, E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 2010, 329, 533–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006-0008. [Google Scholar] [CrossRef] [Green Version]
- Ridley, E.V.; Wong, A.C.-N.; Westmiller, S.; Douglas, A.E. Impact of the Resident Microbiota on the Nutritional Phenotype of Drosophila melanogaster. PLoS ONE 2012, 7, e36765. [Google Scholar] [CrossRef]
- Newell, P.D.; Douglas, A.E. Interspecies Interactions Determine the Impact of the Gut Microbiota on Nutrient Allocation in Drosophila melanogaster. Appl. Environ. Microbiol. 2014, 80, 788–796. [Google Scholar] [CrossRef] [Green Version]
- Koyle, M.L.; Veloz, M.; Judd, A.M.; Wong, A.C.N.; Newell, P.D.; Douglas, A.E.; Chaston, J.M. Rearing the Fruit Fly Drosophila melanogaster Under Axenic and Gnotobiotic Conditions. J. Vis. Exp. 2016, 113, e54219. [Google Scholar] [CrossRef] [PubMed]
- Ja, W.W.; Carvalho, G.B.; Mak, E.M.; De La Rosa, N.N.; Fang, A.Y.; Liong, J.C.; Brummel, T.; Benzer, S. Prandiology of Drosophila and the CAFE assay. Proc. Natl. Acad. Sci. USA 2007, 104, 8253–8256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMullen, J.G.; Peters-Schulze, G.; Cai, J.; Patterson, A.D.; Douglas, A.E. How gut microbiome interactions affect nutritional traits of Drosophila melanogaster. J. Exp. Biol. 2020, 223, jeb227843. [Google Scholar] [CrossRef]
- Burggren, W.; Souder, B.M.; Ho, D.H. Metabolic rate and hypoxia tolerance are affected by group interactions and sex in the fruit fly (Drosophila melanogaster): New data and a literature survey. Biol. Open 2017, 6, 471–480. [Google Scholar] [CrossRef] [Green Version]
- Porter, R.S. The Merck Manual of Diagnosis and Therapy, 20th ed.; Merck: Darmstadt, Germany, 2018; ISBN 0911910425. [Google Scholar]
- Inamine, H.; Ellner, S.P.; Newell, P.D.; Luo, Y.; Buchon, N.; Douglas, A.E. Spatiotemporally heterogeneous population dynamics of gut bacteria inferred from fecal time series data. MBio 2018, 9, e01453-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itskov, P.M.; Moreira, J.M.; Vinnik, E.; Lopes, G.; Safarik, S.; Dickinson, M.H.; Ribeiro, C. Automated monitoring and quantitative analysis of feeding behaviour in Drosophila. Nat. Commun. 2014, 5, 4560. [Google Scholar] [CrossRef] [Green Version]
- Buchon, N.; Broderick, N.A.; Poidevin, M.; Pradervand, S.; Lemaitre, B. Drosophila Intestinal Response to Bacterial Infection: Activation of Host Defense and Stem Cell Proliferation. Cell Host Microbe 2009, 5, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Duneau, D.; Ferdy, J.-B.; Revah, J.; Kondolf, H.; Ortiz, G.A.; Lazzaro, B.P.; Buchon, N. Stochastic variation in the initial phase of bacterial infection predicts the probability of survival in D. melanogaster. eLife 2017, 6, e28298. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Preibisch, S.; Saalfeld, S.; Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 2009, 25, 1463–1465. [Google Scholar] [CrossRef]
- Sharon, G.; Segal, D.; Ringo, J.M.; Hefetz, A.; Zilber-Rosenberg, I.; Rosenberg, E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2010, 107, 20051–20056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thibert, J.; Farine, J.P.; Cortot, J.; Ferveur, J.F. Drosophila food-associated pheromones: Effect of experience, genotype and antibiotics on larval behavior. PLoS ONE 2016, 11, e0151451. [Google Scholar] [CrossRef] [PubMed]
