The Immune Efficacy of Inactivated Pseudorabies Vaccine Prepared from FJ-2012ΔgE/gI Strain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Suckling Pig and Sites
2.2. Experimental Vaccines and Main Reagents
2.3. Experimental Pigs Groups and Immunization Procedures
2.4. Experimental Pigs Challenged Procedures and Clinical Record
2.5. PRV gE, gB Antibody Test, and Neutralizing Antibody Detection
2.6. Necropsy, Tissue Sampling, and Histological Analysis
2.7. Establishment of PRV qPCR Method and Tissue Viral Load Analysis
2.8. Statistical Analysis
2.9. Ethical Statement
3. Results
3.1. PRV gB and gE Antibody Production in Piglets
3.2. Virus Neutralizing Antibodies of PRV Induced by Vaccines in Piglets
3.3. Protection of Vaccinated Piglets against Virulent Challenge
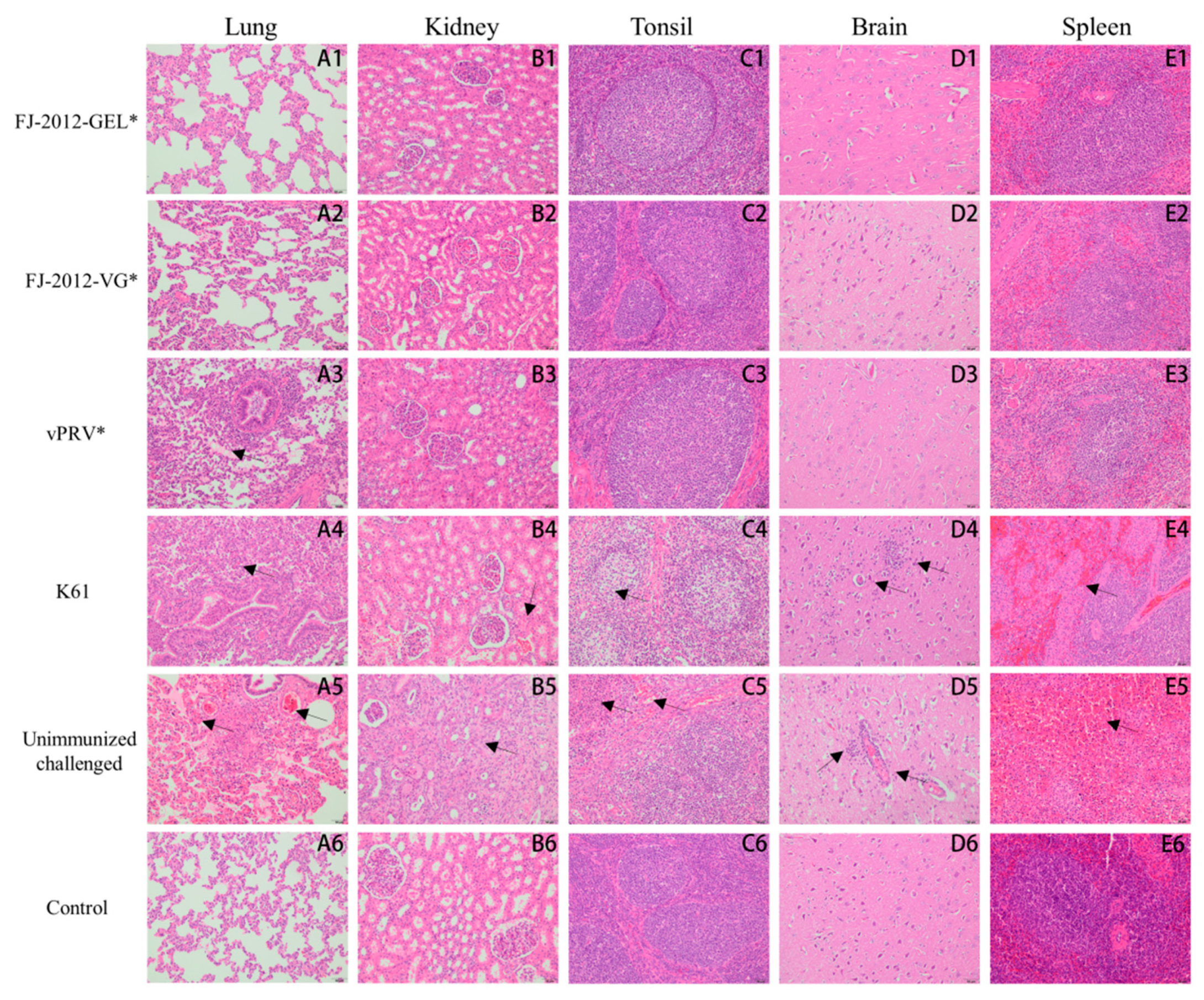
3.4. Necropsy and Histological Analysis
3.5. Tissue Viral Load Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Davison, A.J.; Eberle, R.; Ehlers, B.; Hayward, G.S.; McGeoch, D.J.; Minson, A.C.; Pellett, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The order Herpesvirales. Arch. Virol. 2009, 154, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Brittle, E.E.; Reynolds, A.E.; Enquist, L.W. Two Modes of Pseudorabies Virus Neuroinvasion and Lethality in Mice. J. Virol. 2004, 78, 12951–12963. [Google Scholar] [CrossRef] [PubMed]
- Szpara, M.L.; Tafuri, Y.R.; Parsons, L.; Shamim, S.R.; Verstrepen, K.J.; Legendre, M.; Enquist, L.W. A Wide Extent of Inter-Strain Diversity in Virulent and Vaccine Strains of Alphaherpesviruses. PLoS Pathog. 2011, 7, e1002282. [Google Scholar] [CrossRef] [PubMed]
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular Biology of Pseudorabies Virus: Impact on Neurovirology and Veterinary Medicine. Microbiol. Mol. Biol. Rev. 2005, 69, 462–500. [Google Scholar] [CrossRef]
- Minamiguchi, K.; Kojima, S.; Sakumoto, K.; Kirisawa, R. Isolation and molecular characterization of a variant of Chinese gC-genotype II pseudorabies virus from a hunting dog infected by biting a wild boar in Japan and its pathogenicity in a mouse model. Virus Genes 2019, 55, 322–331. [Google Scholar] [CrossRef]
- Kong, H.; Zhang, K.; Liu, Y.; Shang, Y.; Wu, B.; Liu, X. Attenuated live vaccine (Bartha-K16) caused pseudorabies (Aujeszky’s disease) in sheep. Vet. Res. Commun. 2013, 37, 329–332. [Google Scholar] [CrossRef]
- Cheng, Z.; Kong, Z.; Liu, P.; Fu, Z.; Zhang, J.; Liu, M.; Shang, Y. Natural infection of a variant pseudorabies virus leads to bovine death in China. Transbound. Emerg. Dis. 2020, 67, 518–522. [Google Scholar] [CrossRef]
- Wang, G.-S.; Du, Y.; Wu, J.-Q.; Tian, F.-L.; Yu, X.-J.; Wang, J.-B. Vaccine resistant pseudorabies virus causes mink infection in China. BMC Vet. Res. 2018, 14, 20. [Google Scholar] [CrossRef]
