High Frequency of the EMRSA-15 Clone (ST22-MRSA-IV) in Hospital Wastewater
Abstract
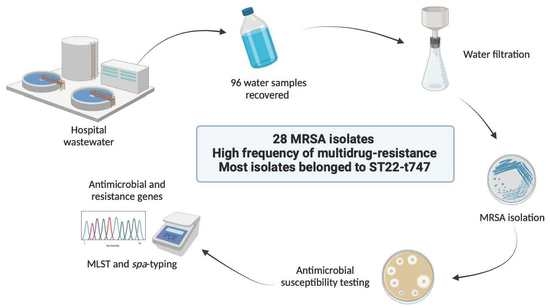
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Bacterial Isolation
2.3. Antimicrobial Resistance Phenotype
2.4. Antimicrobial Resistance and Virulence Genes
2.5. Molecular Typing
3. Results and Discussion
3.1. Presence of MRSA in Hospital Wastewaters
3.2. Antimicrobial Resistance and Virulence
3.3. Molecular Typing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Buelow, E.; Bayjanov, J.R.; Majoor, E.; Willems, R.J.L.; Bonten, M.J.M.; Schmitt, H.; van Schaik, W. Limited influence of hospital wastewater on the microbiome and resistome of wastewater in a community sewerage system. FEMS Microbiol. Ecol. 2018, 94, fiy087. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [Green Version]
- Chagas, T.P.G.; Seki, L.M.; Cury, J.C.; Oliveira, J.A.L.; Dávila, A.M.R.; Silva, D.M.; Asensi, M.D. Multiresistance, beta-lactamase-encoding genes and bacterial diversity in hospital wastewater in Rio de Janeiro, Brazil. J. Appl. Microbiol. 2011, 111, 572–581. [Google Scholar] [CrossRef]
- Grenni, P.; Ancona, V.; Caracciolo, A.B. Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Paulus, G.K.; Hornstra, L.M.; Alygizakis, N.; Slobodnik, J.; Thomaidis, N.; Medema, G. The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int. J. Hyg. Environ. Health 2019, 222, 635–644. [Google Scholar] [CrossRef]
- Łuczkiewicz, A.; Jankowska, K.; Fudala-Książek, S.; Olańczuk-Neyman, K. Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res. 2010, 44, 5089–5097. [Google Scholar] [CrossRef]
- Hocquet, D.; Muller, A.; Bertrand, X. What happens in hospitals does not stay in hospitals: Antibiotic-resistant bacteria in hospital wastewater systems. J. Hosp. Infect. 2016, 93, 395–402. [Google Scholar] [CrossRef]
- Gotkowska-Płachta, A. The Prevalence of Virulent and Multidrug-Resistant Enterococci in River Water and in Treated and Untreated Municipal and Hospital Wastewater. Int. J. Environ. Res. Public Health 2021, 18, 563. [Google Scholar] [CrossRef]
- Papadopoulos, P.; Papadopoulos, T.; Angelidis, A.S.; Boukouvala, E.; Zdragas, A.; Papa, A.; Hadjichristodoulou, C.; Sergelidis, D. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus (MRSA) along the production chain of dairy products in north-western Greece. Food Microbiol. 2018, 69, 43–50. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, P.; Yang, Q. Occurrence and diversity of antibiotic resistance in untreated hospital wastewater. Sci. Total Environ. 2018, 621, 990–999. [Google Scholar] [CrossRef]
- Yao, S.; Ye, J.; Yang, Q.; Hu, Y.; Zhang, T.; Jiang, L.; Munezero, S.; Lin, K.; Cui, C. Occurrence and removal of antibiotics, antibiotic resistance genes, and bacterial communities in hospital wastewater. Environ. Sci. Pollut. Res. 2021, 28, 57321–57333. [Google Scholar] [CrossRef]
