Phylum Gemmatimonadota and Its Role in the Environment
Abstract
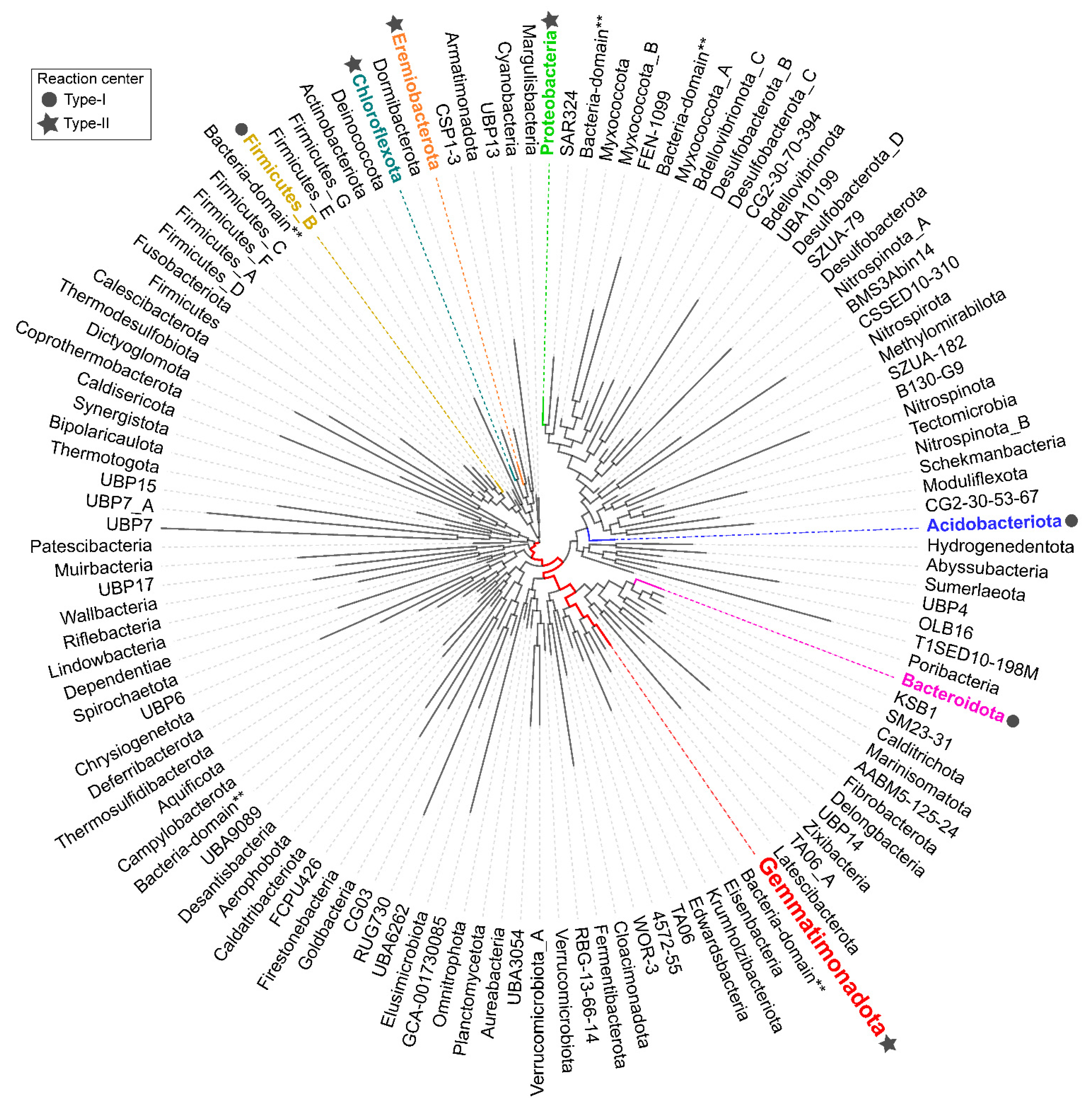
:1. Introduction
2. Cultured Species
2.1. Physiology and Metabolism of Cultured Gemmatimonadota
2.2. Anoxygenic Phototrophy in Gemmatimonadota
3. Environmental Distribution
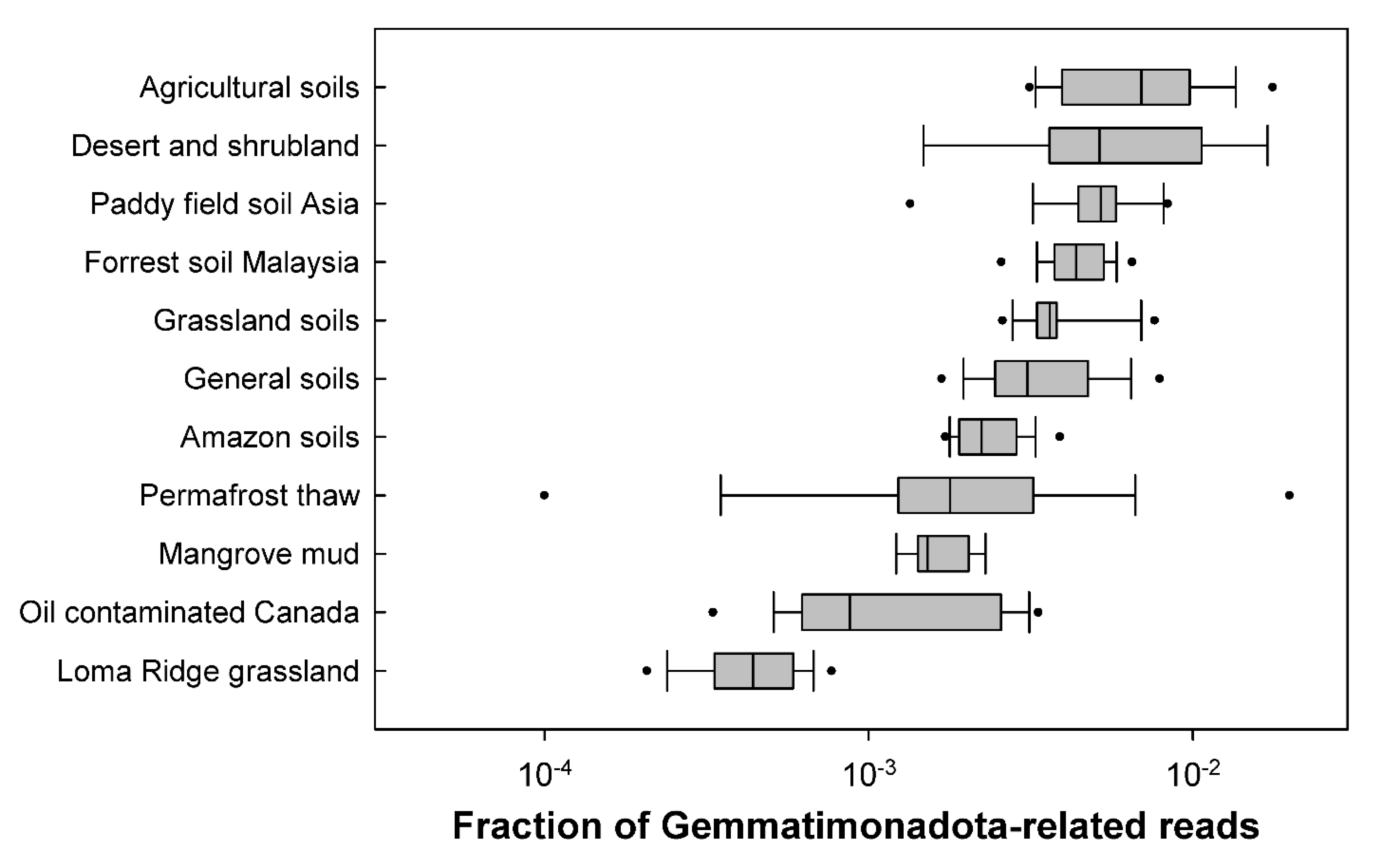
3.1. Distribution in Soils
3.2. Distribution in Aquatic Habitats
3.3. Other Environments
3.4. Distribution of Phototrophic Gemmatimonadota
4. Summary and Perspective
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woese, C.R. Bacterial evolution. Microbiol. Rev. 1987, 51, 221–271. [Google Scholar] [CrossRef]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.A.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017, 2, 1533–1542. [Google Scholar] [CrossRef]
- Hugenholtz, P.; Tyson, G.W.; Webb, R.I.; Wagner, A.M.; Blackall, L.L. Investigation of candidate division tm7, a recently recognized major lineage of the domain bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 2001, 67, 411–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mummey, D.L.; Stahl, P.D. Candidate Division BD: Phylogeny, Distribution and abundance in soil ecosystems. Syst. Appl. Microbiol. 2003, 26, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Kato, C.; Horikoshi, K. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv. 1999, 8, 659–677. [Google Scholar] [CrossRef]
- Madrid, V.M.; Aller, J.Y.; Aller, R.C.; Chistoserdov, A.Y. High prokaryote diversity and analysis of community structure in mobile mud deposits off French Guiana: Identification of two new bacterial candidate divisions. FEMS Microbiol. Ecol. 2001, 37, 197–209. [Google Scholar] [CrossRef]
- Zhang, H.; Sekiguchi, Y.; Hanada, S.; Hugenholtz, P.; Kim, H.; Kamagata, Y.; Nakamura, K. Gemmatimonas aurantiaca gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 1155–1163. [Google Scholar] [CrossRef]
- Zeng, Y.; Feng, F.; Medová, H.; Dean, J.; Koblížek, M. Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadetes. Proc. Natl. Acad. Sci. USA 2014, 111, 7795–7800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, L.M.; Cardona, T.; Holland-Moritz, H. Evolutionary Implications of Anoxygenic Phototrophy in the Bacterial Phylum Candidatus Eremiobacterota (WPS-2). Front. Microbiol. 2019, 10, 1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardona, T. Thinking twice about the evolution of photosynthesis. Open Biol. 2019, 9, 180246. [Google Scholar] [CrossRef] [Green Version]
- Mendler, K.; Chen, H.; Parks, D.H.; Lobb, B.; Hug, L.A.; Doxey, A.C. AnnoTree: Visualization and exploration of a functionally annotated microbial tree of life. Nucleic Acids Res. 2019, 47, 4442–4448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, D.H.; Chuvochina, M.; Rinke, C.; Mussig, A.J.; Chaumeil, P.-A.; Hugenholtz, P. GTDB: An ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2021, 50, 785–794. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Zeng, Y.; Selyanin, V.; Lukeš, M.; Dean, J.; Kaftan, D.; Feng, F.; Koblížek, M. Characterization of the microaerophilic, bacteriochlorophyll a-containing bacterium Gemmatimonas phototrophica sp. nov., and emended descriptions of the genus Gemmatimonas and Gemmatimonas aurantiaca. Int. J. Syst. Evol. Microbiol. 2015, 65, 2410–2419. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Nupur; Wu, N.; Madsen, A.M.; Chen, X.; Gardiner, A.T.; Koblížek, M. Gemmatimonas groenlandica sp. nov. Is an Aerobic Anoxygenic Phototroph in the Phylum Gemmatimonadetes. Front. Microbiol. 2021, 11, 606612. [Google Scholar] [CrossRef]