- Ridley, E.V.; Wong, A.C.-N.; Douglas, A.E. Microbe-dependent and nonspecific effects of procedures to eliminate the resident microbiota from Drosophila melanogaster. Appl. Environ. Microbiol. 2013, 79, 3209–3214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pais, I.S.; Valente, R.S.; Sporniak, M.; Teixeira, L. Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol. 2018, 16, e2005710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, A.C.-N.; Dobson, A.J.; Douglas, A.E. Gut microbiota dictates the metabolic response of Drosophila to diet. J. Exp. Biol. 2014, 217, 1894–1901. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.; Douglas, A.E. Functional traits of the gut microbiome correlated with host lipid content in a natural population of Drosophila melanogaster. Biol. Lett. 2020, 16, 20190803. [Google Scholar] [CrossRef] [Green Version]
- Adair, K.L.; Wilson, M.; Bost, A.; Douglas, A.E. Microbial community assembly in wild populations of the fruit fly Drosophila melanogaster. ISME J. 2018, 12, 959–972. [Google Scholar] [CrossRef]
- Wong, A.C.-N.; Chaston, J.M.; Douglas, A.E. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 2013, 7, 1922–1932. [Google Scholar] [CrossRef] [Green Version]
- Chandler, J.A.; Lang, J.; Bhatnagar, S.; Eisen, J.A.; Kopp, A. Bacterial communities of diverse Drosophila species: Ecological context of a host-microbe model system. PLoS Genet. 2011, 7, e1002272. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lidsky, P.V.; Lukyanov, K.A.; Misra, T.; Handke, B.; Mishin, A.S.; Lehner, C.F. A genetically encoded fluorescent probe for imaging of oxygenation gradients in living Drosophila. Development 2018, 145, dev156257. [Google Scholar] [CrossRef] [Green Version]
- Consuegra, J.; Grenier, T.; Baa-Puyoulet, P.; Rahioui, I.; Akherraz, H.; Gervais, H.; Parisot, N.; Da Silva, P.; Charles, H.; Calevro, F.; et al. Drosophila-Associated Bacteria Differentially Shape the Nutritional Requirements of Their Host during Juvenile Growth. PLoS Biol. 2020, 18, e3000681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaston, J.M.; Newell, P.D.; Douglas, A.E. Metagenome-Wide Association of Microbial Determinants of Host Phenotype in Drosophila melanogaster. mBio 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sibley, C.D.; Duan, K.; Fischer, C.; Parkins, M.D.; Storey, D.G.; Rabin, H.R.; Surette, M.G. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008, 4, e1000184. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Conly, J.; Surette, M.; Sibley, C.; Elsayed, S.; Zhang, K. Assessment of virulence diversity of methicillin-resistant Staphylococcus aureus strains with a Drosophila melanogaster infection model. BMC Microbiol. 2012, 12, 274. [Google Scholar] [CrossRef] [Green Version]
- Obadia, B.; Güvener, Z.T.; Zhang, V.; Ceja-navarro, J.A.; Brodie, E.L.; Ja, W.W.; Ludington, W.B. Probabilistic Invasion Underlies Natural Gut Microbiome Stability. Curr. Biol. 2017, 27, 1999–2006.e8. [Google Scholar] [CrossRef] [Green Version]
- Mulcahy, H.; Sibley, C.D.; Surette, M.G.; Lewenza, S. Drosophila melanogaster as an animal model for the study of Pseudomonas aeruginosa biofilm infections in vivo. PLoS Pathog. 2011, 7, e1002299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoffolano, J.G.; Haselton, A.T. The adult dipteran crop: A unique and overlooked organ. Annu. Rev. Entomol. 2013, 58, 205–225. [Google Scholar] [CrossRef]
