Elioraea tepida, sp. nov., a Moderately Thermophilic Aerobic Anoxygenic Phototrophic Bacterium Isolated from the Mat Community of an Alkaline Siliceous Hot Spring in Yellowstone National Park, WY, USA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain Isolation and Cultivation Conditions
2.2. Microscopy and Analytical Procedures
2.3. Genome Sequencing and Bioinformatic Analyses
3. Results and Discussion
3.1. Isolation and Initial Cultivation
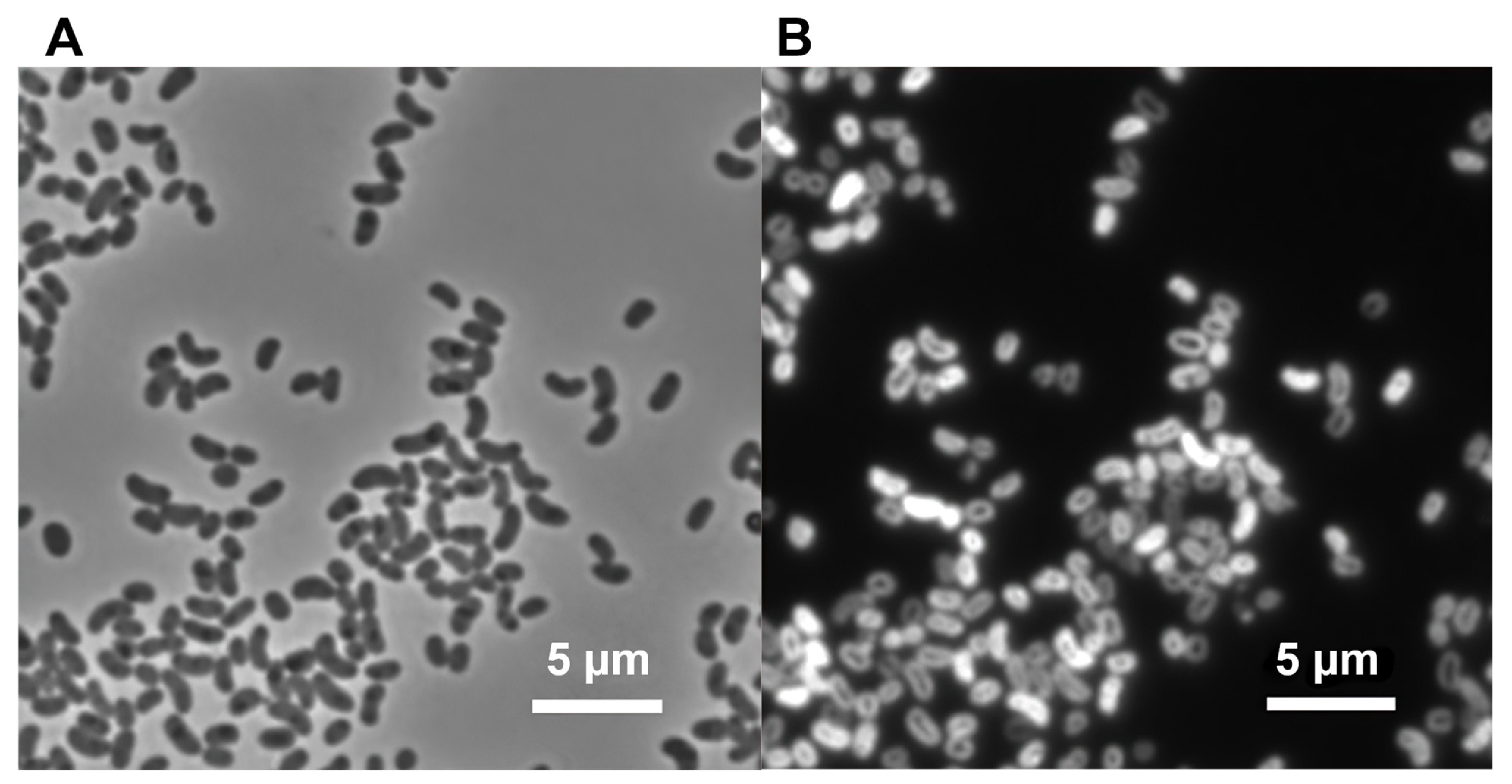
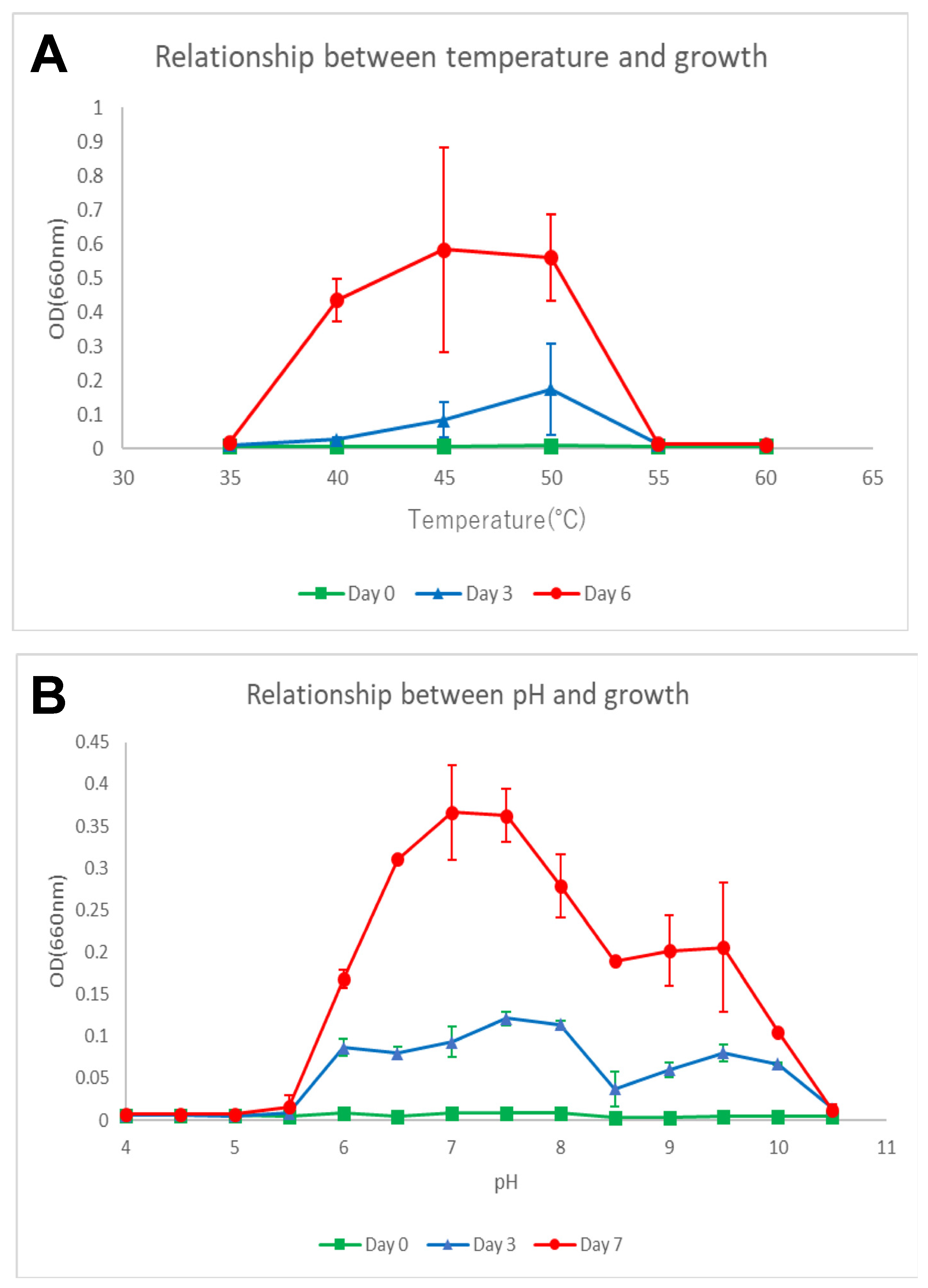
3.2. Phenotypic, Biochemical, and Chemotaxonomic Characterization
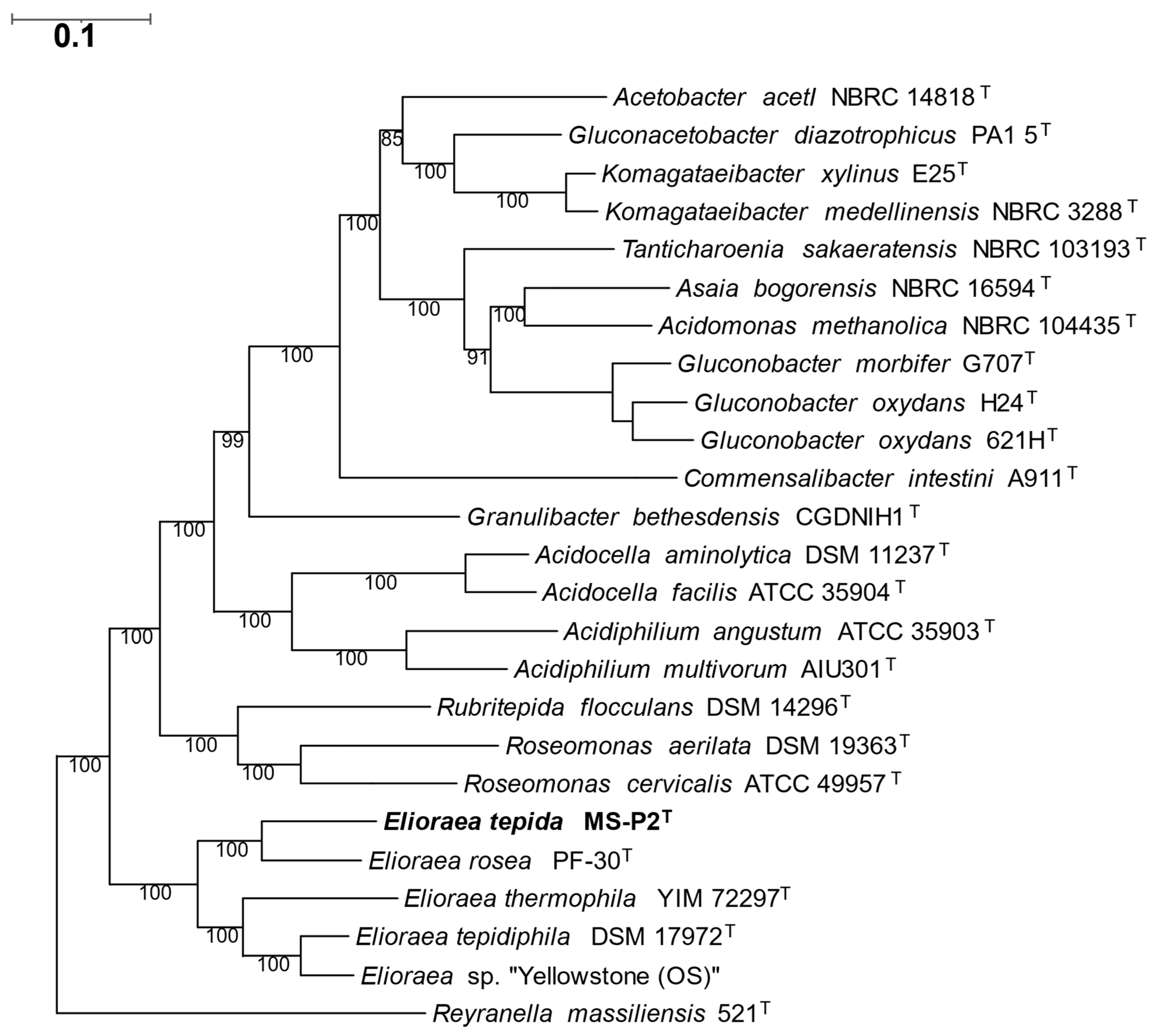
3.3. Genomic Features
3.4. Distribution and Ecological Considerations
3.5. Description of Elioraea tepida sp. nov.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yurkov, V.; Beatty, J.T. Aerobic anoxygenic phototrophic bacteria. Microbiol. Mol. Biol. Rev. 1998, 62, 695–724. [Google Scholar] [CrossRef] [Green Version]
- Yurkov, V.; Hughes, E. Aerobic anoxygenic phototrophs: Four decades of mystery. In Modern Topics in the Phototrophic Prokaryotes: Environmental and Applied Aspects; Hallenbeck, P., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 193–214. [Google Scholar] [CrossRef]
- Koblížek, M. Ecology of aerobic anoxygenic phototrophs in aquatic environments. FEMS Microbiol. Rev. 2015, 39, 854–870. [Google Scholar] [CrossRef] [Green Version]
- Hanada, S.; Kawase, Y.; Hiraishi, A.; Takaichi, S.; Matsuura, K.; Shimada, K.; Nagashima, K.V. Porphyrobacter tepidarius sp. nov., a moderately thermophilic aerobic photosynthetic bacterium isolated from a hot spring. Int. J. Syst. Bacteriol. 1997, 47, 408–413. [Google Scholar] [CrossRef] [Green Version]
- Rainey, F.A.; Silva, J.; Nobre, M.F.; Silva, M.T.; da Costa, M.S. Porphyrobacter cryptus sp. nov., a novel slightly thermophilic, aerobic, bacteriochlorophyll a-containing species. Int. J. Syst. Evol. Microbiol. 2003, 53, 35–41. [Google Scholar] [CrossRef] [Green Version]