- Zeng, Y.; Baumbach, J.; Barbosa, E.G.V.; Azevedo, V.; Zhang, C.; Koblížek, M. Metagenomic evidence for the presence of phototrophic Gemmatimonadetes bacteria in diverse environments. Environ. Microbiol. Rep. 2016, 8, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Mujakić, I.; Andrei, A.-Ş.; Shabarova, T.; Fecskeová, L.K.; Salcher, M.M.; Piwosz, K.; Ghai, R.; Koblížek, M. Common Presence of Phototrophic Gemmatimonadota in Temperate Freshwater Lakes. mSystems 2021, 6, e01241-20. [Google Scholar] [CrossRef] [PubMed]
- Vavourakis, C.; Mehrshad, M.; Balkema, C.; Van Hall, R.; Andrei, A.-Ş.; Ghai, R.; Sorokin, D.Y.; Muyzer, G. Metagenomes and metatranscriptomes shed new light on the microbial-mediated sulfur cycle in a Siberian soda lake. BMC Biol. 2019, 17, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koblížek, M.; Dachev, M.; Bína, D.; Nupur; Piwosz, K.; Kaftan, D. Utilization of light energy in phototrophic Gemmatimonadetes. J. Photochem. Photobiol. B Biol. 2020, 213, 112085. [Google Scholar] [CrossRef]
- DeBruyn, J.M.; Fawaz, M.N.; Peacock, A.D.; Dunlap, J.R.; Nixon, L.T.; Cooper, K.E.; Radosevich, M. Gemmatirosa kalamazoonesis gen. nov., sp. nov., a member of the rarely-cultivated bacterial phylum Gemmatimonadetes. J. Gen. Appl. Microbiol. 2013, 59, 305–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascual, J.; Foesel, B.U.; Geppert, A.; Huber, K.J.; Boedeker, C.; Luckner, M.; Wanner, G.; Overmann, J. Roseisolibacter agri gen. nov., sp. nov., a novel slow-growing member of the under-represented phylum Gemmatimonadetes. Int. J. Syst. Evol. Microbiol. 2018, 68, 1028–1036. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; García-López, M.; Bills, G.F.; Genilloud, O. Longimicrobium terrae gen. nov., sp. nov., an oligotrophic bacterium of the under-represented phylum Gemmatimonadetes isolated through a system of miniaturized diffusion chambers. Int. J. Syst. Evol. Microbiol. 2016, 66, 1976–1985. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Kim, H.; Yoon, S. Nitrous oxide reduction by an obligate. Appl. Environ. Microbiol. 2017, 83, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Chee-Sanford, J.; Tian, D.; Sanford, R. Consumption of N2O and other N-cycle intermediates by Gemmatimonas aurantiaca strain T-27. Microbiology 2019, 165, 1345–1354. [Google Scholar] [CrossRef]
- Hohmann-Marriott, M.F.; Blankenship, R.E. Evolution of Photosynthesis. Annu. Rev. Plant Biol. 2011, 62, 515–548. [Google Scholar] [CrossRef] [Green Version]
- Nagashima, S.; Nagashima, K.V.P. Comparison of Photosynthesis Gene Clusters Retrieved from Total Genome Sequences of Purple Bacteria. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2013; Volume 66, pp. 151–178. ISBN 9780123979230. [Google Scholar]
- Zhaxybayeva, O.; Gogarten, J.P.; Charlebois, R.L.; Doolittle, W.F.; Papke, R.T. Phylogenetic analyses of cyanobacterial genomes: Quantification of horizontal gene transfer events. Genome Res. 2006, 16, 1099–1108. [Google Scholar] [CrossRef] [Green Version]
- Sousa, F.L.; Shavit-Grievink, L.; Allen, J.F.; Martin, W.F. Chlorophyll Biosynthesis Gene Evolution Indicates Photosystem Gene Duplication, Not Photosystem Merger, at the Origin of Oxygenic Photosynthesis. Genome Biol. Evol. 2013, 5, 200–216. [Google Scholar] [CrossRef]
- Igarashi, N.; Harada, J.; Nagashima, S.; Matsuura, K.; Shimada, K.; Nagashima, K.V.P. Horizontal Transfer of the Photosynthesis Gene Cluster and Operon Rearrangement in Purple Bacteria. J. Mol. Evol. 2001, 52, 333–341. [Google Scholar] [CrossRef]
- Ward, L.M.; Hemp, J.; Shih, P.M.; McGlynn, S.E.; Fischer, W.W. Evolution of Phototrophy in the Chloroflexi Phylum Driven by Horizontal Gene Transfer. Front. Microbiol. 2018, 9, 260. [Google Scholar] [CrossRef]
