Effects of Atmospheric Plasma Corona Discharge on Agrobacterium tumefaciens Survival
Abstract
:1. Introduction
2. Materials and Methods
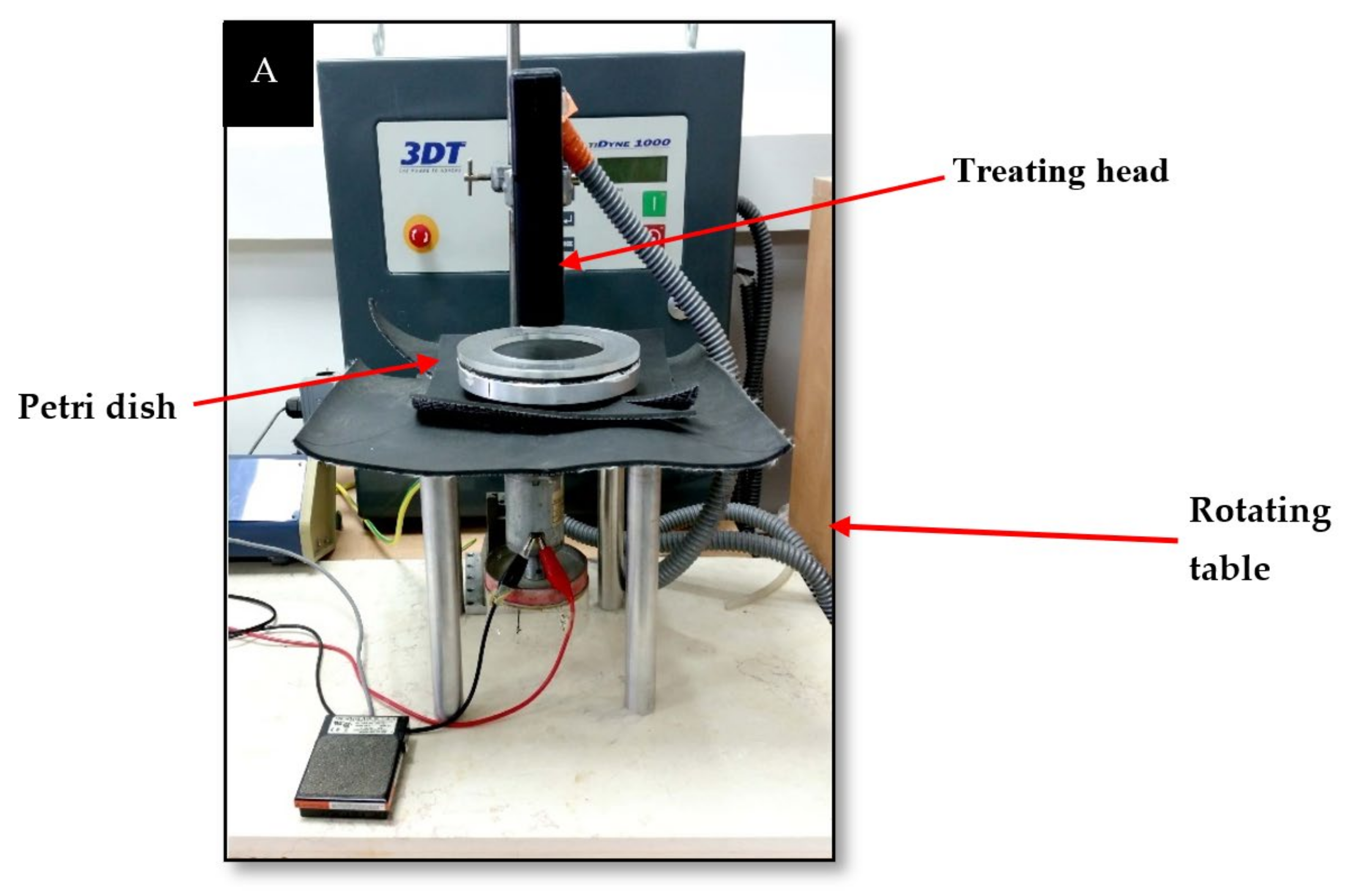
2.1. Plasma Corona Discharge System and Experimental Conditions
2.2. Growth Conditions
2.3. Preparation of the Bacterial Sample for Exposure to Plasma Corona Discharge
2.4. Viable Count Assay
2.5. Cellular ROS Detection
2.6. Examination of Bacterial Membrane Permeability and Cell Size by Flow Cytometry (FCM) Analysis
2.7. Statistics
3. Results
3.1. The Effect of Plasma Corona-Discharge Exposure Duration on the CFU of A. Tumefaciens
3.2. A. Tumefaciens Viability and Possible Recovery of Corona-Treated Bacteria
3.3. A. Tumefaciens Membrane Permeability as a Function of Plasma Treatment and Possible Recovery
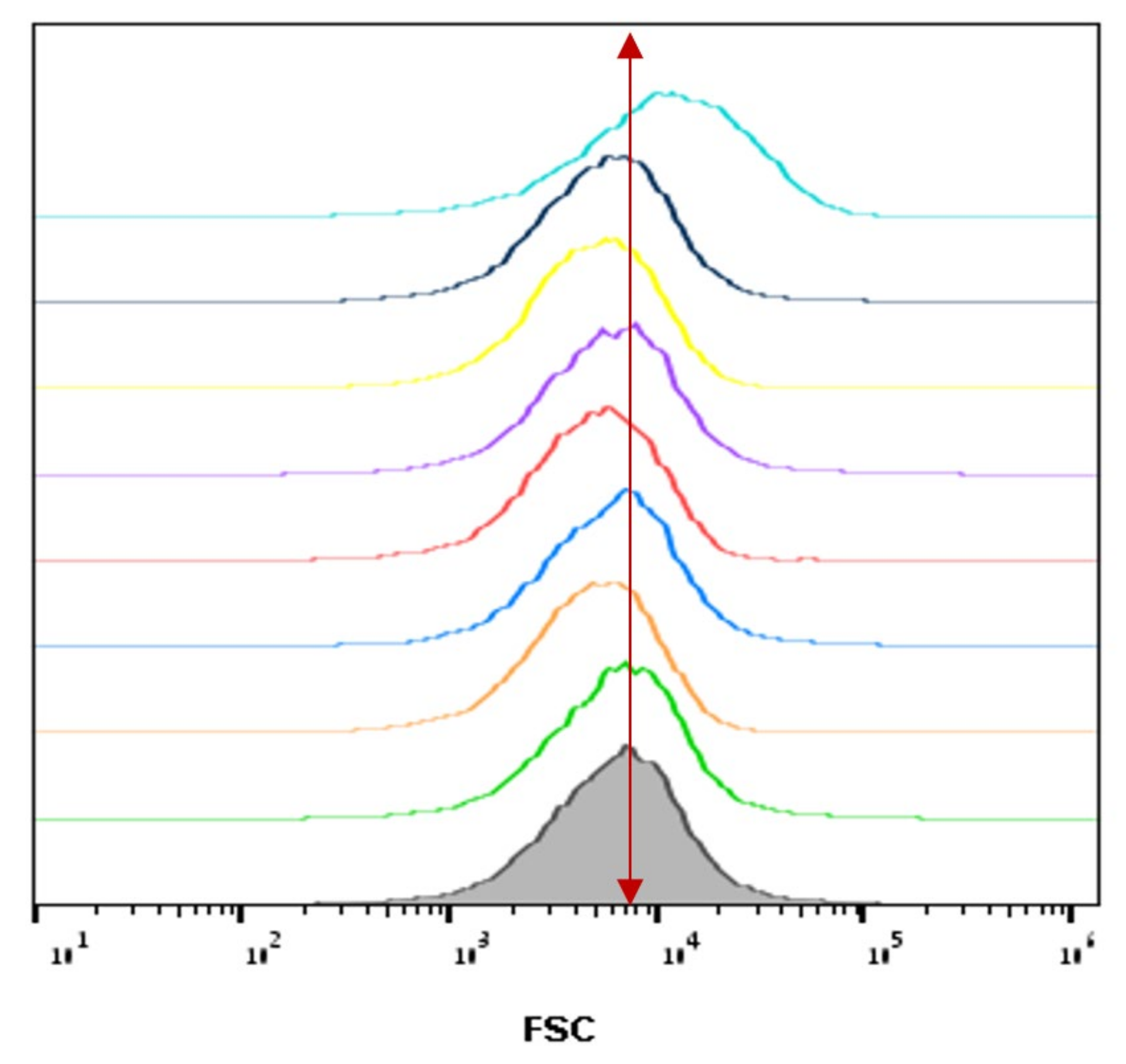
3.4. The Relative Cell Size of A. Tumefaciens as a Function of Plasma Corona Discharge Treatment
3.5. ROS Generation in A. Tumefaciens as a Function of Plasma Treatment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Lucas, P. Diseases caused by soil-borne pathogens. In Epidemiology of Plant Diseases; Springer: Dordrecht, The Netherlands, 2006; pp. 373–386. [Google Scholar] [CrossRef]
- Oerke, E.C. Crop Losses to Pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Nwokolo, N.L.; Enebe, M.C.; Chigor, C.B.; Chigor, V.N.; Dada, O.A. The contributions of biotic lines of defence to improving plant disease suppression in soils: A review. Rhizosphere 2021, 19, 100372. [Google Scholar] [CrossRef]
- Tzfira, T.; Citovsky, V. Agrobacterium-mediated genetic transformation of plants: Biology and biotechnology. Curr. Opin. Biotechnol. 2006, 17, 147–154. [Google Scholar] [CrossRef]
- Morton, E.R.; Platt, T.G.; Fuqua, C.; Bever, J.D. Non-additive costs and interactions alter the competitive dynamics of co-occurring ecologically distinct plasmids. Proc. Biol. Sci. 2014, 281, 20132173. [Google Scholar] [CrossRef]
- Nonaka, S.; Someya, T.; Kadota, Y.; Nakamura, K.; Ezura, H. Super-Agrobacterium Ver. 4: Improving the transformation frequencies and genetic engineering possibilities for crop plants. Front. Plant Sci. 2019, 10, 1204. [Google Scholar] [CrossRef]
- Ragni, L.; Berardinelli, A.; Vannini, L.; Montanari, C.; Sirri, F.; Guerzoni, M.E.; Guarnieri, A. Non-thermal atmospheric gas plasma device for surface decontamination of shell eggs. J. Food Eng. 2010, 100, 125–132. [Google Scholar] [CrossRef]
- Tu, C.; Ristaino, J.B.; Hu, S. Soil microbial biomass and activity in organic tomato farming systems: Effects of organic inputs and straw mulching. Soil Biol. Biochem. 2006, 38, 247–255. [Google Scholar] [CrossRef]