- Baghal Asghari, F.; Dehghani, M.H.; Dehghanzadeh, R.; Farajzadeh, D.; Yaghmaeian, K.; Mahvi, A.H.; Rajabi, A. Antibiotic resistance and antibiotic resistant gens of Pseudomonas spp. and Escherichia coli isolated from untreated hospital wastewater. Water Sci. Technol. 2021, 84, 172–181. [Google Scholar] [CrossRef]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Kourtis, A.P.; Hatfield, K.; Baggs, J.; Mu, Y.; See, I.; Epson, E.; Nadle, J.; Kainer, M.A.; Dumyati, G.; Petit, S.; et al. Vital Signs: Epidemiology and Recent Trends in Methicillin-Resistant and in Methicillin-Susceptible Staphylococcus aureus Bloodstream Infections-United States. MMWR. Morb. Mortal. Wkly. Rep. 2019, 68, 214–219. [Google Scholar] [CrossRef] [Green Version]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- European Centre for Disease Prevention and Control (ECDC). Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report for 2019; European Centre for Disease Prevention and Control (ECDC): Stockholm, Sweden, 2020. [Google Scholar]
- Couto, N.; Belas, A.; Kadlec, K.; Schwarz, S.; Pomba, C. Clonal diversity, virulence patterns and antimicrobial and biocide susceptibility among human, animal and environmental MRSA in Portugal. J. Antimicrob. Chemother. 2015, 70, 2483–2487. [Google Scholar] [CrossRef] [Green Version]
- Silva, V.; Almeida, F.; Carvalho, J.A.; Castro, A.P.; Ferreira, E.; Manageiro, V.; Tejedor-Junco, M.T.; Caniça, M.; Igrejas, G.; Poeta, P. Emergence of community-acquired methicillin-resistant Staphylococcus aureus EMRSA-15 clone as the predominant cause of diabetic foot ulcer infections in Portugal. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 179–186. [Google Scholar] [CrossRef]
- Silva, V.; Hermenegildo, S.; Ferreira, C.; Manaia, C.M.; Capita, R.; Alonso-Calleja, C.; Carvalho, I.; Pereira, J.E.; Maltez, L.; Capelo, J.L. Genetic Characterization of Methicillin-Resistant Staphylococcus aureus Isolates from Human Bloodstream Infections: Detection of MLSB Resistance. Antibiotics 2020, 9, 375. [Google Scholar] [CrossRef]
- Coelho, C.; Torres, C.; Radhouani, H.; Pinto, L.; Lozano, C.; Gómez-Sanz, E.; Zaragaza, M.; Igrejas, G.; Poeta, P. Molecular Detection and Characterization of Methicillin-Resistant Staphylococcus aureus (MRSA) Isolates from Dogs in Portugal. Microb. Drug Resist. 2011, 17, 333–337. [Google Scholar] [CrossRef]
- Silva, V.; Gabriel, S.I.; Borrego, S.B.; Tejedor-Junco, M.T.; Manageiro, V.; Ferreira, E.; Reis, L.; Caniça, M.; Capelo, J.L.; Igrejas, G.; et al. Antimicrobial Resistance and Genetic Lineages of Staphylococcus aureus from Wild Rodents: First Report of mecC-Positive Methicillin-Resistant S. aureus (MRSA) in Portugal. Animals 2021, 11, 1537. [Google Scholar] [CrossRef]
- Silva, V.; Vieira-Pinto, M.; Saraiva, C.; Manageiro, V.; Reis, L.; Ferreira, E.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Prevalence and Characteristics of Multidrug-Resistant Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) CC398 Isolated from Quails (Coturnix Coturnix Japonica) Slaughtered for Human Consumption. Animals 2021, 11, 2038. [Google Scholar] [CrossRef]
- van Wamel, W.J.B.; Rooijakkers, S.H.M.; Ruyken, M.; van Kessel, K.P.M.; van Strijp, J.A.G. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 2006, 188, 1310–1315. [Google Scholar] [CrossRef] [Green Version]
- Shopsin, B.; Mathema, B.; Alcabes, P.; Said-Salim, B.; Lina, G.; Matsuka, A.; Martinez, J.; Kreiswirth, B.N. Prevalence of agr specificity groups among Staphylococcus aureus strains colonizing children and their guardians. J. Clin. Microbiol. 2003, 41, 456–459. [Google Scholar] [CrossRef] [Green Version]