- Brinkmann, H.; Göker, M.; Koblížek, M.; Wagner-Döbler, I.; Petersen, J. Horizontal operon transfer, plasmids, and the evolution of photosynthesis in Rhodobacteraceae. ISME J. 2018, 12, 1994–2010. [Google Scholar] [CrossRef]
- Cardona, T. Origin of Bacteriochlorophyll a and the Early Diversification of Photosynthesis. PLoS ONE 2016, 11, e0151250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dachev, M.; Bína, D.; Sobotka, R.; Moravcová, L.; Gardian, Z.; Kaftan, D.; Šlouf, V.; Fuciman, M.; Polívka, T.; Koblížek, M. Unique double concentric ring organization of light harvesting complexes in Gemmatimonas phototrophica. PLoS Biol. 2017, 15, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Yurkov, V.V.; Beatty, J.T. Aerobic Anoxygenic Phototrophic Bacteria. Microbiol. Mol. Biol. Rev. 1998, 62, 695–724. [Google Scholar] [CrossRef] [Green Version]
- Takaichi, S.; Maoka, T.; Takasaki, K.; Hanada, S. Carotenoids of Gemmatimonas aurantiaca (Gemmatimonadetes): Identification of a novel carotenoid, deoxyoscillol 2-rhamnoside, and proposed biosynthetic pathway of oscillol 2,2′-dirhamnoside. Microbiology 2010, 156, 757–763. [Google Scholar] [CrossRef] [Green Version]
- Yurkov, V.; Csotonyi, J.T. New Light on Aerobic Anoxygenic Phototrophs. In Advances in Photosynthesis and Respiration; Springer: Berlin/Heidelberg, Germany, 2009; pp. 31–55. [Google Scholar]
- Nupur; Kuzma, M.; Hájek, J.; Hrouzek, P.; Gardiner, A.T.; Lukeš, M.; Moos, M.; Šimek, P.; Koblížek, M. Structure elucidation of the novel carotenoid gemmatoxanthin from the photosynthetic complex of Gemmatimonas phototrophica AP64. Sci. Rep. 2021, 11, 15964. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef] [Green Version]
- DeBruyn, J.M.; Nixon, L.T.; Fawaz, M.N.; Johnson, A.M.; Radosevich, M. Global Biogeography and Quantitative Seasonal Dynamics of Gemmatimonadetes in Soil. Appl. Environ. Microbiol. 2011, 77, 6295–6300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef] [Green Version]
- Ren, C.; Chen, J.; Lu, X.; Doughty, R.; Zhao, F.; Zhong, Z.; Han, X.; Yang, G.; Feng, Y.; Ren, G. Responses of soil total microbial biomass and community compositions to rainfall reductions. Soil Biol. Biochem. 2018, 116, 4–10. [Google Scholar] [CrossRef]
- Bakermans, C.; Skidmore, M.L.; Douglas, S.; McKay, C.P. Molecular characterization of bacteria from permafrost of the Taylor Valley, Antarctica. FEMS Microbiol. Ecol. 2014, 89, 331–346. [Google Scholar] [CrossRef]
- Frey, B.; Rime, T.; Phillips, M.; Stierli, B.; Hajdas, I.; Widmer, F.; Hartmann, M. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 2016, 92, fiw018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuorto, S.J.; Darias, P.; McGuinness, L.R.; Panikov, N.; Zhang, T.; Häggblom, M.M.; Kerkhof, L.J. Bacterial genome replication at subzero temperatures in permafrost. ISME J. 2014, 8, 139–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cary, S.C.; McDonald, I.R.; Barrett, J.E.; Cowan, D.A. On the rocks: The microbiology of Antarctic Dry Valley soils. Nat. Rev. Microbiol. 2010, 8, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Breidenbach, B.; Pump, J.; Dumont, M.G. Microbial Community Structure in the Rhizosphere of Rice Plants. Front. Microbiol. 2016, 6, 1537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Wang, N.; Guo, X.; Zhang, Y.; Ye, B. Comparative analysis of bacterial community structure in the rhizosphere of maize by high-throughput pyrosequencing. PLoS ONE 2017, 12, e0178425. [Google Scholar] [CrossRef]