- Mcculloch, A.; Midgley, P.M. Stratospheric chemistry topics: Halogen sources, anthropogenic. In Encyclopedia of Atmospheric Sciences, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2003; pp. 221–227. [Google Scholar]
- Zhang, D.; Yan, D.; Fang, W.; Huang, B.; Wang, X.; Wang, X.; Zhu, J.; Liu, J.; Ouyang, C.; Li, Y. Chloropicrin alternated with biofumigation increases crop yield and modifies soil bacterial and fungal communities in strawberry production. Sci. Total Environ. 2019, 675, 615–622. [Google Scholar] [CrossRef]
- Emanuel, E.; Roman, P.; Cahan, R. Influence of the current density in moderate pulsed electric fields on Pseudomonas putida F1 eradication. Bioelectrochemistry 2019, 126, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Emanuel, E.; Dubrovin, I.; Pogreb, R.; Pinhasi, G.A.; Cahan, R. Resuscitation of pulsed electric field-treated Staphylococcus aureus and Pseudomonas putida in a rich nutrient medium. Foods 2021, 10, 660. [Google Scholar] [CrossRef] [PubMed]
- Lazra, Y.; Dubrovin, I.; Multanen, V.; Bormashenko, E.; Bormashenko, Y.; Cahan, R. Effects of atmospheric plasma corona discharges on soil bacteria viability. Microorganisms 2020, 8, 704. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, M.A.; Michael, A.; Lichtenberg, A.J. Principles of Plasma Discharges and Materials Processing; Wiley-Interscience: Hoboken, NJ, USA, 2005. [Google Scholar]
- Corradini, M.G. Modeling microbial inactivation during cold atmospheric-pressure plasma (CAPP) processing. In Advances in Cold Plasma Applications for Food Safety and Preservation; Elsevier: Amsterdam, The Netherlands, 2019; pp. 93–108. [Google Scholar] [CrossRef]
- Thomas, M.; Mittal, K.L. Atmospheric Pressure Plasma Treatment of Polymers; Scrivener Publishing LLC: Salem, MA, USA, 2013. [Google Scholar]
- Farber, R.; Dabush-Busheri, I.; Chaniel, G.; Rozenfeld, S.; Bormashenko, E.; Multanen, V.; Cahan, R. Biofilm grown on wood waste pretreated with cold low-pressure nitrogen plasma: Utilization for toluene remediation. Int. Biodeterior. Biodegrad. 2019, 139, 62–69. [Google Scholar] [CrossRef]
- Rozenfeld, S.; Hirsch, L.O.; Gandu, B.; Farber, R.; Schechter, A.; Cahan, R. Improvement of microbial electrolysis cell activity by using anode based on combined plasma-pretreated carbon cloth and stainless steel. Energies 2019, 12, 1968. [Google Scholar] [CrossRef] [Green Version]