- Alarico, S.; Rainey, F.; Empadinhas, N.; Schumann, P.; Nobre, M.; da Costa, M.S. Rubritepida flocculans gen. nov., sp. nov., a new slightly thermophilic member of the alpha-1 subclass of the Proteobacteria. Syst. Appl. Microbiol. 2002, 25, 198–206. [Google Scholar] [CrossRef]
- Thiel, V.; Tank, M.; Bryant, D.A. Diversity of chlorophototrophic bacteria revealed in the omics era. Annu. Rev. Plant Biol. 2018, 69, 21–49. [Google Scholar] [CrossRef]
- Yurkov, V.; Stackebrandt, E.; Holmes, A.; Fuerst, J.A.; Hugenholtz, P.; Golecki, J.; Gad’on, N.; Gorlenko, V.M.; Kompantseva, E.I.; Drews, G. Phylogenetic positions of novel aerobic, bacteriochlorophyll-a-containing bacteria and description of Roseococcus thiosulfatophilus gen. nov., sp. nov., Erythromicrobium ramosum gen. nov., sp. nov., and Erythrobacter litoralis sp. nov. Int. J. Syst. Evol. Microbiol. 1994, 44, 427–434. [Google Scholar] [CrossRef]
- Margesin, R.; Zhang, D.C. Humitalea rosea gen. nov., sp. nov., an aerobic bacteriochlorophyll-containing bacterium of the family Acetobacteraceae isolated from soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 1411–1416. [Google Scholar] [CrossRef] [Green Version]
- Saitoh, S.; Suzuki, T.; Nishimura, Y. Proposal of Craurococcus roseus gen. nov., sp. nov. and Paracraurococcus ruber gen. nov., sp. nov., novel aerobic bacteriochlorophyll a-containing bacteria from soil. Int. J. Syst. Bacteriol. 1998, 48, 1043–1047. [Google Scholar] [CrossRef] [Green Version]
- Venkata Ramana, V.; Sasikala, C.; Takaichi, S.; Ramana, C.V. Roseomonas aestuarii sp. nov., a bacteriochlorophyll-a containing alphaproteobacterium isolated from an estuarine habitat of India. Syst. Appl. Microbiol. 2010, 33, 198–203. [Google Scholar] [CrossRef]
- Wakao, N.; Yokoi, N.; Isoyama, N.; Hiraishi, A.; Shimada, K.; Kobayashi, M.; Kise, H.; Iwaki, M.; Itoh, S.; Takaichi, S. Discovery of natural photosynthesis using Zn-containing bacteriochlorophyll in an aerobic bacterium Acidiphilium rubrum. Plant Cell Physiol. 2012, 37, 889–893. [Google Scholar] [CrossRef] [Green Version]