- Jin, H.-L.; Gao, S.-M.; Liu, Y.; Zhang, S.-F.; Hu, R.-L. Pseudorabies in farmed foxes fed pig offal in Shandong province, China. Arch. Virol. 2016, 161, 445–448. [Google Scholar] [CrossRef]
- Lee, J.Y.; Wilson, M.R. A review of pseudorabies (Aujeszky’s disease) in pigs. Can. Vet. J. 1979, 20, 65–69. [Google Scholar]
- Zhu, L.; Guo, W.Z.; Xu, Z.W. Fluctuant rule of colostral antibodies and the date of initial immunization for the piglets from sows inoculated with pseudorabies virus gene-deleted vaccine SA215. Chin. J. Vet. Med. 2004, 24, 320–322. (In Chinese) [Google Scholar]
- He, Q.G.; Chen, H.C.; Fang, L.R.; Wu, B.; Liu, Z.F.; Xiao, S.B.; Jin, M.L. The safety, stability and immunogenicity of double genenegative mutant of pseudorabies virus strain (PRV HB-98). Chin. J. Vet. Med. 2006, 26, 165–168. (In Chinese) [Google Scholar]
- Zheng, H.-H.; Wang, L.-Q.; Fu, P.-F.; Zheng, L.-L.; Chen, H.-Y.; Liu, F. Characterization of a recombinant pseudorabies virus expressing porcine parvovirus VP2 protein and porcine IL-6. Virol. J. 2020, 17, 19. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.-L.; Xia, S.-L.; Wang, Y.; Du, M.; Xiang, G.-T.; Cong, X.; Luo, Y.; Li, L.-F.; Zhang, L.; Yu, J.; et al. Safety and immunogenicity of a gE/gI/TK gene-deleted pseudorabies virus variant expressing the E2 protein of classical swine fever virus in pigs. Immunol. Lett. 2016, 174, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Freuling, C.M.; Müller, T.F.; Mettenleiter, T.C. Vaccines against pseudorabies virus (PrV). Vet. Microbiol. 2017, 206, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, Y.; Chen, Y.; Wang, A.; Feng, H.; Wei, Q.; Zhou, E.; Zhang, G. A single dose glycoprotein D-based subunit vaccine against pseudorabies virus infection. Vaccine 2020, 38, 6153–6161. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Zhang, E.; Zhou, M.; Lin, J.; Yang, Q. Intranasal administration with recombinant Bacillus subtilis induces strong mucosal immune responses against pseudorabies. Microb. Cell Fact. 2019, 18, 103. [Google Scholar] [CrossRef]
- Wang, J.; Guo, R.; Qiao, Y.; Xu, M.; Wang, Z.; Liu, Y.; Gu, Y.; Liu, C.; Hou, J. An inactivated gE-deleted pseudorabies vaccine provides complete clinical protection and reduces virus shedding against challenge by a Chinese pseudorabies variant. BMC Vet. Res. 2016, 12, 277. [Google Scholar] [CrossRef]
- Wang, T.; Xiao, Y.; Yang, Q.; Wang, Y.; Sun, Z.; Zhang, C.; Yan, S.; Wang, J.; Guo, L.; Yan, H.; et al. Construction of a gE-Deleted Pseudorabies Virus and Its Efficacy to the New-Emerging Variant PRV Challenge in the Form of Killed Vaccine. BioMed Res. Int. 2015, 2015, 684945. [Google Scholar] [CrossRef]