- Wolf, D.C.; Dao, T.H.; Scott, H.D.; Lavy, T.L. Influence of sterilization methods on selected soil microbiological, physical and chemical properties. J. Environ. Qual. 1989, 18, 39–44. [Google Scholar] [CrossRef]
- Sakudo, A.; Yagyu, Y.; Onodera, T. Disinfection and sterilization using plasma technology: Fundamentals and future perspectives for biological applications. Int. J. Mol. Sci. 2019, 20, 5216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, P.R.; Pirzada, S.A. Thermal plasma processing of materials: A review. Adv. Perform. Mater. 1994, 1, 35–50. [Google Scholar] [CrossRef]
- Rupf, S.; Lehmann, A.; Hannig, M.; Schäfer, B.; Schubert, A.; Feldmann, U.; Schindler, A. Killing of adherent oral microbes by a non-thermal atmospheric plasma jet. J. Med. Microbiol. 2010, 59, 206–212. [Google Scholar] [CrossRef] [Green Version]
- Meichsner, J.; Schmidt, M.; Schneider, R.; Wagner, H. Nonthermal Plasma Chemistry and Physics; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar] [CrossRef]
- Liu, X.; Hong, F.; Guo, Y.; Zhang, J.; Shi, J. Sterilization of Staphylococcus aureus by an atmospheric non-thermal plasma jet. Plasma Sci. Technol. 2013, 15, 439–442. [Google Scholar] [CrossRef]
- Salgado, B.A.B.; Fabbri, S.; Dickenson, A.; Hasan, M.I.; Walsh, J.L. Surface barrier discharges for escherichia coli biofilm inactivation: Modes of action and the importance of UV radiation. PLoS ONE 2021, 16, e0247589. [Google Scholar] [CrossRef]
- Lukes, P.; Dolezalova, E.; Sisrova, I. Aqueous-phase chemistry and bactericidal effects from an air discharge plasma in contact with water: Evidence for the formation of peroxynitrite through a pseudo-second-order post-discharge reaction of H2O2 and HNO2. Plasma Sources Sci. Technol. 2014, 23, 015019. [Google Scholar] [CrossRef]
- Xu, Z.; Cheng, C.; Shen, J.; Lan, Y.; Hu, S.; Han, W.; Chu, P.K. In vitro antimicrobial effects and mechanisms of direct current air-liquid discharge plasma on planktonic Staphylococcus aureus and Escherichia coli in liquids. Bioelectrochemistry 2018, 121, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Machala, Z.; Tarabova, B.; Hensel, K.; Spetlikova, E.; Sikurova, L.; Lukes, P. Formation of ROS and RNS in water electro-sprayed through transient spark discharge in air and their bactericidal effects. Plasma Processes Polym. 2013, 10, 649–659. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, Z.; Shen, J.; Li, X.; Ding, L.; Ma, J.; Lan, Y.; Xia, W.; Cheng, C.; Sun, Q. Effects and mechanism of atmospheric-pressure dielectric barrier discharge cold plasma on lactate dehydrogenase (LDH) enzyme. Sci. Rep. 2015, 5, 10031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoffels, E.; Sakiyama, Y.; Graves, D.B. Cold atmospheric plasma: Charged species and their interactions with cells and tissues. IEEE Trans. Plasma Sci. 2008, 36, 1441–1457. [Google Scholar] [CrossRef]