- Albuquerque, L.; Rainey, F.A.; Nobre, M.F.; da Costa, M.S. Elioraea tepidiphila gen. nov., sp. nov., a slightly thermophilic member of the Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 2008, 58, 773–778. [Google Scholar] [CrossRef] [Green Version]
- Habib, N.; Khan, I.U.; Xiao, M.; Hejazi, M.S.; Tarhiz, V.; Zhi, X.-Y.; Li, W.-J. Elioraea thermophila sp. nov., a thermophilic bacterium from hot spring of the class Alphaproteobacteria, emended description of the genus Elioraea and proposal of Elioraeaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 1300–1306. [Google Scholar] [CrossRef]
- Hördt, A.; Lopez, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.M.; Tindall, B.J.; Gronow, S.; Kyrpides, N.C.; Woyke, T.; Göker, M. Analysis of 1000+ type-strain genomes substantially improves taxonomic classification of Alphaproteobacteria. Front. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. List of changes in taxonomic opinion no. 32. Notification of changes in taxonomic opinion previously published outside the IJSEM. Int. J. Syst. Evol. Microbiol. 2020, 70, 4061–4090. [Google Scholar] [CrossRef]
- Thiel, V.; Wood, J.M.; Olsen, M.T.; Tank, M.; Klatt, C.G.; Ward, D.M.; Bryant, D.A. The dark side of the mushroom spring microbial mat: Life in the shadow of chlorophototrophs. I. Microbial diversity based on 16S rRNA gene amplicons and metagenomic sequencing. Front. Microbiol. 2016, 7, 919. [Google Scholar] [CrossRef] [Green Version]
- Tank, M.; Thiel, V.; Ward, D.M.; Bryant, D.A. A panoply of phototrophs: An overview of the thermophilic chlorophototrophs of the microbial mats of alkaline siliceous hot springs in Yellowstone National Park, WY, USA. In Modern Topics in the Phototrophic Prokaryotes: Environmental and Applied Aspects; Hallenbeck, P., Ed.; Springer International Publishing: Zürich, Switzerland, 2017; pp. 87–137. [Google Scholar] [CrossRef]
- Lee, H.-J.; Whang, K.-S. Elioraea rosea sp. nov., a plant promoting bacterium isolated from floodwater of a paddy field. Int. J. Syst. Evol. Microbiol. 2020, 70, 2132–2136. [Google Scholar] [CrossRef]