- Gkarmiri, K.; Mahmood, S.; Ekblad, A.; Alström, S.; Högberg, N.; Finlay, R. Identifying the Active Microbiome Associated with Roots and Rhizosphere Soil of Oilseed Rape. Appl. Environ. Microbiol. 2017, 83, e01938-17. [Google Scholar] [CrossRef] [Green Version]
- Ren, N.; Wang, Y.; Ye, Y.; Zhao, Y.; Huang, Y.; Fu, W.; Chu, X. Effects of Continuous Nitrogen Fertilizer Application on the Diversity and Composition of Rhizosphere Soil Bacteria. Front. Microbiol. 2020, 11, 1948. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.T.F.; Valverde, N.B.; Lagurara, P.; Revale, S.; De Souza, J.A.M.; Vilaro, M.D.R. Soil and Rhizosphere Bacterial Diversity in Maize Agro- Ecosystem. Sustain. Agric. Res. 2017, 6, 35. [Google Scholar] [CrossRef] [Green Version]
- Na, X.; Xu, T.; Li, M.; Zhou, Z.; Ma, S.; Wang, J.; He, J.; Jiao, B.; Ma, F. Variations of Bacterial Community Diversity Within the Rhizosphere of Three Phylogenetically Related Perennial Shrub Plant Species Across Environmental Gradients. Front. Microbiol. 2018, 9, 709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gugliandolo, C.; Michaud, L.; Giudice, A.L.; Lentini, V.; Rochera, C.; Camacho, A.; Maugeri, T.L. Prokaryotic Community in Lacustrine Sediments of Byers Peninsula (Livingston Island, Maritime Antarctica). Microb. Ecol. 2016, 71, 387–400. [Google Scholar] [CrossRef]
- Sheng, P.; Yu, Y.; Zhang, G.; Huang, J.; He, L.; Ding, J. Bacterial diversity and distribution in seven different estuarine sediments of Poyang Lake, China. Environ. Earth Sci. 2016, 75, 479. [Google Scholar] [CrossRef]
- Röske, K.; Sachse, R.; Scheerer, C.; Röske, I. Microbial diversity and composition of the sediment in the drinking water reservoir Saidenbach (Saxonia, Germany). Syst. Appl. Microbiol. 2012, 35, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Li, Z.; Du, B.; Wang, G.; Ding, Y. Bacterial communities in sediments of the shallow Lake Dongping in China. J. Appl. Microbiol. 2012, 112, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, J.; Zhao, L.; Zhang, X.; Xie, S. Spatial distribution of bacterial communities in high-altitude freshwater wetland sediment. Limnology 2014, 15, 249–256. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Y.; Yuzhao, L.; Li, Y.; Xie, S.; Liu, Y. Distribution of sediment bacterial and archaeal communities in plateau freshwater lakes. Appl. Microbiol. Biotechnol. 2015, 99, 3291–3302. [Google Scholar] [CrossRef]
- Hanada, S.; Sekiguchi, Y. The phylum Gemmatimonadetes. In The Prokaryotes, 4th ed.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 11, pp. 677–681. [Google Scholar]
- Qin, H.; Ji, B.; Zhang, S.; Kong, Z. Study on the bacterial and archaeal community structure and diversity of activated sludge from three wastewater treatment plants. Mar. Pollut. Bull. 2018, 135, 801–807. [Google Scholar] [CrossRef]
- Durbin, A.M.; Teske, A. Microbial diversity and stratification of South Pacific abyssal marine sediments. Environ. Microbiol. 2011, 13, 3219–3234. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, T.; Pinho, D.; Egas, C.; Froufe, H.; Altermark, B.; Candeias, C.; Santos, R.S.; Bettencourt, R. Microbial diversity in deep-sea sediments from the Menez Gwen hydrothermal vent system of the Mid-Atlantic Ridge. Mar. Genom. 2015, 24, 343–355. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, Q.-L.; Zeng, Z.-G.; Chen, S.; Sun, L. Microbial diversity in the deep-sea sediments of Iheya North and Iheya Ridge, Okinawa Trough. Microbiol. Res. 2015, 177, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Kang, M.; Xu, J.; Xu, J.; Shuai, Y.; Zhou, X.; Yang, Z.; Ma, K. Bacterial and archaeal communities in the deep-sea sediments of inactive hydrothermal vents in the Southwest India Ridge. Sci. Rep. 2016, 6, 25982. [Google Scholar] [CrossRef]