- Gu, Z.; Dong, J.; Wang, J.; Hou, C.; Sun, H.; Yang, W.; Bai, J.; Jiang, P. A novel inactivated gE/gI deleted pseudorabies virus (PRV) vaccine completely protects pigs from an emerged variant PRV challenge. Virus Res. 2015, 195, 57–63. [Google Scholar] [CrossRef]
- Wang, C.-H.; Yuan, J.; Qin, H.-Y.; Luo, Y.; Cong, X.; Li, Y.; Chen, J.; Li, S.; Sun, Y.; Qiu, H.-J. A novel gE-deleted pseudorabies virus (PRV) provides rapid and complete protection from lethal challenge with the PRV variant emerging in Bartha-K61-vaccinated swine population in China. Vaccine 2014, 32, 3379–3385. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Li, G.; Liang, C.; Liu, F.; Tian, Q.; Cao, Y.; Li, L.; Zheng, X.; Zheng, H.; Tong, G. A live, attenuated pseudorabies virus strain JS-2012 deleted for gE/gI protects against both classical and emerging strains. Antivir. Res. 2016, 130, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Tong, W.; Zheng, H.; Liu, F.; Wu, J.; Li, G.; Zhou, E.-M.; Tong, G. A high-temperature passaging attenuated Pseudorabies vaccine protects piglets completely against emerging PRV variant. Res. Vet. Sci. 2017, 112, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Xu, Z.; Liu, X.; Li, P.; Yang, F.; Zhao, J.; Fan, Y.; Sun, X.; Zhu, L. A live gI/gE-deleted pseudorabies virus (PRV) protects weaned piglets against lethal variant PRV challenge. Virus Genes 2017, 53, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, Z.; Ge, A.; Guo, R.; Qiao, Y.; Xu, M.; Wang, Z.; Liu, Y.; Zheng, Y.; Fan, H.; et al. Safety and immunogenicity of an attenuated Chinese pseudorabies variant by dual deletion of TK&gE genes. BMC Vet. Res. 2018, 14, 287. [Google Scholar] [CrossRef]
- Dong, J.; Bai, J.; Sun, T.; Gu, Z.; Wang, J.; Sun, H.; Jiang, P. Comparative pathogenicity and immunogenicity of triple and double gene-deletion pseudorabies virus vaccine candidates. Res. Vet. Sci. 2017, 115, 17–23. [Google Scholar] [CrossRef]
- Hu, R.-M.; Zhou, Q.; Song, W.-B.; Sun, E.-C.; Zhang, M.-M.; He, Q.-G.; Chen, H.-C.; Wu, B.; Liu, Z.-F. Novel pseudorabies virus variant with defects in TK, gE and gI protects growing pigs against lethal challenge. Vaccine 2015, 33, 5733–5740. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, L.; Jia, X.; Wang, T.; Wang, J.; Sun, Z.; Wang, L.; Li, X.; Tan, F.; Tian, K. Construction of a triple gene-deleted Chinese Pseudorabies virus variant and its efficacy study as a vaccine candidate on suckling piglets. Vaccine 2015, 33, 2432–2437. [Google Scholar] [CrossRef]