- Robertson, S.; Ramaley, R.F.; Meyer, T.; Kyndt, J.A. Whole-genome sequence of a unique Elioraea species strain isolated from a Yellowstone National Park hot spring. Microbiol. Res. Announc. 2019, 8, e0907-19. [Google Scholar] [CrossRef] [Green Version]
- Tank, M.; Bryant, D.A. Chloracidobacterium thermophilum gen. nov., sp. nov.: An anoxygenic microaerophilic chlorophotoheterotrophic acidobacterium. Int. J. Syst. Evol. Microbiol. 2015, 65, 1426–1430. [Google Scholar] [CrossRef] [Green Version]
- Tank, M.; Bryant, D.A. Nutrient requirements and growth physiology of the photoheterotrophic Acidobacterium, Chloracidobacterium thermophilum. Front. Microbiol. 2015, 6, 226. [Google Scholar] [CrossRef] [Green Version]
- Saini, M.K.; Sebastian, A.; Siratori, Y.; Soulier, N.T.; Garcia Costas, A.M.; Drautz-Moses, D.I.; Schuster, S.C.; Albert, I.; Harruta, S.; Hanada, S.; et al. Genomic and phenotypic characterization of Chloracidobacterium species isolates provides evidence for multiple species. Front. Microbiol. 2021, 12, 704168. [Google Scholar] [CrossRef] [PubMed]
- Frigaard, N.-U.; Maresca, J.A.; Yunker, C.E.; Jones, A.D.; Bryant, D.A. Genetic manipulation of carotenoid biosynthesis in the green sulfur bacterium Chlorobium Tepidum. J. Bacteriol. 2004, 186, 5210–5220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanada, S.; Takaichi, S.; Matsuura, K.; Nakamura, K. Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int. J. Syst. Evol. Microbiol. 2002, 52, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Minnikin, D.E.; Collins, M.D.; Goodfellow, M. Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J. Appl. Bacteriol. 1979, 47, 87–95. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Saini, M.K.; Weng, C.-C.; Soulier, N.; Sebastian, A.; Albert, I.; Thiel, V.; Bryant, D.A.; Hanada, S.; Tank, M. Caldichromatium japonicum gen. nov., sp. nov., a novel thermophilic phototrophic purple sulphur bacterium of the Chromatiaceae isolated from Nakabusa hot springs, Japan. Int. J. Syst. Evol. Microbiol. 2020, 70, 5701–5710. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Tatusova, T.; Di Cuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Haft, D.H.; DiCuccio, M.; Badretdin, A.; Brover, V.; Chetvernin, V.; Badretin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: An update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018, 46, D851–D860. [Google Scholar] [CrossRef]
