Isolation and Characterization of Effective Bacteria That Reduce Ammonia Emission from Livestock Manure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation, Maintenance, and Identification of Microbial Cultures
2.2. Changes in the pH during Bacterial Culturing
2.3. Ammonia Removal from the Liquid Medium
2.4. Urease Inhibition Activity of the Isolated Strains
2.5. Manure Samples and the Growth Conditions of Microbes
2.6. The Rate of Ammonia Removal in Minimal Salt Medium
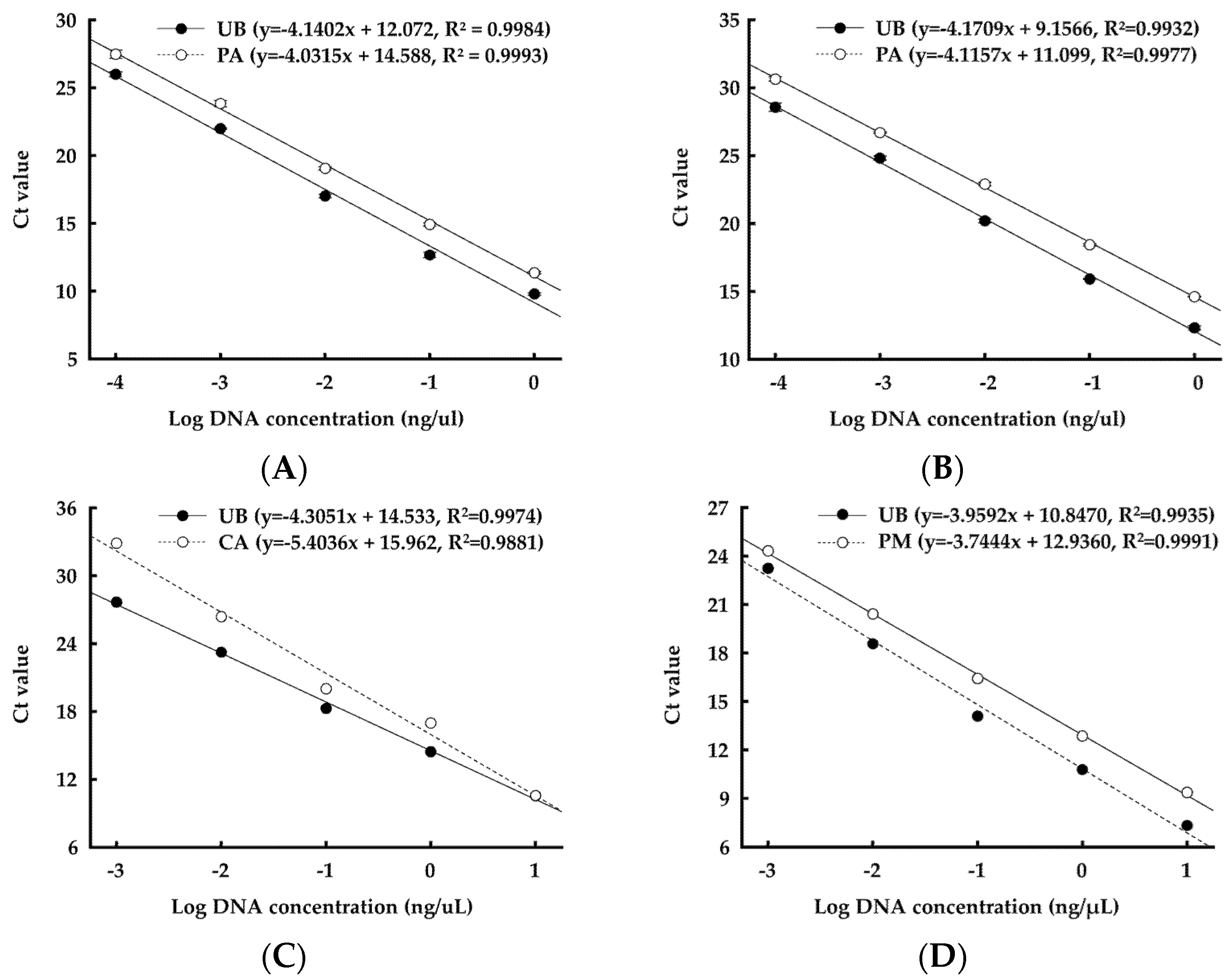
2.7. DNA Extraction and Real-Time Quantitative Polymerase Chain Reaction (qPCR) Analysis
2.8. Abundance of the Selected Strains in Manure under Different Culture Conditions
2.9. Determination of pH Change and Growth of HAB in Manure
2.10. Monitoring Ammonia Gas Emission and the Chemical Properties of Manure