- Harmsen, D.; Claus, H.; Witte, W.; Rothgänger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of Methicillin-Resistant Staphylococcus aureus in a University Hospital Setting by Using Novel Software for spa Repeat Determination and Database Management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; McClure, J.-A.; Elsayed, S.; Louie, T.; Conly, J.M. Novel Multiplex PCR Assay for Characterization and Concomitant Subtyping of Staphylococcal Cassette Chromosome mec Types I to V in Methicillin-Resistant Staphylococcus aureus. J. Clin. Microbiol. 2005, 43, 5026–5033. [Google Scholar] [CrossRef] [Green Version]
- Witte, W.; Strommenger, B.; Stanek, C.; Cuny, C. Methicillin-resistant Staphylococcus aureus ST398 in Humans and Animals, Central Europe. Emerg. Infect. Dis. 2007, 13, 255–258. [Google Scholar] [CrossRef]
- Kondo, Y.; Ito, T.; Ma, X.X.; Watanabe, S.; Kreiswirth, B.N.; Etienne, J.; Hiramatsu, K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 2007, 51, 264–274. [Google Scholar] [CrossRef] [Green Version]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef] [Green Version]
- Serwecińska, L. Antimicrobials and Antibiotic-Resistant Bacteria: A Risk to the Environment and to Public Health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Amirsoleimani, A.; Brion, G.M.; Diene, S.M.; François, P.; Richard, E.M. Prevalence and characterization of Staphylococcus aureus in wastewater treatment plants by whole genomic sequencing. Water Res. 2019, 158, 193–202. [Google Scholar] [CrossRef]
- Goldstein, R.E.R.; Micallef, S.A.; Gibbs, S.G.; Davis, J.A.; He, X.; George, A.; Kleinfelter, L.M.; Schreiber, N.A.; Mukherjee, S.; Sapkota, A.; et al. Methicillin-Resistant Staphylococcus aureus (MRSA) Detected at Four U.S. Wastewater Treatment Plants. Environ. Health Perspect. 2012, 120, 1551–1558. [Google Scholar] [CrossRef] [Green Version]
- Gómez, P.; Lozano, C.; Benito, D.; Estepa, V.; Tenorio, C.; Zarazaga, M.; Torres, C. Characterization of staphylococci in urban wastewater treatment plants in Spain, with detection of methicillin resistant Staphylococcus aureus ST. Environ. Pollut. 2016, 212, 71–76. [Google Scholar] [CrossRef]
- Boopathy, R. Presence of Methicillin Resistant Staphylococcus aureus (MRSA) in sewage treatment plant. Bioresour. Technol. 2017, 240, 144–148. [Google Scholar] [CrossRef]
- Thompson, J.M.; Gündoğdu, A.; Stratton, H.M.; Katouli, M. Antibiotic resistant S taphylococcus aureus in hospital wastewaters and sewage treatment plants with special reference to methicillin-resistant S taphylococcus aureus (MRSA). J. Appl. Microbiol. 2013, 114, 44–54. [Google Scholar] [CrossRef] [Green Version]
- Martin, V.; Dorte, F.; Hanne, I. Antibiotic Resistance and the MRSA Problem. Microbiol. Spectr. 2019, 7, 1–23. [Google Scholar] [CrossRef]
- Boyle-Vavra, S.; Daum, R.S. Molecular Strategies of Staphylococcus aureus for Resisting Antibiotics. Staphylococcus Genet. Physiol. 2016, 249–300. [Google Scholar]