- Kamke, J.; Taylor, M.W.; Schmitt, S. Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. ISME J. 2010, 4, 498–508. [Google Scholar] [CrossRef] [PubMed]
- Montalvo, N.F.; Hill, R.T. Sponge-Associated Bacteria Are Strictly Maintained in Two Closely Related but Geographically Distant Sponge Hosts. Appl. Environ. Microbiol. 2011, 77, 7207–7216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souza, D.T.; Genuário, D.B.; Silva, F.S.P.; Pansa, C.C.; Kavamura, V.N.; Moraes, F.C.; Taketani, R.G.; Melo, I.S. Analysis of bacterial composition in marine sponges reveals the influence of host phylogeny and environment. FEMS Microbiol. Ecol. 2017, 93, fiw204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, J.; Flemer, B.; Jackson, S.A.; Morrissey, J.P.; O’Gara, F.; Dobson, A.D.W. Evidence of a Putative Deep Sea Specific Microbiome in Marine Sponges. PLoS ONE 2014, 9, e91092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gołębiewski, M.; Całkiewicz, J.; Creer, S.; Piwosz, K. Tideless estuaries in brackish seas as possible freshwater-marine transition zones for bacteria: The case study of the Vistula river estuary. Environ. Microbiol. Rep. 2017, 9, 129–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hentschel, U.; Hopke, J.; Horn, M.; Friedrich, A.B.; Wagner, M.; Hacker, J.; Moore, B.S. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 2002, 68, 4431–4440. [Google Scholar] [CrossRef] [Green Version]
- Dunbar, J.; Barns, S.M.; Ticknor, L.O.; Kuske, C.R. Empirical and theoretical bacterial diversity in four arizona soils. Appl. Environ. Microbiol. 2002, 68, 3035–3045. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. Model Finder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bajerski, F.; Wagner, D. Bacterial succession in Antarctic soils of two glacier forefields on Larsemann Hills, East Antarctica. FEMS Microbiol. Ecol. 2013, 85, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Niederberger, T.D.; McDonald, I.R.; Hacker, A.L.; Soo, R.M.; Barrett, J.E.; Wall, D.H.; Cary, S.C. Microbial community composition in soils of Northern Victoria Land, Antarctica. Environ. Microbiol. 2008, 10, 1713–1724. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Martínez, V.; Dowd, S.; Sun, Y.; Allen, V. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 2008, 40, 2762–2770. [Google Scholar] [CrossRef]
- Kim, J.-S.; Dungan, R.S.; Crowley, D. Microarray analysis of bacterial diversity and distribution in aggregates from a desert agricultural soil. Biol. Fertil. Soils 2008, 44, 1003–1011. [Google Scholar] [CrossRef]
- Ahmed, V.; Verma, M.K.; Gupta, S.; Mandhan, V.; Chauhan, N.S. Metagenomic Profiling of Soil Microbes to Mine Salt Stress Tolerance Genes. Front. Microbiol. 2018, 9, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Liu, J.J.; Banerjee, S.; Zhou, N.; Zhao, Z.Y.; Zhang, K.; Hu, M.F.; Tian, C.Y. Biogeographical distribution of bacterial communities in saline agricultural soil. Geoderma 2020, 361, 114095. [Google Scholar] [CrossRef]