2.11. Statistical Analysis
3. Results and Discussion
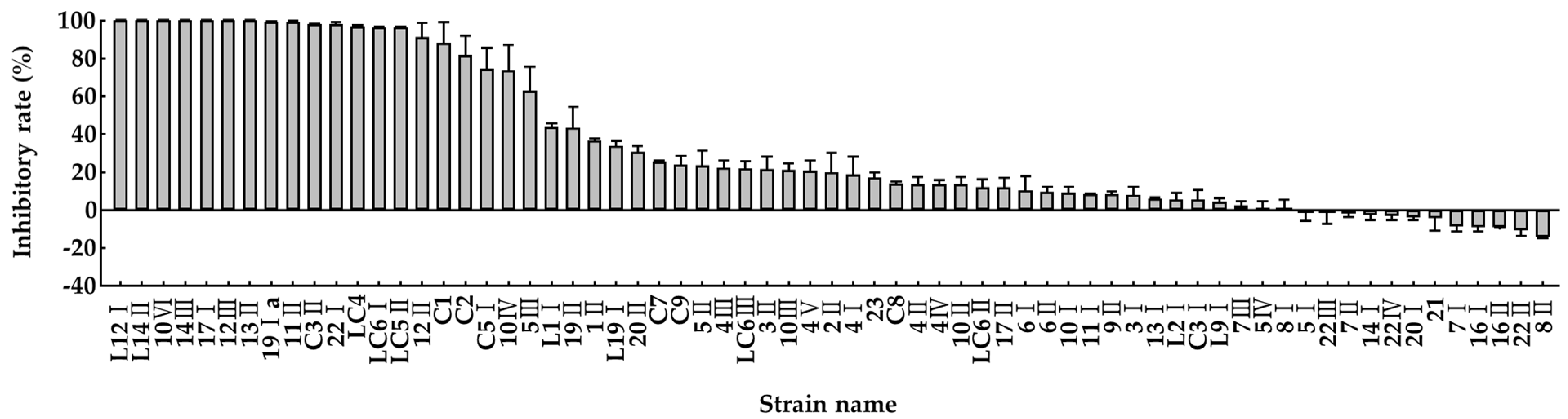
3.1. Screening and Identification of Potential Effective Bacterial Strains for Reducing Ammonia Emission
3.2. Identification of Selected Strains by Using API 50 CHL Test
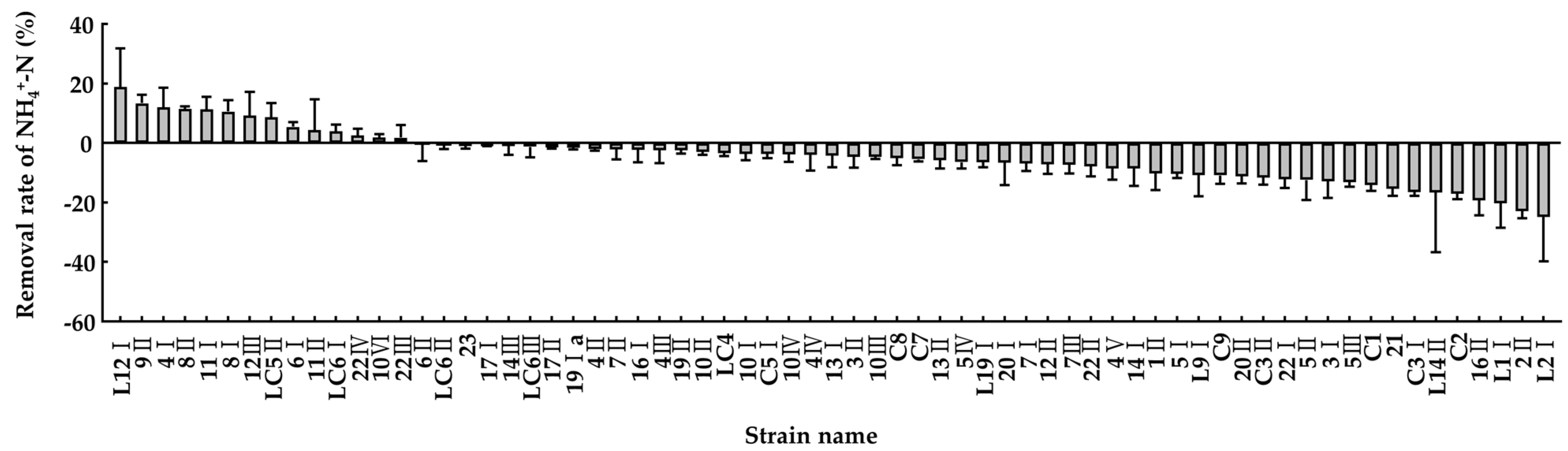
3.3. Ammonia Removal Capacity of the Isolated Bacteria in the Minimal Salt Medium
3.4. DNA Abundance of Selected Strains in Manure
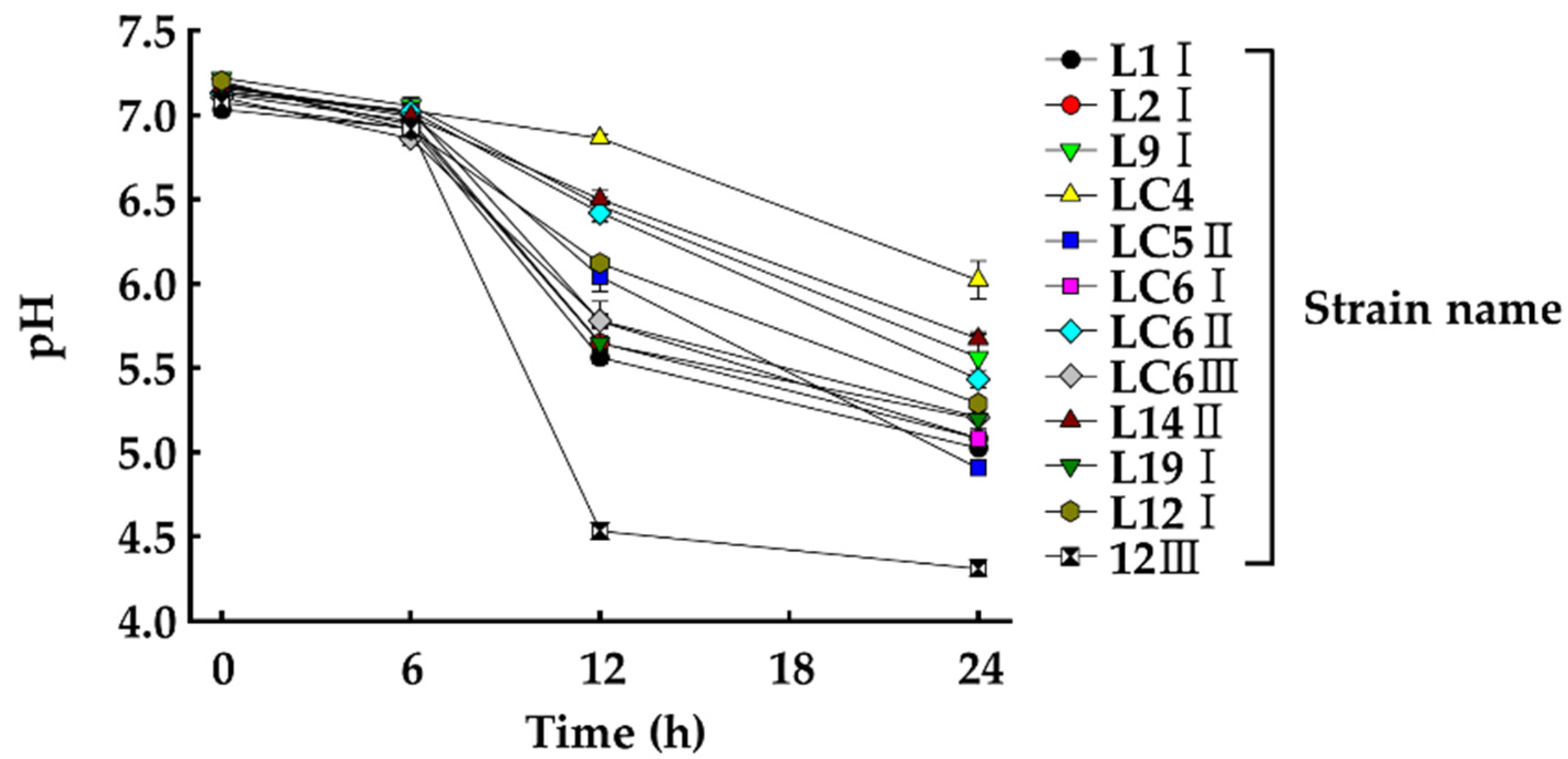
3.5. Changes in pH and Growth Inhibition of HAB in Manure by the Selected Strains
| Time (h) | 0 | 24 | ||||
|---|---|---|---|---|---|---|
| Conditions | Group | Average | SE | Average | SE | |
| Aerobic | Control | 7.22 | 0.03 | 7.34 * | 0.04 | |
| L12I | (L) | 7.28 | 0.02 | 7.25 | 0.02 | |