- Li, W.; O’Neill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the Prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2015, 66, 1100–1103. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Göker, M.; Spröer, C.; Klenk, H.-P. When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 2013, 195, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2 approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Bhaya, D.; Grossman, A.R.; Steunou, A.S.; Khuri, N.; Cohan, F.M.; Hamamura, N.; Melendrez, M.C.; Bateson, M.M.; Ward, D.M.; Heidelberg, J.F. Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. ISME J. 2007, 1, 703–713. [Google Scholar] [CrossRef] [Green Version]
- Klatt, C.G.; Wood, J.M.; Rusch, D.B.; Bateson, M.M.; Hamamura, N.; Heidelberg, J.F.; Grossman, A.R.; Bhaya, D.; Cohan, F.M.; Kühl, M.; et al. Community ecology of hot spring cyanobacterial mats: Predominant populations and their functional potential. ISME J. 2011, 5, 1262–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Klatt, C.G.; Wood, J.M.; Rusch, D.B.; Ludwig, M.; Wittekindt, M.; Tomsho, L.P.; Schuster, S.C.; Ward, D.M.; Bryant, D.A. Metatranscriptomic analyses of chlorophototrophs of a hot-spring microbial mat. ISME J. 2011, 5, 1279–1290. [Google Scholar] [CrossRef] [Green Version]
- Klatt, C.G.; Liu, Z.; Ludwig, M.; Kühl, M.; Jensen, S.I.; Bryant, D.A.; Ward, D.M. Temporal metatranscriptomic patterning in phototrophic Chloroflexi inhabiting microbial mat in a geothermal spring. ISME J. 2013, 7, 1775–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Klatt, C.G.; Ludwig, M.; Rusch, D.B.; Jensen, S.I.; Kühl, M.; Ward, D.M.; Bryant, D.A. “Candidatus Thermochlorobacter aerophilum”: An aerobic chlorophotoheterotrophic member of the phylum Chlorobi. ISME