- Tang, Y.-D.; Liu, J.-T.; Wang, T.-Y.; An, T.-Q.; Sun, M.-X.; Wang, S.-J.; Fang, Q.-Q.; Hou, L.-L.; Tian, Z.-J.; Cai, X.-H. Live attenuated pseudorabies virus developed using the CRISPR/Cas9 system. Virus Res. 2016, 225, 33–39. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, L.-Q.; Zheng, H.-H.; Yang, Y.-R.; Liu, F.; Zheng, L.-L.; Jin, Y.; Chen, H.-Y. Construction and immunogenicity of a gE/gI/TK-deleted PRV based on porcine pseudorabies virus variant. Mol. Cell. Probes 2020, 53, 101605. [Google Scholar] [CrossRef]
- Cong, X.; Lei, J.-L.; Xia, S.-L.; Wang, Y.-M.; Li, Y.; Li, S.; Luo, Y.; Sun, Y.; Qiu, H.-J. Pathogenicity and immunogenicity of a gE/gI/TK gene-deleted pseudorabies virus variant in susceptible animals. Vet. Microbiol. 2016, 182, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.F.; Lai, Z.; Shu, Y.H.; Qi, S.H.; Ma, J.J.; Wu, B.Q.; Gong, J.P. Isolation and identification of porcine pseudorabies virus (PRV) C strain. Acta Agric. Shanghai 2015, 31, 32–36. (In Chinese) [Google Scholar]
- Sun, Y.; Luo, Y.; Wang, C.-H.; Yuan, J.; Li, N.; Song, K.; Qiu, H.-J. Control of swine pseudorabies in China: Opportunities and limitations. Vet. Microbiol. 2016, 183, 119–124. [Google Scholar] [CrossRef]
- Delva, J.L.; Nauwynck, H.J.; Mettenleiter, T.C.; Favoreel, H.W. The Attenuated Pseudorabies Virus Vaccine Strain Bartha K61: A Brief Review on the Knowledge Gathered During 60 Years of Research. Pathogens 2020, 9, 897. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, S.; Wang, X.; Zou, M.; Gao, S. Bartha-k61 vaccine protects growing pigs against challenge with an emerging variant pseudorabies virus. Vaccine 2017, 35, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, K.V.; Michailidou, M.; Grivas, I.; Petridou, E.; Stamelou, E.; Efraimidis, K.; Chen, L.; Drew, T.W.; Kritas, S.K. Bartha-K61 vaccine protects nursery pigs against challenge with novel european and asian strains of suid herpesvirus 1. Vet. Res. 2022, 53, 47. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Liu, F.; Zheng, H.; Liang, C.; Zhou, Y.-J.; Jiang, Y.-F.; Shan, T.-L.; Gao, F.; Li, G.-X.; Tong, G.-Z. Emergence of a Pseudorabies virus variant with increased virulence to piglets. Vet. Microbiol. 2015, 181, 236–240. [Google Scholar] [CrossRef]
- Wu, X.-M.; Chen, Q.-Y.; Chen, R.-J.; Che, Y.-L.; Wang, L.-B.; Wang, C.-Y.; Yan, S.; Liu, Y.-T.; Xiu, J.-S.; Zhou, L.-J. Pathogenicity and Whole Genome Sequence Analysis of a Pseudorabies Virus Strain FJ-2012 Isolated from Fujian, Southern China. Can. J. Infect. Dis. Med Microbiol. 2017, 2017, 9073172. [Google Scholar] [CrossRef]
- Gerdts, V.; Jöns, A.; Mettenleiter, T.C. Potency of an experimental DNA vaccine against Aujeszky’s disease in pigs. Vet. Microbiol. 1999, 66, 1–13. [Google Scholar] [CrossRef]
- Song, C.; Huang, X.; Gao, Y.; Zhang, X.; Wang, Y.; Zhang, Y.; Lv, T.; Zhang, Z.; Zhang, Y.; Pan, Q.; et al. Histopathology of brain functional areas in pigs infected by porcine pseudorabies virus. Res. Vet. Sci. 2021, 141, 203–211. [Google Scholar] [CrossRef]