- Ludington, W.B.; Ja, W.W. Drosophila as a model for the gut microbiome. PLoS Pathog. 2020, 16, 1–6. [Google Scholar] [CrossRef]
- Zimmermann, M.; Patil, K.R.; Typas, A.; Maier, L. Towards a mechanistic understanding of reciprocal drug–microbiome interactions. Mol. Syst. Biol. 2021, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; He, L.; Cui, P.; Wang, W.; Yuan, Y.; Liu, S.; Xu, T.; Zhang, S.; Wu, J.; Zhang, W.; et al. Ranking of persister genes in the same Escherichia coli genetic background demonstrates varying importance of individual persister genes in tolerance to different antibiotics. Front. Microbiol. 2015, 6, 1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, S.H.; Wang, X.; O’Connor, H.F.; Benedik, M.J.; Wood, T.K. Bacterial persistence increases as environmental fitness decreases. Microb. Biotechnol. 2012, 5, 509–522. [Google Scholar] [CrossRef]
- Wilmaerts, D.; Herpels, P.; Michiels, J.; Verstraeten, N.A. Genetic determinants of persistence in Escherichia coli. In Persister Cells and Infectious Disease; Springer: Berlin/Heidelberg, Germany, 2019; pp. 133–180. ISBN 9783030252410. [Google Scholar]
- Aranda-Díaz, A.; Obadia, B.; Dodge, R.; Thomsen, T.; Hallberg, Z.F.; Güvener, Z.T.; Ludington, W.B.; Huang, K.C. Bacterial interspecies interactions modulate pH-mediated antibiotic tolerance. eLife 2020, 9, 1–26. [Google Scholar] [CrossRef] [PubMed]

| Name | Remarks | Reference |
| Bacillus subtilis indicator strain | ATCC 6051 | [87] |
| hipA7 | MG1655 zde-264::Tn10 hipA7, mutant of MG21 with increased persistence | [88] |
| MG21 | MG1655 zde-264::Tn10, parental strain of hipA7; also called wild type in figures/text | [88] |
| hipA7 pTimer | hipA7 chemically transformed with pTimer | This study |
| MG21 pTimer | MG21 chemically transformed with pTimer | This study |
| SX4 | A BW25513 related strain that contains a tsr-venus tag in the lacZ gene along with a KmR cassette; also called wild type in figures/text | [89] |
| SX43 | SX4 strain where the KmR-cassette was removed via expression of FLP recombinase; also called wild type in figures/text | [24] |
| BW25113 | The ancestor of the Keio collection, a derivative of the K12 BD792 strain | [90] |
| ΔrpoS | E. coli BW25113 rpoS::KmR (JW5437-1) that is cured from its KmR cassette | [49,90] |
| ΔrelAΔspoT | E. coli BW25113 relA::KmR (JW2755-1) that is cured from its KmR cassette and in which spoT is subsequently deleted | [49,90] |
| pTimer | pBR322_Timer, expressing DsRed.T3_S4T from a constitutive promoter that encodes a green-to-red maturing fluorophore | [58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van den Bergh, B. Bugs on Drugs: A Drosophila melanogaster Gut Model to Study In Vivo Antibiotic Tolerance of E. coli. Microorganisms 2022, 10, 119. https://doi.org/10.3390/microorganisms10010119
Van den Bergh B. Bugs on Drugs: A Drosophila melanogaster Gut Model to Study In Vivo Antibiotic Tolerance of E. coli. Microorganisms. 2022; 10(1):119. https://doi.org/10.3390/microorganisms10010119
Chicago/Turabian StyleVan den Bergh, Bram. 2022. "Bugs on Drugs: A Drosophila melanogaster Gut Model to Study In Vivo Antibiotic Tolerance of E. coli" Microorganisms 10, no. 1: 119. https://doi.org/10.3390/microorganisms10010119
APA StyleVan den Bergh, B. (2022). Bugs on Drugs: A Drosophila melanogaster Gut Model to Study In Vivo Antibiotic Tolerance of E. coli. Microorganisms, 10(1), 119. https://doi.org/10.3390/microorganisms10010119