| (H) | 7.26 | 0.06 | 7.31 | 0.01 | ||
| 12III | (L) | 7.25 | 0.03 | 7.27 | 0.03 | |
| (H) | 7.23 | 0.04 | 7.21 | 0.05 | ||
| Anaerobic | Control | 7.24 | 0.09 | 6.94 a,* | 0.03 | |
| L12I | (L) | 7.28 | 0.05 | 6.56 b,* | 0.01 | |
| (H) | 7.22 | 0.03 | 6.53 b,* | 0.02 | ||
| 12III | (L) | 7.20 | 0.01 | 6.55 b,* | 0.01 | |
| (H) | 7.22 | 0.03 | 6.52 b,* | 0.01 | ||
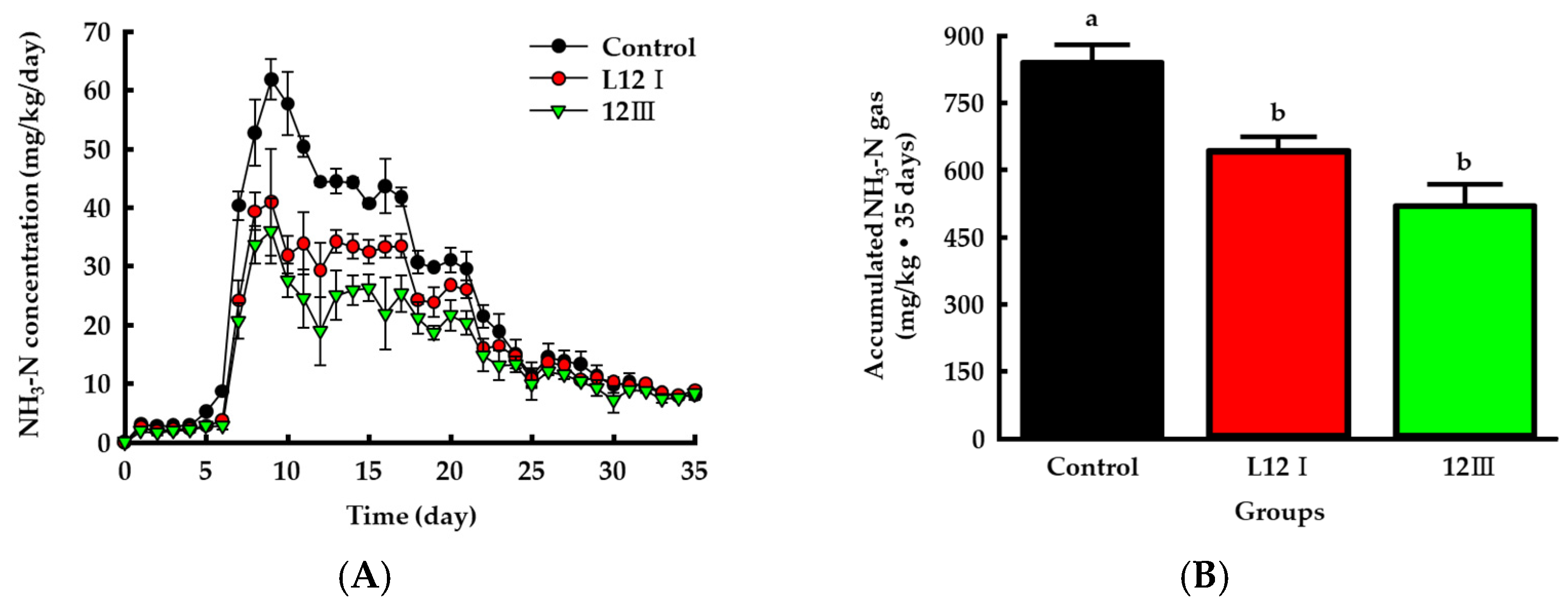
3.6. Ammonia Emission from Manure and Changes in the Chemical Properties of the Manure
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Institute of Environmental Research (NIER). Statistics of Air Pollution in OECD Countries; National Institute for Environmental Research (NIER): Incheon, Korea, 2016.
- Gong, J.; Zhu, T.; Kipen, H.; Wang, G.; Hu, M.; Guo, Q.; Ohman-Strickland, P.; Lu, S.-E.; Wang, Y.; Zhu, P.; et al. Comparisons of Ultrafine and Fine Particles in Their Associations with Biomarkers Reflecting Physiological Pathways. Environ. Sci. Technol. 2014, 48, 5264–5273. [Google Scholar] [CrossRef]
- Shin, D.; Joo, H.; Seo, E.; Kim, C. Management Strategies to Reduce PM2.5 Emission: Emphasis-Ammonia; Korea Environment Institute: Sejong, Korea, 2017. [Google Scholar]
- Bai, Z.; Dong, Y.; Wang, Z.; Zhu, T. Emission of ammonia from indoor concrete wall and assessment of human exposure. Environ. Int. 2006, 32, 303–311. [Google Scholar] [CrossRef]
- National Institute for Environmental Research (NIER). National Air Pollutants Emission Service, Air Pollutant Emission Statistics; National Institute for Environmental Research (NIER): Incheon, Korea, 2017.