- Neilson, J.W.; Califf, K.; Cardona, C.; Copeland, A.; van Treuren, W.; Josephson, K.L.; Knight, R.; Gilbert, J.A.; Quade, J.; Caporaso, J.G.; et al. Significant Impacts of Increasing Aridity on the Arid Soil Microbiome. mSystems 2017, 2, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Mummey, D.; Holben, W.; Six, J.; Stahl, P. Spatial Stratification of Soil Bacterial Populations in Aggregates of Diverse Soils. Microb. Ecol. 2006, 51, 404–411. [Google Scholar] [CrossRef]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- Mendez, M.O.; Neilson, J.W.; Maier, R.M. Characterization of a Bacterial Community in an Abandoned Semiarid Lead-Zinc Mine Tailing Site. Appl. Environ. Microbiol. 2008, 74, 3899–3907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malard, L.A.; Anwar, M.Z.; Jacobsen, C.S.; Pearce, D.A. Biogeographical patterns in soil bacterial communities across the Arctic region. FEMS Microbiol. Ecol. 2019, 95, fiz128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, Y.; Jiang, N.; Wu, Y.; Yang, Z.; Bello, A.; Yang, W. Disentangling the role of salinity-sodicity in shaping soil microbiome along a natural saline-sodic gradient. Sci. Total Environ. 2021, 765, 142738. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, X.; Zhu, R.; Chen, N.; Ding, L.; Chen, C. Vegetation richness, species identity and soil nutrients drive the shifts in soil bacterial communities during restoration process. Environ. Microbiol. Rep. 2021, 13, 1758–2229. [Google Scholar] [CrossRef]
- Deng, J.; Bai, X.; Zhou, Y.; Zhu, W.; Yin, Y. Variations of soil microbial communities accompanied by different vegetation restoration in an open-cut iron mining area. Sci. Total Environ. 2020, 704, 135243. [Google Scholar] [CrossRef]
- Ye, W.; Liu, X.; Lin, S.; Tan, J.; Pan, J.; Li, D.; Yang, H. The vertical distribution of bacterial and archaeal communities in the water and sediment of Lake Taihu. FEMS Microbiol. Ecol. 2009, 70, 263–276. [Google Scholar] [CrossRef] [Green Version]
- Traving, S.J.; Rowe, O.; Jakobsen, N.M.; Sørensen, H.; Dinasquet, J.; Stedmon, C.A.; Andersson, A.; Riemann, L. The Effect of Increased Loads of Dissolved Organic Matter on Estuarine Microbial Community Composition and Function. Front. Microbiol. 2017, 8, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Morrison, J.M.; Baker, K.D.; Zamor, R.M.; Nikolai, S.; Elshahed, M.S.; Youssef, N.H. Spatiotemporal analysis of microbial community dynamics during seasonal stratification events in a freshwater lake (Grand Lake, OK, USA). PLoS ONE 2017, 12, e0177488. [Google Scholar] [CrossRef] [Green Version]
- Cabello-Yeves, P.J.; Zemskaya, T.; Rosselli, R.; Coutinho, F.H.; Zakharenko, A.S.; Blinov, V.V.; Rodriguez-Valera, F. Genomes of Novel Microbial Lineages Assembled from the Sub-Ice Waters of Lake Baikal. Appl. Environ. Microbiol. 2018, 84, e02132-17. [Google Scholar] [CrossRef] [Green Version]
- Shia, L.; Cai, Y.; Wang, X.; Li, P.; Yu, Y.; Kong, F. Community Structure of Bacteria Associated withMicrocystisColonies from Cyanobacterial Blooms. J. Freshw. Ecol. 2010, 25, 193–203. [Google Scholar] [CrossRef]
- Vavourakis, C.D.; Andrei, A.S.; Mehrshad, M.; Ghai, R.; Sorokin, D.Y.; Muyzer, G. A metagenomics roadmap to the uncultured genome diversity in hypersaline soda lake sediments 06 Biological Sciences 0605 Microbiology 06 Biological Sciences 0604 Genetics. Microbiome 2018, 6, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Çınar, S.; Mutlu, M.B. Prokaryotic community compositions of the hypersaline sediments of tuz lake demonstrated by cloning and high-throughput sequencing. Microbiology 2020, 89, 756–768. [Google Scholar] [CrossRef]
- Cui, G.; Li, J.; Gao, Z.; Wang, Y. Spatial variations of microbial communities in abyssal and hadal sediments across the Challenger Deep. PeerJ 2019, 7, e6961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peoples, L.M.; Grammatopoulou, E.; Pombrol, M.; Xu, X.; Osuntokun, O.; Blanton, J.C.; Allen, E.E.; Nunnally, C.C.; Drazen, J.; Mayor, D.J.; et al. Microbial Community Diversity Within Sediments from Two Geographically Separated Hadal Trenches. Front. Microbiol. 2019, 10, 347. [Google Scholar] [CrossRef]