- Guo, L.; Yang, L.; Qi, Y.; Niyazi, G.; Huang, L.; Gou, L.; Wang, Z.; Zhang, L.; Liu, D.; Wang, X. Cold atmospheric-pressure plasma caused protein damage in methicillin-resistant Staphylococcus aureus cells in biofilms. Microorganisms 2021, 9, 1072. [Google Scholar] [CrossRef]
- Mošovská, S.; Medvecká, V.; Halászová, N.; Ďurina, P.; Valík, Ľ.; Mikulajová, A.; Zahoranová, A. Cold atmospheric pressure ambient air plasma inhibition of pathogenic bacteria on the surface of black pepper. Food Res. Int. 2018, 106, 862–869. [Google Scholar] [CrossRef]
- Calvo, T.; Álvarez-Ordóñez, A.; Prieto, M.; González-Raurich, M.; López, M. Influence of processing parameters and stress adaptation on the inactivation of Listeria monocytogenes by non-thermal atmospheric plasma (NTAP). Food Res. Int. 2016, 89, 631–637. [Google Scholar] [CrossRef]
- Arserim, E.H.; Salvi, D.; Fridman, G.; Schaffner, D.W.; Karwe, M. Microbial inactivation by non-equilibrium short-pulsed atmospheric pressure dielectric barrier discharge (cold plasma): Numerical and experimental studies. Food Eng. Rev. 2021, 13, 136–147. [Google Scholar] [CrossRef]
- Lee, S.H.I.; Fröhling, A.; Schlüter, O.; Corassin, C.H.; de Martinis, E.C.P.; Alves, V.F.; Pimentel, T.C.; Oliveira, C.A.F. Cold atmospheric pressure plasma inactivation of dairy associated planktonic cells of Listeria monocytogenes and Staphylococcus aureus. LWT 2021, 146, 111452. [Google Scholar] [CrossRef]
- Ermis, E.; Ogurlu, M.; Muhammed, Z.D. Combination of vaporized ethyl pyruvate and non-thermal atmospheric pressure plasma for the inactivation of bacteria on lettuce surfaces. Innov. Food Sci. Emerg. Technol. 2021, 73, 102795. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, J.; Yuan, Y.; Chen, H.; Dai, L.; Wang, X.; Yue, T. Bactericidal effect of cold plasma on microbiota of commercial fish balls. Innov. Food Sci. Emerg. Technol. 2019, 52, 394–405. [Google Scholar] [CrossRef]
- Mai-Prochnow, A.; Clauson, M.; Hong, J.; Murphy, A.B. Gram positive and gram negative bacteria differ in their sensitivity to cold plasma. Sci. Rep. 2016, 6, 38610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Händel, N.; Schuurmans, J.M.; Brul, S.; Ter Kuile, B.H. Compensation of the metabolic costs of antibiotic resistance by physiological adaptation in Escherichia coli. Antimicrob. Agents Chemother. 2013, 57, 3752–3762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.; Cheng, J.H.; Lv, X.; Sun, D.W. Assessing the inactivation efficiency of Ar/O2 plasma treatment against Listeria monocytogenes cells: Sublethal injury and inactivation kinetics. LWT 2019, 111, 318–327. [Google Scholar] [CrossRef]