- European Environment Agency (EEA). EMEP/EEA Air Pollutant Emission Inventory Guidebook; European Environment Agency (EEA): Copenhagen, Denmark, 2016.
- McCrory, D.F.; Hobbs, P.J. Additives to Reduce Ammonia and Odor Emissions from Livestock Wastes: A Review. J. Environ. Qual. 2001, 30, 345–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigurdarson, J.J.; Svane, S.; Karring, H. The molecular processes of urea hydrolysis in relation to ammonia emissions from agriculture. Rev. Environ. Sci. Bio/Technol. 2018, 17, 241–258. [Google Scholar] [CrossRef] [Green Version]
- Vince, A.; Dawson, A.M.; Park, N.; O’Grady, F. Ammonia production by intestinal bacteria. Gut 1973, 14, 171–177. [Google Scholar] [CrossRef] [Green Version]
- Whitehead, T.R.; Cotta, M.A. Isolation and Identification of Hyper-Ammonia Producing Bacteria from Swine Manure Storage Pits. Curr. Microbiol. 2004, 48, 20–26. [Google Scholar] [CrossRef]
- Zhou, S.; Zhang, X.; Liao, X.; Wu, Y.; Mi, J.; Wang, Y. Effect of Different Proportions of Three Microbial Agents on Ammonia Mitigation during the Composting of Layer Manure. Molecules 2019, 24, 2513. [Google Scholar] [CrossRef] [Green Version]
- Susan, W.G.; Knowlton, K. Ammonia emissions and animal agriculture. Va. Coop. Ext. Biol. Syst. Eng. 2005, 442, 1–5. [Google Scholar]
- Pinder, R.W.; Adams, P.J.; Pandis, S.N. Ammonia Emission Controls as a Cost-Effective Strategy for Reducing Atmospheric Particulate Matter in the Eastern United States. Environ. Sci. Technol. 2007, 41, 380–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.-H.; Ha, D.-M.; Lee, J.-Y.; Shin, H.-S.; Song, J.-I.; Kim, D.-H. Effects of Physico-Chemical Treatment on Odor Reduction of Swine Farm. Ann. Anim. Resour. Sci. 2017, 28, 64–71. [Google Scholar] [CrossRef]
- Li, H.; Xin, H.; Liang, Y.; Burns, R.T. Reduction of Ammonia Emissions from Stored Laying Hen Manure Through Topical Application of Zeolite, Al+ Clear, Ferix-3, or Poultry Litter Treatment. J. Appl. Poult. Res. 2008, 17, 421–431. [Google Scholar] [CrossRef]
- Suryawan, I.W.K.; Prajati, G.; Afifah, A.S.; Apritama, M.R.; Adicita, Y. Continuous Piggery Wastewater Treatment With Anaerobic Baffled Reactor (Abr) By Bio-Activator Effective Microorganisms (Em4). Indones. J. Urban Environ. Technol. 2019, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Bastami, M.S.B.; Jones, D.L.; Chadwick, D.R. Reduction of Methane Emission during Slurry Storage by the Addition of Effective Microorganisms and Excessive Carbon Source from Brewing Sugar. J. Environ. Qual. 2016, 45, 2016–2022. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-P.; Wang, S.-M.; Zhang, D.-W.; Zhou, L.-X. Isolation and nitrogen removal characteristics of an aerobic heterotrophic nitrifying-denitrifying bacterium, Bacillus subtilis A1. Bioresour. Technol. 2011, 102, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhu, C.; Sun, D.; Guo, P.; Tian, Y. Removal of ammonium-N from ammonium-rich sewage using an immobilized Bacillus subtilis AYC bioreactor system. J. Environ. Sci. 2011, 23, 1279–1285. [Google Scholar] [CrossRef]
- Sasaki, H.; Yano, H.; Sasaki, T.; Nakai, Y. A survey of ammonia-assimilating micro-organisms in cattle manure composting. J. Appl. Microbiol. 2005, 99, 1356–1363. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, J.; Li, Q.X.; Wang, Y.; Li, S.; Ren, T.; Wang, L. Simultaneous heterotrophic nitrification and aerobic denitrification by bacterium Rhodococcus sp. CPZ24. Bioresour. Technol. 2012, 116, 266–270. [Google Scholar] [CrossRef]