- Kato, S.; Nakawake, M.; Kita, J.; Yamanaka, T.; Utsumi, M.; Okamura, K.; Ishibashi, J.-I.; Ohkuma, M.; Yamagishi, A. Characteristics of Microbial Communities in Crustal Fluids in a Deep-Sea Hydrothermal Field of the Suiyo Seamount. Front. Microbiol. 2013, 4, 85. [Google Scholar] [CrossRef] [Green Version]
- Nunoura, T.; Nishizawa, M.; Hirai, M.; Shimamura, S.; Harnvoravongchai, P.; Koide, O.; Morono, Y.; Fukui, T.; Inagaki, F.; Miyazaki, J.; et al. Microbial Diversity in Sediments from the Bottom of the Challenger Deep, the Mariana Trench. Microbes Environ. 2018, 33, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Thiel, V.; Neulinger, S.C.; Staufenberger, T.; Schmaljohann, R.; Imhoff, J.F. Spatial distribution of sponge-associated bacteria in the Mediterranean sponge Tethya aurantium. FEMS Microbiol. Ecol. 2007, 59, 47–63. [Google Scholar] [CrossRef] [Green Version]
- Slaby, B.M.; Hackl, T.; Horn, H.; Bayer, K.; Hentschel, U. Metagenomic binning of a marine sponge microbiome reveals unity in defense but metabolic specialization. ISME J. 2017, 11, 2465–2478. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Yela, A.C.A.; Mosquera-Rendón, J.; Noreña-P, A.; Cristancho, M.; López-Alvarez, D. Microbial Diversity Exploration of Marine Hosts at Serrana Bank, a Coral Atoll of the Seaflower Biosphere Reserve. Front. Mar. Sci. 2019, 6, 338. [Google Scholar] [CrossRef]
- Liu, R.; Wang, Z.; Wang, L.; Li, Z.; Fang, J.; Wei, X.; Wei, W.; Cao, J.; Wei, Y.; Xie, Z. Bulk and Active Sediment Prokaryotic Communities in the Mariana and Mussau Trenches. Front. Microbiol. 2020, 11, 1521. [Google Scholar] [CrossRef]
- Vipindas, P.V.; Mujeeb, R.K.M.; Jabir, T.; Thasneem, T.R.; Mohamed Hatha, A.A. Diversity of sediment bacterial communities in the South Eastern Arabian Sea. Reg. Stud. Mar. Sci. 2020, 35, 101153. [Google Scholar] [CrossRef]
- Herlemann, D.P.R.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.O.O.; Karlson, B.; Charvet, S.; Andersson, A.F. Diversity of Pico- to Mesoplankton along the 2000 km Salinity Gradient of the Baltic Sea. Front. Microbiol. 2016, 7, 679. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, H.; Chen, Z.; Hu, H.; Deng, C.; Wang, X. The benefits of autotrophic nitrogen removal from high concentration of urea wastewater through a process of urea hydrolysis and partial nitritation in sequencing batch reactor. J. Environ. Manag. 2021, 292, 112762. [Google Scholar] [CrossRef]
- Hu, H.; Deng, C.; Wang, X.; Chen, Z.; Zhong, Z.; Wang, R. Performance and mechanism of urea hydrolysis in partial nitritation system based on SBR. Chemosphere 2020, 258, 127228. [Google Scholar] [CrossRef]
- Chen, L.; Feng, Q.; Li, C.; Wei, Y.; Zhao, Y.; Feng, Y.; Zheng, H.; Li, F.; Li, H. Impacts of aquaculture wastewater irrigation on soil microbial functional diversity and community structure in arid regions. Sci. Rep. 2017, 7, 11193. [Google Scholar] [CrossRef] [PubMed]
- Ziganshina, E.E.; Ibragimov, E.M.; Ilinskaya, O.N.; Ziganshin, A.M. Bacterial communities inhabiting toxic industrial wastewater generated during nitrocellulose production. Biologia 2016, 71, 70–78. [Google Scholar] [CrossRef]
- Correa-Galeote, D.; Roibás-Rozas, A.; Mosquera-Corral, A.; Juárez-Jiménez, B.; González-López, J.; Rodelas, B. Revealing the dissimilar structure of microbial communities in different WWTPs that treat fish-canning wastewater with different NaCl content. J. Water Process. Eng. 2021, 44, 102328. [Google Scholar] [CrossRef]
- Rampadarath, S.; Bandhoa, K.; Puchooa, D.; Jeewon, R.; Bal, S. Early bacterial biofilm colonizers in the coastal waters of Mauritius. Electron. J. Biotechnol. 2017, 29, 13–21. [Google Scholar] [CrossRef]