- Liao, X.; Liu, D.; Xiang, Q.; Ahn, J.; Chen, S.; Ye, X.; Ding, T. Inactivation mechanisms of non-thermal plasma on microbes: A review. Food Control. 2017, 75, 83–91. [Google Scholar] [CrossRef]
- Bermudez-Aguirre, D.; Wemlinger, E.; Pedrow, P.; Barbosa-Cánovas, G.; Garcia-Perez, M. Effect of atmospheric pressure cold plasma (APCP) on the inactivation of Escherichia coli in fresh produce. Food Control. 2013, 34, 149–157. [Google Scholar] [CrossRef]
- Yusupov, M. Plasma-induced destruction of bacterial cell wall components: A reactive molecular dynamics simulation. J. Phys. Chem. 2013, 117, 5993–5998. [Google Scholar] [CrossRef]
- Huang, H.; Lung, H.M.; Yang, B.B.; Wang, C.Y. Responses of microorganisms to high hydrostatic pressure processing. Food Control. 2014, 40, 250–259. [Google Scholar] [CrossRef]
- Joshi, S.G.; Cooper, M.; Yost, A.; Paff, M.; Ercan, Y.K.; Fridman, G.; Fridman, G.; Friedman, G.; Fridman, A.; Brooks, A.D. Nonthermal dielectric-barrier discharge plasma-induced inactivation involves oxidative DNA damage and membrane lipid peroxidation in Escherichia coli. Antimicrob. Agents Chemother. 2011, 55, 1053–1062. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, G.; Helmann, J.D. A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol. Microbiol. 2010, 45, 997–1005. [Google Scholar] [CrossRef]
- Eiamphungporn, W.; Nakjarung, K.; Prapagdee, B.; Vattanaviboon, P.; Mongkolsuk, S. Oxidantinducible resistance to hydrogen peroxide killing in Agrobacterium tumefaciens requires the global peroxide sensor-regulator OxyR and KatA. FEMS Microbiol. Lett. 2003, 225, 167–172. [Google Scholar] [CrossRef] [Green Version]
- Storz, G.; Imlay, J.A. Oxidative stress. Curr. Opin. Microbiol. 1999, 2, 188–194. [Google Scholar] [CrossRef]


 ) and LB (
) and LB (  ), compared to the controls of the nontreated bacteria in PBS (
), compared to the controls of the nontreated bacteria in PBS (  ) and LB (
) and LB (  ). Results are the mean of SD of three independent experiments. P-value significance is presented by t-test of CFU/ml of the plasma-treated A. tumefaciens in LB, compared to the treated sample in PBS on the same selected time ## p < 0.01. p-value of the CFU/mL count in each examined time related to its control * p < 0.05, ** p < 0.01, and *** p < 0.001.