- Opriessnig, T.; Thacker, E.L.; Yu, S.; Fenaux, M.; Meng, X.-J.; Halbur, P.G. Experimental Reproduction of Postweaning Multisystemic Wasting Syndrome in Pigs by Dual Infection with Mycoplasma hyopneumoniae and Porcine Circovirus Type 2. Vet. Pathol. 2004, 41, 624–640. [Google Scholar] [CrossRef] [PubMed]
- An, T.-Q.; Peng, J.-M.; Tian, Z.-J.; Zhao, H.-Y.; Li, N.; Liu, Y.-M.; Chen, J.-Z.; Leng, C.-L.; Sun, Y.; Chang, D.; et al. Pseudorabies Virus Variant in Bartha-K61–Vaccinated Pigs, China, 2012. Emerg. Infect. Dis. 2013, 19, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Bai, C.; Sun, J.; Chang, S.; Zhang, X. Emergence of virulent pseudorabies virus infection in Northern China. J. Vet. Sci. 2013, 14, 363–365. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shi, Z.; Liu, C.; Wang, P.; Wang, M.; Wang, S.; Liu, Z.; Wei, L.; Sun, Z.; He, X.; et al. Implication of the Identification of an Earlier Pseudorabies Virus (PRV) Strain HLJ-2013 to the Evolution of Chinese PRVs. Front. Microbiol. 2020, 11, 612474. [Google Scholar] [CrossRef]
- Ye, C.; Zhang, Q.-Z.; Tian, Z.-J.; Zheng, H.; Zhao, K.; Liu, F.; Guo, J.-C.; Tong, W.; Jiang, C.-G.; Wang, S.-J.; et al. Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: Evidence for the existence of two major genotypes. Virology 2015, 483, 32–43. [Google Scholar] [CrossRef]
- Yu, T.; Chen, F.; Ku, X.; Fan, J.; Zhu, Y.; Ma, H.; Li, S.; Wu, B.; He, Q. Growth characteristics and complete genomic sequence analysis of a novel pseudorabies virus in China. Virus Genes 2016, 52, 474–483. [Google Scholar] [CrossRef]
- Husak, P.J.; Kuo, T.; Enquist, L.W. Pseudorabies Virus Membrane Proteins gI and gE Facilitate Anterograde Spread of Infection in Projection-Specific Neurons in the Rat. J. Virol. 2000, 74, 10975–10983. [Google Scholar] [CrossRef]
- Mettenleiter, T.C.; Klupp, B.G.; Weiland, F.; Visser, N. Characterization of a quadruple glycoprotein-deleted pseudorabies virus mutant for use as a biologically safe live virus vaccine. J. Gen. Virol. 1994, 75 Pt 7, 1723–1733. [Google Scholar] [CrossRef]
- Mettenleiter, T.C. Aujeszky’s disease (pseudorabies) virus: The virus and molecular pathogenesis—State of the art, June 1999. Vet. Res. 2000, 31, 99–115. [Google Scholar] [CrossRef]
- McKee, A.S.; Munks, M.W.; Marrack, P. How Do Adjuvants Work? Important Considerations for New Generation Adjuvants. Immunity 2007, 27, 687–690. [Google Scholar] [CrossRef]
- Xu, B. Innovative polymeric adjuvant for PCV2 vaccination. In Proceedings of the 2016 American Association of Swine Veterinarians Annual Meeting, New Orleans, LA, USA, 27 February–1 March 2016. [Google Scholar]