- Silva, V.; Miranda, C.; Bezerra, M.; Antão, H.S.; Guimarães, J.; Prada, J.; Pires, I.; Maltez, L.; Pereira, J.E.; Capelo, J.L. Anti-biofilm activity of dalbavancin against methicillin-resistant Staphylococcus aureus (MRSA) isolated from human bone infection. J. Chemother. 2021, 33, 469–475. [Google Scholar] [CrossRef]
- Akya, A.; Chegenelorestani, R.; Shahvaisi-Zadeh, J.; Bozorgomid, A. Antimicrobial Resistance of Staphylococcus aureus Isolated from Hospital Wastewater in Kermanshah, Iran. Risk Manag. Healthc. Policy 2020, 13, 1035–1042. [Google Scholar] [CrossRef]
- Torabi, M.; Rahimi, F. Characteristics of Methicillin Resistant Staphylococcus aureus Strains Isolated from Hospital Wastewater in Tehran, Iran. Infect. Epidemiol. Microb. 2021, 7, 215–227. [Google Scholar] [CrossRef]
- Lim, K.T.; Hanifah, Y.A.; Yusof, M.Y.M.; Thong, K.L. ermA, ermC, tetM and tetK are essential for erythromycin and tetracycline resistance among methicillin-resistant Staphylococcus aureus strains isolated from a tertiary hospital in Malaysia. Indian J. Med. Microbiol. 2012, 30, 203–207. [Google Scholar] [CrossRef]
- Safarpoor Dehkordi, F.; Gandomi, H.; Basti, A.A.; Misaghi, A.; Rahimi, E. Phenotypic and genotypic characterization of antibiotic resistance of methicillin-resistant Staphylococcus aureus isolated from hospital food. Antimicrob. Resist. Infect. Control 2017, 6, 104. [Google Scholar] [CrossRef]
- Bidell, M.R.; Lodise, T.P. Use of oral tetracyclines in the treatment of adult outpatients with skin and skin structure infections: Focus on doxycycline, minocycline, and omadacycline. Pharmacotherapy 2021, 41, 915–931. [Google Scholar] [CrossRef]
- Dale, G.E.; Langen, H.; Page, M.G.; Then, R.L.; Stüber, D. Cloning and characterization of a novel, plasmid-encoded trimethoprim-resistant dihydrofolate reductase from Staphylococcus haemolyticus MUR313. Antimicrob. Agents Chemother. 1995, 39, 1920–1924. [Google Scholar] [CrossRef] [Green Version]
- Nurjadi, D.; Olalekan, A.O.; Layer, F.; Shittu, A.O.; Alabi, A.; Ghebremedhin, B.; Schaumburg, F.; Hofmann-Eifler, J.; Van Genderen, P.J.J.; Caumes, E. Emergence of trimethoprim resistance gene dfrG in Staphylococcus aureus causing human infection and colonization in sub-Saharan Africa and its import to Europe. J. Antimicrob. Chemother. 2014, 69, 2361–2368. [Google Scholar] [CrossRef] [Green Version]
- Trieu-Cuot, P.; De Cespedes, G.; Bentorcha, F.; Delbos, F.; Gaspar, E.; Horaud, T. Study of heterogeneity of chloramphenicol acetyltransferase (CAT) genes in streptococci and enterococci by polymerase chain reaction: Characterization of a new CAT determinant. Antimicrob. Agents Chemother. 1993, 37, 2593–2598. [Google Scholar] [CrossRef] [Green Version]
- Conceição, T.; Martins, H.; Rodrigues, S.; de Lencastre, H.; Aires-de-Sousa, M. Staphylococcus aureus nasal carriage among homeless population in Lisbon, Portugal. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2037–2044. [Google Scholar] [CrossRef]
- Santos, V.; Gomes, A.; Ruiz-Ripa, L.; Mama, O.M.; Sabença, C.; Sousa, M.; Silva, V.; Sousa, T.; Vieira-Pinto, M.; Igrejas, G.; et al. Methicillin-Resistant Staphylococcus aureus CC398 in Purulent Lesions of Piglets and Fattening Pigs in Portugal. Microb. Drug Resist. 2020, 26, 850–856. [Google Scholar] [CrossRef]