). Results are the mean of SD of three independent experiments. P-value significance is presented by t-test of CFU/ml of the plasma-treated A. tumefaciens in LB, compared to the treated sample in PBS on the same selected time ## p < 0.01. p-value of the CFU/mL count in each examined time related to its control * p < 0.05, ** p < 0.01, and *** p < 0.001.
 ) and LB (
) and LB (  ), compared to the controls of the nontreated bacteria in PBS (
), compared to the controls of the nontreated bacteria in PBS (  ) and LB (
) and LB (  ). Results are the mean of SD of three independent experiments. P-value significance is presented by t-test of CFU/ml of the plasma-treated A. tumefaciens in LB, compared to the treated sample in PBS on the same selected time ## p < 0.01. p-value of the CFU/mL count in each examined time related to its control * p < 0.05, ** p < 0.01, and *** p < 0.001.
). Results are the mean of SD of three independent experiments. P-value significance is presented by t-test of CFU/ml of the plasma-treated A. tumefaciens in LB, compared to the treated sample in PBS on the same selected time ## p < 0.01. p-value of the CFU/mL count in each examined time related to its control * p < 0.05, ** p < 0.01, and *** p < 0.001.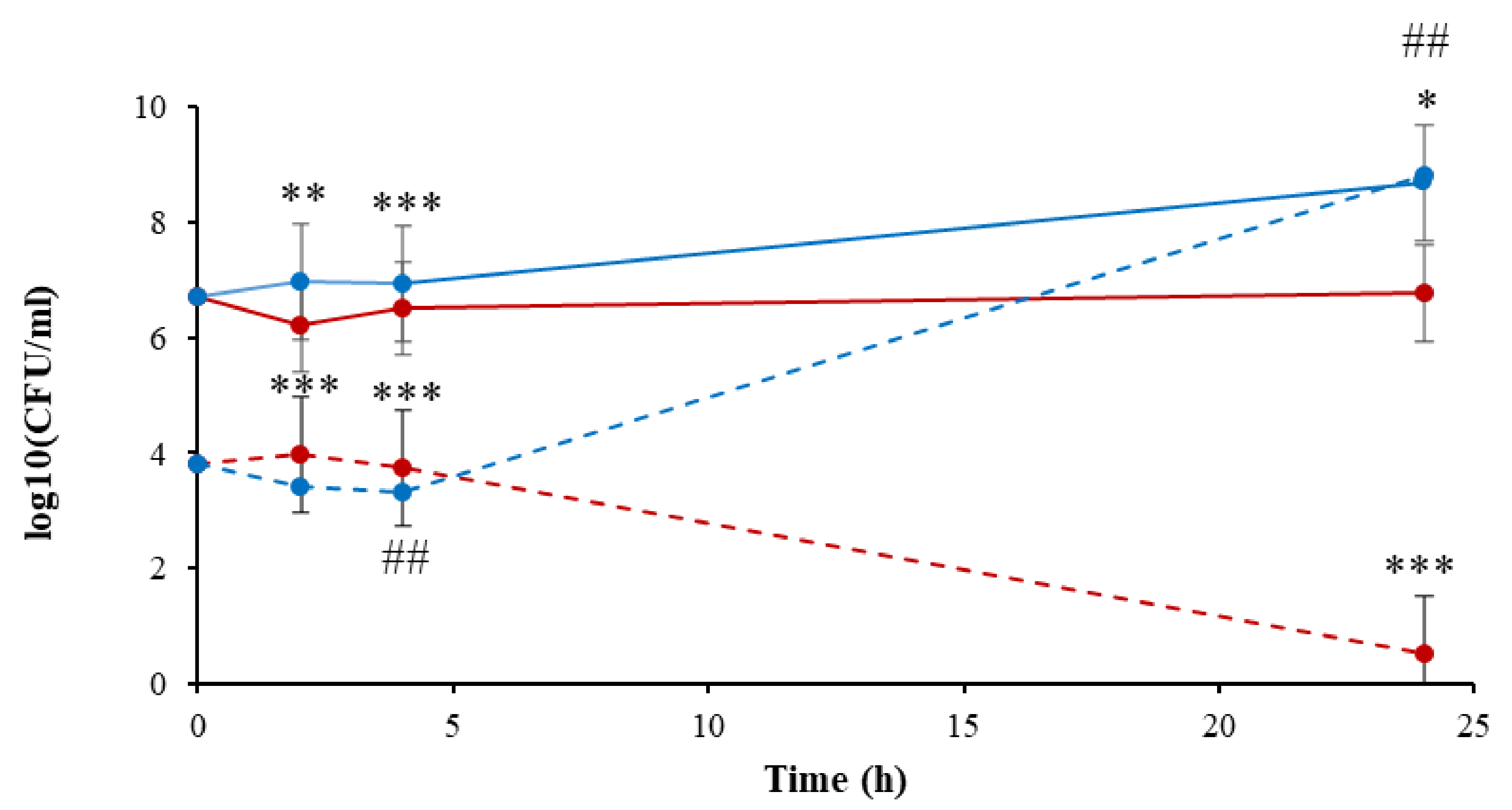