- Pérez, R.A.; González, E.A.; Guerra, A.T.; Guerra, N.P. A Review on Some Important Factors Affecting Bacteriocin Production by Lactococci, Lactobacilli and Pediococci. Curr. Biochem. Eng. 2014, 1, 9–24. [Google Scholar] [CrossRef]
- Anastasiadou, S.; Papagianni, M.; Ambrosiadis, I.; Koidis, P. Rapid quantifiable assessment of nutritional parameters influencing pediocin production by Pediococcus acidilactici NRRL B5627. Bioresour. Technol. 2008, 99, 6646–6650. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Jeong, S.-J.; Kim, J.H. Characterization of a glutamate decarboxylase (GAD) gene from Lactobacillus zymae. Biotechnol. Lett. 2014, 36, 1791–1799. [Google Scholar] [CrossRef]
- Satokari, R.; Mattila-Sandholm, T.; Suihko, M.-L. Identification of pediococci by ribotyping. J. Appl. Microbiol. 2000, 88, 260–265. [Google Scholar] [CrossRef] [Green Version]
- Abubakr, M.A.S.; Al-Adiwish, W.M. Isolation and Identification of Lactic Acid Bacteria from Different Fruits with Proteolytic Activity. Int. J. Microbiol. Biotechnol. 2017, 2, 58. [Google Scholar] [CrossRef]
- National Institute for Environmental Research (NIER). Standard Methods for the Examination of Water Pollution ES 04355.1c, Ammonium Nitrogen; National Institute for Environmental Research (NIER): Incheon, Korea, 2017.
- Kobashi, K.; Kumaki, K.; Hase, J.I. Effect of acyl residues of hydroxamic acids on urease inhibition. Biochim. Biophys. Acta 1971, 227, 429–441. [Google Scholar] [CrossRef]
- Lee, J.; Um, H.; Hahm, K.; Park, C.; Kang, D. Natural Urease Inhibitors. J. Cancer Prev. 2006, 11, 129–136. [Google Scholar]
- Uesato, S.; Hashimoto, Y.; Nishino, M.; Nagaoka, Y.; Kuwajima, H. N-substituted hydroxyureas as urease inhibitors. Chem. Pharm. Bull. 2002, 50, 1280–1282. [Google Scholar] [CrossRef] [Green Version]
- Kabeerdoss, J.; Ferdous, S.; Balamurugan, R.; Mechenro, J.; Vidya, R.; Santhanam, S.; Jana, A.K.; Ramakrishna, B.S. Development of the gut microbiota in southern Indian infants from birth to 6 months: A molecular analysis. J. Nutr. Sci. 2013, 2, e18. [Google Scholar] [CrossRef] [Green Version]
- Mora, D.; Fortina, M.G.; Parini, C.; Manachini, P.L. Identification of Pediococcus acidilactici and Pediococcus pentosaceus based on 16S rRNA and ldhD gene-targeted multiplex PCR analysis. FEMS Microbiol. Lett. 1997, 151, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Liu, Z.; Yu, Z.; Zhu, W. Monensin and Nisin Affect Rumen Fermentation and Microbiota Differently In Vitro. Front. Microbiol. 2017, 8, 1111. [Google Scholar] [CrossRef]
- Zhang, W.; Niu, Z.; Yin, K.; Liu, P.; Chen, L. Quick identification and quantification of Proteus mirabilis by polymerase chain reaction (PCR) assays. Ann. Microbiol. 2013, 63, 683–689. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Ohta, M.; Richardson, P.M.; Mills, D.A. Monitoring Seasonal Changes in Winery-Resident Microbiota. PLoS ONE 2013, 8, e66437. [Google Scholar] [CrossRef]
- Park, J.-H.; Lee, S.-L.; Hwang, S.-W.; Eom, J.-H.; Kim, S.-H.; Kang, S.-W.; Cho, J.-S.; Seo, D.-C. Characteristics of ammonia gas emissions from soybean cultivation soils treated with mixed microorganisms. Appl. Biol. Chem. 2020, 63, 20. [Google Scholar] [CrossRef]
- National Academy of Agricultural Science (NAAS). Method of Soil Chemical Analysis; National Academy of Agricultural Science (NAAS): Wanju-gun, Korea, 2010.