- Bouchami, O.; Fraqueza, M.J.; Faria, N.A.; Alves, V.; Lawal, O.U.; de Lencastre, H.; Miragaia, M. Evidence for the Dissemination to Humans of Methicillin-Resistant Staphylococcus aureus ST398 through the Pork Production Chain: A Study in a Portuguese Slaughterhouse. Microorganisms 2020, 8, 1892. [Google Scholar] [CrossRef]
- Rhee, C.H.; Woo, G.J. Emergence and characterization of foodborne methicillin-resistant Staphylococcus aureus in Korea. J. Food Prot. 2010, 73, 2285–2290. [Google Scholar] [CrossRef]
- Sauer, P.; Síla, J.; Štosová, T.; Večeřová, R.; Hejnar, P.; Vágnerová, I.; Kolář, M.; Raclavský, V.; Petrželová, J.; Lovečková, Y. Prevalence of genes encoding extracellular virulence factors among meticillin-resistant Staphylococcus aureus isolates from the University Hospital, Olomouc, Czech Republic. J. Med. Microbiol. 2008, 57, 403–410. [Google Scholar] [CrossRef]
- Argudín, M.A.; Argumosa, V.; Mendoza, M.C.; Guerra, B.; Rodicio, M.R. Population structure and exotoxin gene content of methicillin-susceptible Staphylococcus aureus from Spanish healthy carriers. Microb. Pathog. 2013, 54, 26–33. [Google Scholar] [CrossRef]
- Coombs, G.W.; Daley, D.A.; Lee, Y.T.; Pang, S. Australian group on antimicrobial resistance (AGAR) Australian Staphylococcus aureus sepsis outcome programme (ASSOP) annual report Commun. Dis. Intell. 2019, 43, 1–18. [Google Scholar]
- Richardson, J.F.; Reith, S. Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J. Hosp. Infect. 1993, 25, 45–52. [Google Scholar] [CrossRef]
- Dhawan, B.; Rao, C.; Udo, E.E.; Gadepalli, R.; Visnhnubhatla, S.; Kapil, A. Dissemination of methicillin-resistant Staphylococcus aureus SCCmec type IV and SCCmec type V epidemic clones in a tertiary hospital: Challenge to infection control. Epidemiol. Infect. 2015, 143, 343–353. [Google Scholar] [CrossRef] [Green Version]
- Holden, M.T.G.; Hsu, L.-Y.; Kurt, K.; Weinert, L.A.; Mather, A.E.; Harris, S.R.; Strommenger, B.; Layer, F.; Witte, W.; de Lencastre, H.; et al. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013, 23, 653–664. [Google Scholar] [CrossRef] [Green Version]
- Viana, A.S.; Nunes Botelho, A.M.; Moustafa, A.M.; Boge, C.L.K.; Pires Ferreira, A.L.; da Silva Carvalho, M.C.; Guimarães, M.A.; Costa, B. de S.S.; de Mattos, M.C.; Maciel, S.P.; et al. Multidrug-Resistant Methicillin-Resistant Staphylococcus aureus Associated with Bacteremia and Monocyte Evasion, Rio de Janeiro, Brazil. Emerg. Infect. Dis. 2021, 27, 2825–2835. [Google Scholar] [CrossRef]
- Espadinha, D.; Faria, N.A.; Miragaia, M.; Lito, L.M.; Melo-Cristino, J.; de Lencastre, H.; Network, M.S. Extensive Dissemination of Methicillin-Resistant Staphylococcus aureus (MRSA) between the Hospital and the Community in a Country with a High Prevalence of Nosocomial MRSA. PLoS ONE 2013, 8, e59960. [Google Scholar] [CrossRef] [Green Version]
- Faria, N.A.; Miragaia, M.; de Lencastre, H.; The Multi Laboratory Project Collaborators. Massive Dissemination of Methicillin Resistant Staphylococcus aureus in Bloodstream Infections in a High MRSA Prevalence Country: Establishment and Diversification of EMRSA-15. Microb. Drug Resist. 2013, 19, 483–490. [Google Scholar] [CrossRef]