J. 2012, 6, 1869–1882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryant, D.A.; Hunter, C.N.; Warren, M.J. Biosynthesis of the modified tetrapyrroles—The pigments of life. J. Biol. Chem. 2020, 295, 6888–6925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Keswani, J.; Whitman, W.B. Relationship of 16S rRNA sequence similarity to DNA hybridization in prokaryotes. Int. J. Syst. Evol. Microbiol. 2001, 51, 667–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, D.M.; Bateson, M.M.; Weller, R.; Ruff-Roberts, A.L. Ribosomal RNA analysis of microorganisms as they occur in nature. In Advances in Microbial Ecology; Arshall, K., Ed.; Springer: Boston, MA, USA, 1992; Volume 12, pp. 219–286. [Google Scholar] [CrossRef]
- Thiel, V.; Hügler, M.; Ward, D.M.; Bryant, D.A. The dark side of the Mushroom Spring microbial mat: Life in the shadow of chlorophototrophs. II. Metabolic functions of abundant community members predicted from metagenomic analyses. Front. Microbiol. 2017, 8, 943. [Google Scholar] [CrossRef] [Green Version]
- Thiel, V.; Garcia Costas, A.M.; Fortney, N.W.W.; Martinez, J.N.; Roden, E.E.; Boyd, E.S.; Ward, D.M.; Bryant, D.A. “Candidatus Thermonerobacter thiotrophicus,” a non-phototrophic, sulfate-reducing member of the phylum Chlorobi that inhabits hot-spring communities. Front. Microbiol. 2019, 9, 3159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becraft, E.; Wood, J.M.; Rusch, D.B.; Kühl, M.; Jensen, S.I.; Bryant, D.A.; Roberts, D.W.; Cohan, F.M.; Ward, D.M. The molecular dimension of microbial species. 1. Ecological distinctions among, and homogeneity within, putative ecotypes of Synechococcus inhabiting the cyanobacterial mat of Mushroom Spring, Yellowstone National Park. Front. Microbiol. 2015, 6, 590. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Property | 1E. tepidiphila DSM17972T | 2E. thermophila YIM 72297T | 3E. rosea PF-30AT | E. tepida MS-P2T |
|---|---|---|---|---|
| Cell morphology | Rods | Curved rods | Rods | Curved rods |
| Cell size (width × length (µm)) | 0.5–1.0 × 1.0–1.5 | 0.7–0.9 × 2.2–3.2 | 1.0–1.2 × 2.2–2.6 | 0.5–1.0 × 1.0–1.5 |