- Rural Development Administration (RDA). Fertilizer Quality Method and Sample Collection Guideline, Physicochemical Method of Fertilizer; Rural Development Administration (RDA): Jeonju-si, Korea, 2019. [Google Scholar]
- Rahmatullah, M.; Boyde, T.R. Improvements in the determination of urea using diacetyl monoxime; methods with and without deproteinisation. Clin. Chim. Acta 1980, 107, 3–9. [Google Scholar] [CrossRef]
- Mathys, S.; von Ah, U.; Lacroix, C.; Staub, E.; Mini, R.; Cereghetti, T.; Meile, L. Detection of the pediocin gene pedA in strains from human faeces by real-time PCR and characterization of Pediococcus acidilactici UVA1. BMC Biotechnol. 2007, 7, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimaya, C.; Hashimoto, T. Isolation and characterization of novel thermophilic nitrifying Bacillus sp. from compost. Soil Sci. Plant Nutr. 2011, 57, 150–156. [Google Scholar] [CrossRef] [Green Version]
- Toe, C.J.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdul Rahim, R.; Idrus, Z. Extracellular Proteolytic Activity and Amino Acid Production by Lactic Acid Bacteria Isolated from Malaysian Foods. Int. J. Mol. Sci. 2019, 20, 1777. [Google Scholar] [CrossRef] [Green Version]
- Lim, Y.H.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdul Rahim, R. Rapid Evaluation and Optimization of Medium Components Governing Tryptophan Production by Pediococcus acidilactici TP-6 Isolated from Malaysian Food via Statistical Approaches. Molecules 2020, 25, 779. [Google Scholar] [CrossRef] [Green Version]
- Mehta, C.M.; Sirari, K. Comparative study of aerobic and anaerobic composting for better understanding of organic waste management: A mini review. Plant Arch. 2018, 18, 44–48. [Google Scholar]
- Othman, M.; Ariff, A.B.; Wasoh, H.; Kapri, M.R.; Halim, M. Strategies for improving production performance of probiotic Pediococcus acidilactici viable cell by overcoming lactic acid inhibition. AMB Express 2017, 7, 215. [Google Scholar] [CrossRef] [Green Version]
- Bansal, P.; Kumar, R.; Singh, J.; Dhanda, S. Next generation sequencing, biochemical characterization, metabolic pathway analysis of novel probiotic Pediococcus acidilactici NCDC 252 and it’s evolutionary relationship with other lactic acid bacteria. Mol. Biol. Rep. 2019, 46, 5883–5895. [Google Scholar] [CrossRef]
- Arogo, J.; Westerman, P.W.; Heber, A.J.; Robarge, W.P.; Classen, J.J. Ammonia Emissions from Animal Feeding Operations; ASABE: St. Joseph, MI, USA, 2006; ISBN 1892769514.
- van Kempen, T.A. Dietary adipic acid reduces ammonia emission from swine excreta. J. Anim. Sci. 2001, 79, 2412–2417. [Google Scholar] [CrossRef]
- Kavanagh, I.; Burchill, W.; Healy, M.G.; Fenton, O.; Krol, D.J.; Lanigan, G.J. Mitigation of ammonia and greenhouse gas emissions from stored cattle slurry using acidifiers and chemical amendments. J. Clean. Prod. 2019, 237, 117822. [Google Scholar] [CrossRef]
- Porto, M.C.W.; Kuniyoshi, T.M.; Azevedo, P.O.S.; Vitolo, M.; Oliveira, R.P.S. Pediococcus spp.: An important genus of lactic acid bacteria and pediocin producers. Biotechnol. Adv. 2017, 35, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Mugita, Y.; Nakagami, G.; Minematsu, T.; Kitamura, A.; Sanada, H. Combination of urease inhibitor and antiseptic inhibits urea decomposition-induced ammonia production by Proteus mirabilis. Int. Wound J. 2020, 17, 1558–1565. [Google Scholar] [CrossRef] [PubMed]
- Asing, J.; Saggar, S.; Singh, J.; Bolan, N. Assessment of nitrogen losses from urea and an organic manure with and without nitrification inhibitor, dicyandiamide, applied to lettuce under glasshouse conditions. Aust. J. Soil Res. 2008, 46, 535–541. [Google Scholar] [CrossRef]