 ) (0–24 h); plasma-treated A. tumefaciens in PBS (0–24 h) (
) (0–24 h); plasma-treated A. tumefaciens in PBS (0–24 h) (  ); nontreated A. tumefaciens in LB (2–24 h) (
); nontreated A. tumefaciens in LB (2–24 h) (  ); and plasma-treated A. tumefaciens in LB (2–24 h) (
); and plasma-treated A. tumefaciens in LB (2–24 h) (  ). Results are mean SD of four independent experiments; p-value significance is presented by t-test of treatment compared to control: * p < 0.05, ** p < 0.001, *** p < 0.0001.
). Results are mean SD of four independent experiments; p-value significance is presented by t-test of treatment compared to control: * p < 0.05, ** p < 0.001, *** p < 0.0001.
 ) (0–24 h); plasma-treated A. tumefaciens in PBS (0–24 h) (
) (0–24 h); plasma-treated A. tumefaciens in PBS (0–24 h) (  ); nontreated A. tumefaciens in LB (2–24 h) (
); nontreated A. tumefaciens in LB (2–24 h) (  ); and plasma-treated A. tumefaciens in LB (2–24 h) (
); and plasma-treated A. tumefaciens in LB (2–24 h) (  ). Results are mean SD of four independent experiments; p-value significance is presented by t-test of treatment compared to control: * p < 0.05, ** p < 0.001, *** p < 0.0001.