- Wangkaghart, E.; Deville, S.; Wang, B.; Srisapoome, P.; Wang, T.; Secombes, C.J. Immune Response and Protective Efficacy of Two New Adjuvants, MontanideTM ISA 763B VG and MontanideTM GEL02, Administered with a Streptococcus agalactiae Ghost Vaccine in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2021, 116, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.-J.; Pei, S.-X.; Song, J.-J.; Zhan, P.-F.; Han, Y.-N.; Xue, Y.; Ding, K.; Zhao, Z.-Q. Screening immune adjuvants for an inactivated vaccine against Erysipelothrix rhusiopathiae. Front. Vet. Sci. 2022, 9, 922867. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, L.; Ascarateil, S.; Aucouturier, J.; Ganne, V. SEPPIC Vaccine Adjuvants for Poultry. Ann. N. Y. Acad. Sci. 2006, 1081, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Bidart, J.; Mignaqui, A.; Kornuta, C.; Lupi, G.; Gammella, M.; Soria, I.; Galarza, R.; Ferella, A.; Cardillo, S.; Langellotti, C.; et al. FMD empty capsids combined with the Immunostant Particle Adjuvant-ISPA or ISA206 induce protective immunity against foot and mouth disease virus. Virus Res. 2021, 297, 198339. [Google Scholar] [CrossRef] [PubMed]
- van Oirschot, J.T.; Kaashoek, M.J.; Rijsewijk, F.A.M.; Stegeman, J.A. The use of marker vaccines in eradication of herpesviruses. J. Biotechnol. 1996, 44, 75–81. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, S.; Xu, Q.; Wu, J.; Zhai, X.; Li, S.; Wang, J.; Ni, J.; Yuan, L.; Song, X.; et al. Investigation on pseudorabies prevalence in Chinese swine breeding farms in 2013–2016. Trop. Anim. Health Prod. 2018, 50, 1279–1285. [Google Scholar] [CrossRef]
- Xia, L.; Sun, Q.; Wang, J.; Chen, Q.; Liu, P.; Shen, C.; Sun, J.; Tu, Y.; Shen, S.; Zhu, J.; et al. Epidemiology of pseudorabies in intensive pig farms in Shanghai, China: Herd-level prevalence and risk factors. Prev. Vet. Med. 2018, 159, 51–56. [Google Scholar] [CrossRef]
- Maresch, C.; Lange, E.; Teifke, J.P.; Fuchs, W.; Klupp, B.; Müller, T.; Mettenleiter, T.C.; Vahlenkamp, T.W. Oral immunization of wild boar and domestic pigs with attenuated live vaccine protects against Pseudorabies virus infection. Vet. Microbiol. 2012, 161, 20–25. [Google Scholar] [CrossRef]
- Yan, Z.; Chen, M.; Tang, D.; Wu, X.; Ren, X.; Pan, H.; Li, Y.; Ji, Q.; Luo, Y.; Fan, H.; et al. Better immune efficacy triggered by the inactivated gI/gE-deleted pseudorabies virus with the additional insertion of gC gene in mice and weaned pigs. Virus Res. 2021, 296, 198353. [Google Scholar] [CrossRef]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef]




| Vaccine Type | Gene-Deleted Vaccines | Vaccine Strain | Phenotype | References |
|---|---|---|---|---|
| Inactivated | Single gene-deleted | AH02LA, HN1201 (licensed) | gE-deleted | [18,19] |
| Double gene-deleted | ZJ01 | gE/gI-deleted | [20] | |