- Besemer, K.; Hödl, I.; Singer, G.; Battin, T.J. Architectural differentiation reflects bacterial community structure in stream biofilms. ISME J. 2009, 3, 1318–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Pan, J.; Li, M.; Li, Y.; Bartlam, M.; Wang, Y. Selective enrichment of bacterial pathogens by microplastic biofilm. Water Res. 2019, 165, 114979. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, D.C.; Cretoiu, M.S.; Stal, L.J.; Bolhuis, H. Seasonal development of a coastal microbial mat. Sci. Rep. 2019, 9, 9035. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Suzuki, M.T.; Rosenberg, M.; Rotem, D.; Madigan, M.T.; Süling, J.; Imhoff, J.F.; Béjà, O. BchY-Based Degenerate Primers Target All Types of Anoxygenic Photosynthetic Bacteria in a Single PCR. Appl. Environ. Microbiol. 2009, 75, 7556–7559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Zeng, Y.; Lu, H.; Feng, H.; Zeng, Y.; Koblížek, M. Novel acsF Gene Primers Revealed a Diverse Phototrophic Bacterial Population, Including Gemmatimonadetes, in Lake Taihu (China). Appl. Environ. Microbiol. 2016, 82, 5587–5594. [Google Scholar] [CrossRef] [Green Version]
- Boldareva-Nuianzina, E.N.; Bláhová, Z.; Sobotka, R.; Koblížek, M. Distribution and Origin of Oxygen-Dependent and Oxygen-Independent Forms of Mg-Protoporphyrin Monomethylester Cyclase among Phototrophic Proteobacteria. Appl. Environ. Microbiol. 2013, 79, 2596–2604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fecskeová, L.K.; Piwosz, K.; Hanusová, M.; Nedoma, J.; Znachor, P.; Koblížek, M. Diel changes and diversity of pufM expression in freshwater communities of anoxygenic phototrophic bacteria. Sci. Rep. 2019, 9, 18766. [Google Scholar] [CrossRef]
- Zorz, J.K.; Sharp, C.; Kleiner, M.; Gordon, P.M.K.; Pon, R.T.; Dong, X.; Strous, M. A shared core microbiome in soda lakes separated by large distances. Nat. Commun. 2019, 10, 4230. [Google Scholar] [CrossRef] [Green Version]
- Ashida, H.; Saito, Y.; Kojima, C.; Kobayashi, K.; Ogasawara, N.; Yokota, A. A Functional Link Between RuBisCO-like Protein of Bacillus and Photosynthetic RuBisCO. Science 2003, 302, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Ashida, H.; Danchin, A.; Yokota, A. Was photosynthetic RuBisCO recruited by acquisitive evolution from RuBisCO-like proteins involved in sulfur metabolism? Res. Microbiol. 2005, 156, 611–618. [Google Scholar] [CrossRef]
- Gudelj, I.; Weitz, J.S.; Ferenci, T.; Horner-Devine, M.; Marx, C.J.; Meyer, J.R.; Forde, S.E. An integrative approach to understanding microbial diversity: From intracellular mechanisms to community structure. Ecol. Lett. 2010, 13, 1073–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piwosz, K.; Shabarova, T.; Tomasch, J.; Šimek, K.; Kopejtka, K.; Kahl, S.; Pieper, D.H.; Koblížek, M. Determining lineage-specific bacterial growth curves with a novel approach based on amplicon reads normalization using internal standard (ARNIS). ISME J. 2018, 12, 2640–2654. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mujakić, I.; Piwosz, K.; Koblížek, M. Phylum Gemmatimonadota and Its Role in the Environment. Microorganisms 2022, 10, 151. https://doi.org/10.3390/microorganisms10010151
Mujakić I, Piwosz K, Koblížek M. Phylum Gemmatimonadota and Its Role in the Environment. Microorganisms. 2022; 10(1):151. https://doi.org/10.3390/microorganisms10010151
Chicago/Turabian StyleMujakić, Izabela, Kasia Piwosz, and Michal Koblížek. 2022. "Phylum Gemmatimonadota and Its Role in the Environment" Microorganisms 10, no. 1: 151. https://doi.org/10.3390/microorganisms10010151