). Results are mean SD of four independent experiments; p-value significance is presented by t-test of treatment compared to control: * p < 0.05, ** p < 0.001, *** p < 0.0001.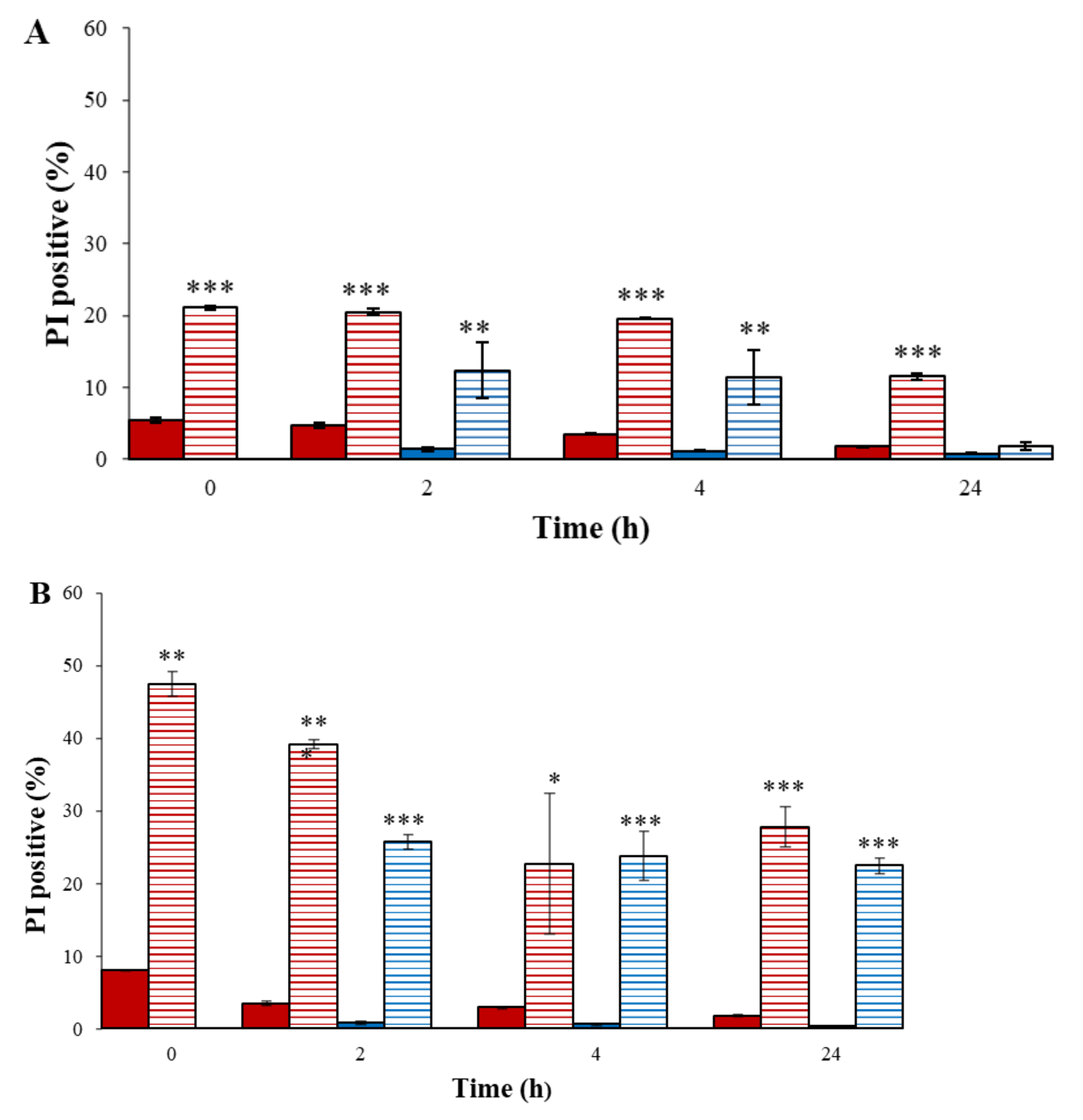

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazra, Y.; Gandu, B.; Amar, I.D.; Emanuel, E.; Cahan, R. Effects of Atmospheric Plasma Corona Discharge on Agrobacterium tumefaciens Survival. Microorganisms 2022, 10, 32. https://doi.org/10.3390/microorganisms10010032
Lazra Y, Gandu B, Amar ID, Emanuel E, Cahan R. Effects of Atmospheric Plasma Corona Discharge on Agrobacterium tumefaciens Survival. Microorganisms. 2022; 10(1):32. https://doi.org/10.3390/microorganisms10010032
Chicago/Turabian StyleLazra, Yulia, Bharath Gandu, Irina Dubrovin Amar, Efrat Emanuel, and Rivka Cahan. 2022. "Effects of Atmospheric Plasma Corona Discharge on Agrobacterium tumefaciens Survival" Microorganisms 10, no. 1: 32. https://doi.org/10.3390/microorganisms10010032
APA StyleLazra, Y., Gandu, B., Amar, I. D., Emanuel, E., & Cahan, R. (2022). Effects of Atmospheric Plasma Corona Discharge on Agrobacterium tumefaciens Survival. Microorganisms, 10(1), 32. https://doi.org/10.3390/microorganisms10010032