- Takadama, S.; Nakaminami, H.; Sato, A.; Shoshi, M.; Fujii, T.; Noguchi, N. Dissemination of Panton-Valentine leukocidin–positive methicillin-resistant Staphylococcus aureus USA300 clone in multiple hospitals in Tokyo, Japan. Clin. Microbiol. Infect. 2018, 24, 1211.e1. [Google Scholar] [CrossRef] [Green Version]
- Planet, P.J.; LaRussa, S.J.; Dana, A.; Smith, H.; Xu, A.; Ryan, C.; Uhlemann, A.-C.; Boundy, S.; Goldberg, J.; Narechania, A. Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. MBio 2013, 4, e00889-13. [Google Scholar] [CrossRef] [Green Version]
- Klein, S.; Boutin, S.; Heeg, K.; Zanger, P.; Nurjadi, D. Genomic structure of ST8-t008 USA300 and USA300-LV MRSA in the Rhine-Neckar Region, Germany, 2012–2018. Int. J. Antimicrob. Agents 2021, 57, 106312. [Google Scholar] [CrossRef]
- Enström, J.; Fröding, I.; Giske, C.G.; Ininbergs, K.; Bai, X.; Sandh, G.; Tollström, U.-B.; Ullberg, M.; Fang, H. USA300 methicillin-resistant Staphylococcus aureus in Stockholm, Sweden, from 2008 to 2016. PLoS ONE 2018, 13, e0205761. [Google Scholar] [CrossRef]
- Argudín, M.A.; Deplano, A.; Nonhoff, C.; Yin, N.; Michel, C.; Martiny, D.; De Keersmaecker, S.C.J.; Hallin, M. Epidemiology of the Staphylococcus aureus CA-MRSA USA300 in Belgium. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2335–2347. [Google Scholar] [CrossRef]
- Börjesson, S.; Matussek, A.; Melin, S.; Löfgren, S.; Lindgren, P.E. Methicillin-resistant Staphylococcus aureus (MRSA) in municipal wastewater: An uncharted threat? J. Appl. Microbiol. 2010, 108, 1244–1251. [Google Scholar] [CrossRef]
| Isolate | Hospital (Treated/Untreated Wastewater) | Antimicrobial Resistance | Virulence Factors | Molecular Typing | |||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | Genotype | IEC Type | Other Genes | ST (CC) | spa | SCCmec | agr | ||
| VS2932 | Vila Real (Untreated) | BEN,CXI, CIP, ERY | mecA, blaZ, ermA | B | hld | 105 | t10682 | II | II |
| VS2933 | Vila Real (Untreated) | BEN, CXI, CIP, GEN, ERY, CLI, FUS | mecA, blaZ, aph(3′)-IIIa, ermC | B | tst | 22 (22) | t747 | IV | I |
| VS2934 | Vila Real (Untreated) | BEN, CXI, CIP, GEN, ERY | mecA, blaZ, aph(3′)-IIIa, ermC | B | hld | 22 (22) | t747 | IV | I |
| VS2935 | Vila Real (Untreated) | BEN, CXI, CIP, GEN, TOB, ERY, CLI, FUS | mecA, blaZ, aph(3′)-IIIa, ermC | B | hld, tst | 22 (22) | t6966 | IV | I |
| VS2936 | Vila Real (Untreated) | BEN, CXI, CIP, GEN, KAN, TOB, ERY, CLI, TET, CHL, FUS, TRS | mecA, blaZ, aph(3′)-IIIa, ermC, tetL, catpC221, dfrA | B | hld | 22 (22) | t747 | IV | I |
| VS2937 | Vila Real (Untreated) | BEN, CXI, CIP | mecA, blaZ | B | hld | 22 (22) | t020 | IV | I |
| VS2938 | Vila Real (Untreated) | BEN, CXI, CIP, ERY | mecA, blaZ, ermC | B | hld | 22 (22) | t747 | IV | I |
| VS2939 | Vila Real (Treated) | BEN, CXI, CIP, ERY | mecA, blaZ, ermC | - | hld | 22 (22) | t020 | IV | I |
| VS2940 | Chaves (Untreated) | BEN, CXI, CIP, GEN, ERY | mecA, blaZ, aac(6′)-Ie-aph(2′’)-Ia, aph(3′)-IIIa, ermB, ermC | B | hld | 22 (22) | t747 | IV | I |
| VS2941 | Chaves (Untreated) | BEN, CXI, CIP, LNZ, GEN | mecA, blaZ, aph(3′)-IIIa, tetL | - | hld, tst | 22 (22) | t19963 | IV | I |