| Motility (flagella genes present) | + | − | + | + |
| Temperature optimum (range) °C | 45–50 (30–50) | 55 (45–60) | 28 (20–40) | 45–50 (>35–<55) |
| pH optimum (range) | 8.0–8.5 (6.0–9.8) | 7.0–7.5 (5.0–9.0) | 7.0 (5.0–9.0) | 7.0–7.5 (6.0–10.0) |
| Catalase | + | + | –(but katG is present) 4 | + |
| Photoautotrophic growth | − | + | − | − |
| Substrate Utilization 5 | ||||
| D-Fructose | − | + | − | + |
| D-Fucose | − | − | − | + |
| D-Galactose | − | − | − | + |
| D-Mannose | − | − | − | + |
| D-Melibiose | − | − | − | + |
| D-Psicose | − | − | ND | + |
| Glutamate | + | ND | ND | − |
| Glutamine | + | ND | ND | − |
| Proline | + | ND | ND | + |
| Compounds Hydrolyzed | ||||
| Starch | + | − | − | − |
| Gelatin | − | − | − | + |
| Aesculin | − | − | + | + |
| Pectin | − | ND | ND | + |
| Casein | − | − | − | ND |
| Fatty Acid | 1E. tepidiphila DSM17972T | 2E. thermophila YIM 72297T | 3E. rosea PF-30AT | E. tepida MS-P2T |
|---|---|---|---|---|
| 14:0 | 0.3% | – | – | – |
| 15:0 iso | – | – | – | 7.8% |
| 16:0 | 5.4% | 12.6% | 3.6% | 12.3% |
| 17:0 | 0.4% | – | 1.6% | – |
| 16:0 2-OH | 1.9% | 5.4% | 2.7% | – |
| 18:1 ω7c | 19.0% 4 | 30.1% | 35.7% | 9.9% |
| 18:0 | 24.8% | 35.8% | 22.7% | 30.0% |
| 18:1 ω7c 11-methyl | 8.0% | – | 3.4% | 22.0% |
| 19:0 cyclo ω8c | 12.4% | 4.2% | 5.7% | 13.0% |
| 18:0 2-OH | 0.6% | 1.5% | 2.7% | – |
| 18:0 3-OH | 3.8% | 2.5% | 2.9% | 4.7% |
| 19:0 2-OH cyclo ω8 (Δ11:12) | 18.6% | – | 7.9% | – |
| Genomic Properties | 1E. tepidiphila DSM17972T | 2E. thermophila YIM 72297T | 3E. rosea PF-30AT | 4Elioraea sp. “Yellowstone” | E. tepida MS-P2T |
|---|---|---|---|---|---|
| Size (bp) | >4,304,240 | 3,029,970 | >4,487,660 | >3,824,070 | 3,367,643 |
| Proteins (CDS) | >4014 | 2845 | >4189 | >3647 | 3083 |
| rRNA genes | 3 | 3 | 3 | 3 | 3 |
| tRNAs | 48 | 46 | 46 | 46 | 46 |
| DNA Mol % G + C | 71.3 | 70.9 | 69.9 | 72.4 | 70.6 |
| Average nucleotide identity (%) 5 | 75.06 | 73.34 | 78.59 | 75.84 | –– |
| Digital DNA-DNA hybridization (%) 5 | 19.8 | 18.7 | 21.9 | 20.5 | –– |