| Live-attenuated | Single gene-deleted | TJ | gE-deleted | [21] |
| Double gene-deleted | JS-2012 | gE/gI-deleted | [22] | |
| Double gene-deleted | JS-2012 | gE/US2-deleted | [23] | |
| Double gene-deleted | XJ, AH02LA | gE/TK-deleted | [24,25] | |
| Triple-gene-deleted | ZJ01, MX, HN1201, HeN1, NY, and TJ | gE/gI/TK-deleted | [26,27,28,29,30,31] | |
| Four-gene-deleted (natural losses) | C (licensed) | gE/gI/US9/US2-deleted | [32] |
| Groups | Immunization Time | Challenge Time | |
|---|---|---|---|
| 0 Days | 21 Days | 42 Days | |
| FJ-2012-GEL* | √ | √ | √ |
| FJ-2012-VG* | √ | √ | √ |
| vPRV* | √ | √ | √ |
| K61 | √ | × | √ |
| Unimmunized challenged | √ | √ | √ |
| Control | × | × | × |
| Groups | Lung | Kidney | Tonsil | Brain | Spleen |
|---|---|---|---|---|---|
| FJ-2012-GEL* | 0 a/5 b | 0/5 | 0/5 | 0/5 | 0/5 |
| FJ-2012-VG* | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| vPRV* | 2/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| K61 | 3/5 | 0/5 | 2/5 | 1/5 | 0/5 |
| Unimmunized challenged | 5/5 | 3/5 | 3/5 | 5/5 | 2/5 |
| Control | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Groups | MLS ± SD § | ||||
|---|---|---|---|---|---|
| Lung † | Kidney ‡ | Tonsil † | Brain † | Spleen † | |
| FJ-2012-GEL* | 0.00 ± 0.00 Bc‖ | 0.00 ± 0.00 Bc | 0.00 ± 0.00 Cc | 0.00 ± 0.00 Bc | 0.00 ± 0.00 Bc |
| FJ-2012-VG* | 0.00 ± 0.00 Bc | 0.00 ± 0.00 Bc | 0.00 ± 0.00 Cc | 0.00 ± 0.00 Bc | 0.00 ± 0.00 Bc |
| vPRV* | 2.00 ± 1.00 BCb | 0.00 ± 0.00 Bc | 0.00 ± 0.00 Cc | 0.00 ± 0.00 Bc | 0.00 ± 0.00 Bc |
| K61 | 3.67 ± 1.15 ACb | 1.33 ± 0.58 Ab | 2.33 ± 0.58 Bb | 1.67 ± 1.16 Bb | 2.33 ± 1.53 ABb |
| Unimmunized challenged | 5.67 ± 0.58 Aa | 2.33 ± 0.58 Aa | 5.00 ± 1.00 Aa | 4.67 ± 1.16 Aa | 4.33 ± 0.58 Aa |
| Control | 0.00 ± 0.00 Bc | 0.00 ± 0.00 Bc | 0.00 ± 0.00 Cc | 0.00 ± 0.00 Bc | 0.00 ± 0.00 Bc |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.-Y.; Wu, X.-M.; Che, Y.-L.; Chen, R.-J.; Hou, B.; Wang, C.-Y.; Wang, L.-B.; Zhou, L.-J. The Immune Efficacy of Inactivated Pseudorabies Vaccine Prepared from FJ-2012ΔgE/gI Strain. Microorganisms 2022, 10, 1880. https://doi.org/10.3390/microorganisms10101880
Chen Q-Y, Wu X-M, Che Y-L, Chen R-J, Hou B, Wang C-Y, Wang L-B, Zhou L-J. The Immune Efficacy of Inactivated Pseudorabies Vaccine Prepared from FJ-2012ΔgE/gI Strain. Microorganisms. 2022; 10(10):1880. https://doi.org/10.3390/microorganisms10101880
Chicago/Turabian StyleChen, Qiu-Yong, Xue-Min Wu, Yong-Liang Che, Ru-Jing Chen, Bo Hou, Chen-Yan Wang, Long-Bai Wang, and Lun-Jiang Zhou. 2022. "The Immune Efficacy of Inactivated Pseudorabies Vaccine Prepared from FJ-2012ΔgE/gI Strain" Microorganisms 10, no. 10: 1880. https://doi.org/10.3390/microorganisms10101880