| VS2942 | Chaves (Untreated) | BEN, CXI, CIP, GEN, KAN, ERY, CLI, FUS | mecA, blaZ, aph(3′)-IIIa, ermC | B | hld | 22 (22) | t747 | IVc | I |
| VS2943 | Chaves (Untreated) | BEN, CXI, CIP, ERY | mecA, blaZ, ermC | B | hld, tst | 22 (22) | t747 | IVc | I |
| VS2944 | Chaves (Untreated) | BEN, CXI, CIP, ERY, CLI, FUS | mecA, blaZ, ermC | - | hlb, hld, tst | 22 (22) | t747 | IVc | I |
| VS2945 | Chaves (Untreated) | BEN, CXI, CIP, ERY, TET | mecA, blaZ, ermC, tetM | B | hld | 22 (22) | t747 | IV | I |
| VS2946 | Chaves (Untreated) | BEN, CXI, CIP, GEN, KAN, TOB, ERY, CLI, TET, FUS, TRS | mecA, blaZ, aac(6′)-Ie-aph(2′’)-Ia, aph(3′)-IIIa, ermB, ermC, tetL, dfrG | - | hlb, hld | 22 (22) | t6966 | IV | I |
| VS2947 | Chaves (Untreated) | BEN, CXI, CIP | mecA, blaZ | B | hld | 22 (22) | t747 | IV | I |
| VS2948 | Lamego (Untreated) | BEN, CXI, CIP, ERY | mecA, blaZ, ermC | B | hld | 22 (22) | t747 | IVc | I |
| VS2949 | Lamego (Untreated) | BEN, CXI, CIP | mecA, blaZ | B | hld | 22 (22) | t747 | IV | I |
| VS2950 | Lamego (Untreated) | BEN, CXI, CIP, GEN, ERY | mecA, blaZ, aph(3′)-IIIa, ermC | B | hld | 22 (22) | t747 | IV | I |
| VS2951 | Lamego (Untreated) | BEN, CXI, CIP, GEN, TOB, ERY | mecA, blaZ, aac(6′)-Ie-aph(2′’)-Ia, aph(3′)-IIIa, ermB, ermC | B | hld | 22 (22) | t1302 | IVc | I |
| VS2952 | Lamego (Untreated) | BEN, CXI, CIP, GEN, KAN, TOB, ERY, CLI, TET, CHL, FUS, TRS | mecA, blaZ, aph(3′)-IIIa, ermB, ermC, tetL, dfrG, dfrA,catpC221 | B | hld | 22 (22) | t747 | IVc | I |
| VS2953 | Lamego (Untreated) | BEN, CXI, CIP | mecA, blaZ | B | hld, tst | 22 (22) | t747 | IVc | I |
| VS2954 | Lamego (Untreated) | BEN, CXI, CIP, GEN, ERY | mecA, blaZ, aph(3′)-IIIa, ermC | B | hld, tst | 22 (22) | t747 | IVc | I |
| VS2955 | Lamego (Untreated) | BEN, CXI, CIP, ERY | mecA, blaZ, ermC | B | hld | 22 (22) | t747 | IVc | I |
| VS2956 | Lamego (Untreated) | BEN, CXI, CIP, ERY, FUS | mecA, blaZ, ermC | B | tst | 22 (22) | t747 | IV | I |
| VS2957 | Lamego (Untreated) | BEN, CXI, CIP, GEN, KAN, ERY, CLI, FUS | mecA, blaZ, aac(6′)-Ie-aph(2′’)-Ia, aph(3′)-IIIa, ermC, ermA | D | hld, tst | 8 | t008 | IV | I |
| VS2958 | Lamego (Untreated) | BEN, CXI, CIP | mecA, blaZ | B | hld | 22 (22) | t747 | IV | I |
| VS2959 | Lamego (Untreated) | BEN, CXI, CIP | mecA, blaZ | B | tst | 22 (22) | t747 | IV | I |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, V.; Ribeiro, J.; Rocha, J.; Manaia, C.M.; Silva, A.; Pereira, J.E.; Maltez, L.; Capelo, J.L.; Igrejas, G.; Poeta, P. High Frequency of the EMRSA-15 Clone (ST22-MRSA-IV) in Hospital Wastewater. Microorganisms 2022, 10, 147. https://doi.org/10.3390/microorganisms10010147
Silva V, Ribeiro J, Rocha J, Manaia CM, Silva A, Pereira JE, Maltez L, Capelo JL, Igrejas G, Poeta P. High Frequency of the EMRSA-15 Clone (ST22-MRSA-IV) in Hospital Wastewater. Microorganisms. 2022; 10(1):147. https://doi.org/10.3390/microorganisms10010147
Chicago/Turabian StyleSilva, Vanessa, Jessica Ribeiro, Jaqueline Rocha, Célia M. Manaia, Adriana Silva, José Eduardo Pereira, Luís Maltez, José Luis Capelo, Gilberto Igrejas, and Patrícia Poeta. 2022. "High Frequency of the EMRSA-15 Clone (ST22-MRSA-IV) in Hospital Wastewater" Microorganisms 10, no. 1: 147. https://doi.org/10.3390/microorganisms10010147