- Zhang, Q.; Widmer, G.; Tzipori, S. A pig model of the human gastrointestinal tract. Gut Microbes 2013, 4, 193–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, D.; Adams, S.; Wei, C.; Gui-Xin, Q.; Atiba, E.M.; Hailong, J. Effects of Astragalus membranaceus fiber on growth performance, nutrient digestibility, microbial composition, VFA production, gut pH, and immunity of weaned pigs. Microbiologyopen 2019, 8, e00712. [Google Scholar] [CrossRef] [Green Version]



| Target Strain | Primer Name | Sequence (5′–3′) | Product Size (bp) | Reference |
|---|---|---|---|---|
| Total bacteria | UB-F | CGGCAACGAGCGCAACCC | 161 | [31] |
| UB-R | CCATTGTAGCACGTGTGTAGCC | |||
| Pediococcus acidilactici | IdhDF | GGACTTGATAACGTACCCGC | 449 | [32] |
| IdhDR | GTTCCGTCTTGCATTTGACC | |||
| Clostridium aminophilum | 57F | ACGGAAATTACAGAAGGAAG | 560 | [33] |
| 616R | GTTTCCAAAGCAATTCCAC | |||
| Proteus mirabilis | ureF | GGTGAGATTTGTATTAATGG | 225 | [34] |
| ureR | ATAATCTGGAAGATGACGAG |
| Carbohydrates | L12 I | 12 III | Carbohydrates | L12 I | 12 III |
|---|---|---|---|---|---|
| Control | − | − | Esculin | − | + |
| Glycerol | − | − | Salicin | + | + |
| Erythritol | − | − | D-Cellobiose | + | + |
| D-Arabinose | − | − | D-Maltose | − | − |
| L-Arabinose | + | + | Lactose | − | − |
| D-Ribose | + | + | D-Melibiose | − | − |
| D-Xylose | + | + | Sucrose | − | − |
| L-Xylose | − | − | Trehalose | + | + |
| D-Adonitol | − | − | Inulin | − | − |
| Methyl-β D-xylopyranoside | − | − | D-Melezitose | − | − |
| D-Galactose | + | + | Raffinose | − | − |
| D-Glucose | + | + | Starch | − | − |
| D-Fructose | + | + | Glycogen | − | − |
| D-Mannose | + | + | Xylitol | − | − |
| L-Sorbose | − | − | Gentiobiose | − | + |
| L-Rhamnose | + | + | D-Turanose | − | − |
| Dulcitol | − | − | D-Lyxose | − | − |
| Inositol | − | − | D-Tagatose | + | + |
| Mannitol | − | − | D-Fucose | − | − |
| Sorbitol | − | − | L-Fucose | − | − |
| Methyl-α D-Mannopyranoside | − | − | D-Arabitol | − | − |
| Methyl-α D-Glucopyranoside | − | − | L-Arabitol | − | − |
| N-Acetylglucosamine | + | + | Potassium gluconate | − | − |
| Amygdalin | − | − | Potassium 2-ketogluconate | − | − |
| Arbutin | + | − | Potassium 5-ketogluconate | − | − |
| Group | Control | L12 I | 12 III | |||
|---|---|---|---|---|---|---|
| Time (Day) | 0 | 35 | 0 | 35 | 0 | 35 |
| OM (%) | 41.66 ± 0.86 | 43.03 ± 1.06 | 42.69 ± 0.99 | 40.61 ± 0.54 | 42.21 ± 0.55 | 40.38 ± 1.16 |
| T-N (%) | 1.94 ± 0.05 | 1.51 ± 0.02 b | 2.01 ± 0.13 | 1.71 ± 0.16 b | 2.24 ± 0.10 | 2.01 ± 0.09 a |
| OM/T-N ratio | 21.51 ± 0.44 a | 28.47 ± 0.70 a,* | 21.26 ± 0.49 a | 23.70 ± 0.31 b,* | 18.88 ± 0.25 b | 20.12 ± 0.58 c |
| Urea (mg/100g) | 14.78 ± 1.04 | 5.59 ± 0.23 b,* | 15.29 ± 0.50 | 7.71 ± 0.12 a,* | 13.42 ± 0.24 | 7.38 ± 0.41 a,* |
| pH | 7.49 ± 0.02 | 8.46 ± 0.04 * | 7.46 ± 0.02 | 8.49 ± 0.02 * | 7.48 ± 0.02 | 8.48 ± 0.01 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-I.; Heo, W.; Lee, S.-J.; Kim, Y.-J. Isolation and Characterization of Effective Bacteria That Reduce Ammonia Emission from Livestock Manure. Microorganisms 2022, 10, 77. https://doi.org/10.3390/microorganisms10010077
Kim S-I, Heo W, Lee S-J, Kim Y-J. Isolation and Characterization of Effective Bacteria That Reduce Ammonia Emission from Livestock Manure. Microorganisms. 2022; 10(1):77. https://doi.org/10.3390/microorganisms10010077
Chicago/Turabian StyleKim, Sun-Il, Wan Heo, So-Jung Lee, and Young-Jun Kim. 2022. "Isolation and Characterization of Effective Bacteria That Reduce Ammonia Emission from Livestock Manure" Microorganisms 10, no. 1: 77. https://doi.org/10.3390/microorganisms10010077
APA StyleKim, S. -I., Heo, W., Lee, S. -J., & Kim, Y. -J. (2022). Isolation and Characterization of Effective Bacteria That Reduce Ammonia Emission from Livestock Manure. Microorganisms, 10(1), 77. https://doi.org/10.3390/microorganisms10010077