| 16S rRNA % identity 6 | 97.26 | 96.58 | 97.99 | 97.52 | –– |
| Nitrate reductase (NarG) /growth with nitrate | +/− | −/− | +/− | +/ND | −/− |
| RubisCO (RbcLS) and Phosphoribulokinase (PRK) | − | + | − | − | − |
| Urease (UreABCDEFGJ) | − | + | + | − | + |
| Thiosulfate oxidation (SoxBCDYZ) | + | + | − | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saini, M.K.; Yoshida, S.; Sebastian, A.; Hara, E.; Tamaki, H.; Soulier, N.T.; Albert, I.; Hanada, S.; Tank, M.; Bryant, D.A. Elioraea tepida, sp. nov., a Moderately Thermophilic Aerobic Anoxygenic Phototrophic Bacterium Isolated from the Mat Community of an Alkaline Siliceous Hot Spring in Yellowstone National Park, WY, USA. Microorganisms 2022, 10, 80. https://doi.org/10.3390/microorganisms10010080
Saini MK, Yoshida S, Sebastian A, Hara E, Tamaki H, Soulier NT, Albert I, Hanada S, Tank M, Bryant DA. Elioraea tepida, sp. nov., a Moderately Thermophilic Aerobic Anoxygenic Phototrophic Bacterium Isolated from the Mat Community of an Alkaline Siliceous Hot Spring in Yellowstone National Park, WY, USA. Microorganisms. 2022; 10(1):80. https://doi.org/10.3390/microorganisms10010080
Chicago/Turabian StyleSaini, Mohit Kumar, Shohei Yoshida, Aswathy Sebastian, Eri Hara, Hideyuki Tamaki, Nathan T. Soulier, Istvan Albert, Satoshi Hanada, Marcus Tank, and Donald A. Bryant. 2022. "Elioraea tepida, sp. nov., a Moderately Thermophilic Aerobic Anoxygenic Phototrophic Bacterium Isolated from the Mat Community of an Alkaline Siliceous Hot Spring in Yellowstone National Park, WY, USA" Microorganisms 10, no. 1: 80. https://doi.org/10.3390/microorganisms10010080
APA StyleSaini, M. K., Yoshida, S., Sebastian, A., Hara, E., Tamaki, H., Soulier, N. T., Albert, I., Hanada, S., Tank, M., & Bryant, D. A. (2022). Elioraea tepida, sp. nov., a Moderately Thermophilic Aerobic Anoxygenic Phototrophic Bacterium Isolated from the Mat Community of an Alkaline Siliceous Hot Spring in Yellowstone National Park, WY, USA. Microorganisms, 10(1), 80. https://doi.org/10.3390/microorganisms10010080






