First Detection of the SARS-CoV-2 Omicron BA.5/22B in Monaco
Abstract
:1. Introduction
2. Materials and Methods
2.1. Diagnosis of SARS-CoV-2 Infection by Real-Time Reverse Transcription-PCR
2.2. SARS-CoV-2 Serology and SARS-CoV-2 Seroneutralisation Assay
2.3. Reverse Transcription PCR-Based SARS-CoV-2 Genotyping
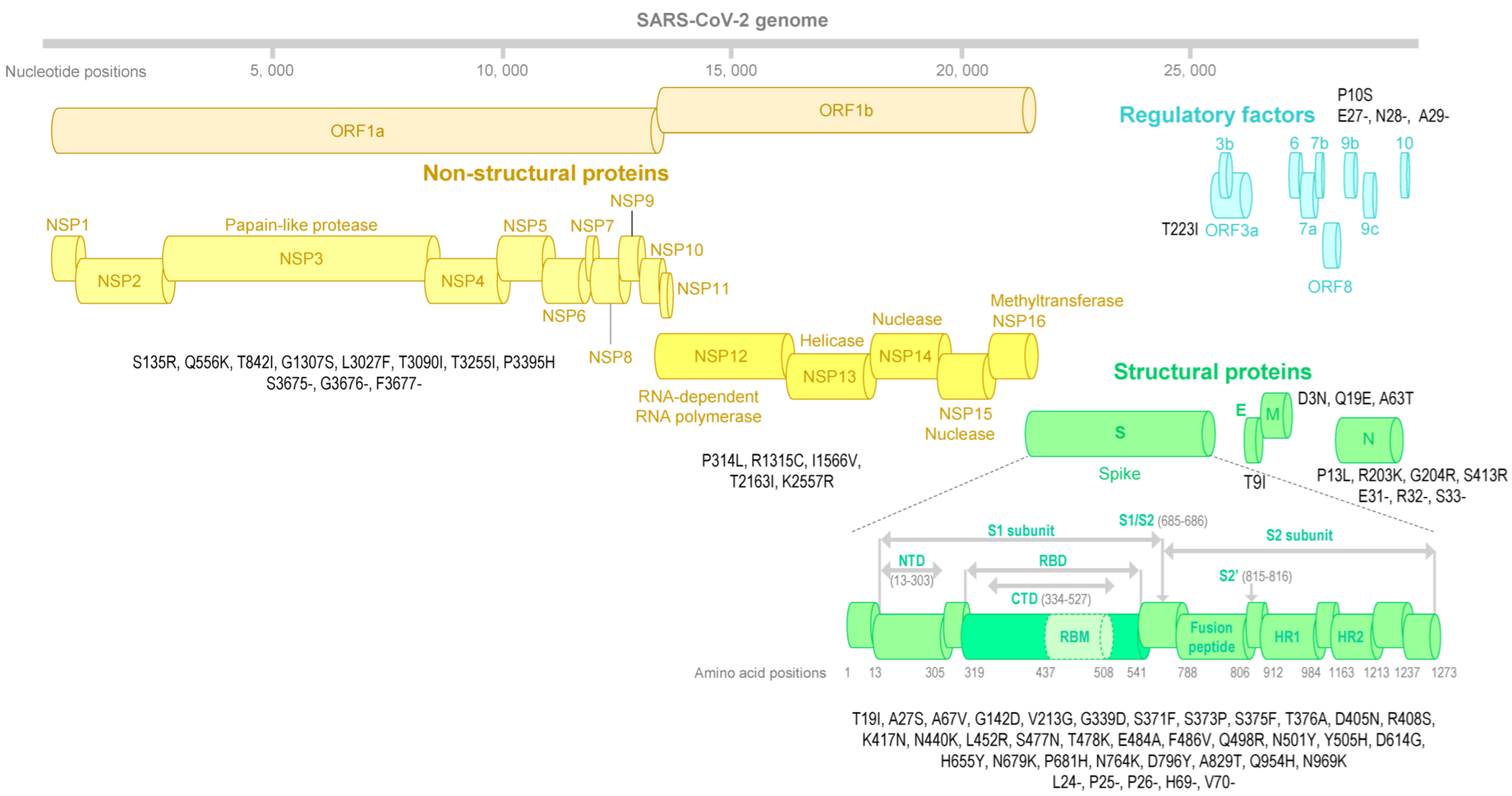
2.4. SARS-CoV-2 Genome Sequencing and Analysis
2.5. SARS-CoV-2 Spike Structure Analyses
3. Results
3.1. Cases’ Reports
3.2. SARS-CoV-2 Serology and Seroneutralization Assay
3.3. SARS-CoV-2 Genotyping
3.4. SARS-CoV-2 Spike Structure Analyses
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Colson, P.; Fournier, P.-E.; Chaudet, H.; Delerce, J.; Giraud-Gatineau, A.; Houhamdi, L.; Andrieu, C.; Brechard, L.; Bedotto, M.; Prudent, E.; et al. Analysis of SARS-CoV-2 Variants From 24,181 Patients Exemplifies the Role of Globalization and Zoonosis in Pandemics. Front. Microbiol. 2022, 12, 786233. [Google Scholar] [CrossRef] [PubMed]
- Lemey, P.; Ruktanonchai, N.; Hong, S.L.; Colizza, V.; Poletto, C.; Broeck, F.V.D.; Gill, M.S.; Ji, X.; Levasseur, A.; Munnink, B.B.O.; et al. Untangling introductions and persistence in COVID-19 resurgence in Europe. Nature 2021, 595, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Rochman, N.D.; Wolf, Y.I.; Faure, G.; Mutz, P.; Zhang, F.; Koonin, E.V. Ongoing global and regional adaptive evolution of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2021, 118, e2104241118. [Google Scholar] [CrossRef]
- Aksamentov, I.; Roemer, C.; Hodcroft, E.B.; Neher, R.A. Nextclade: Clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021, 6, 3773. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Hodcroft, E. CoVariants: SARS-CoV-2 Mutations and Variants of Interest. 2021. Available online: https://covariants.org/https://covariants.org/ (accessed on 19 July 2022).
- Sharma, V.; Rai, H.; Gautam, D.N.S.; Prajapati, P.K.; Sharma, R. Emerging evidence on Omicron (B.1.1.529) SARS-CoV-2 variant. J. Med. Virol. 2022, 94, 1876–1885. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, L.B.; Foster, C.; Rawlinson, W.; Tedla, N.; Bull, R.A. Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission. Rev. Med. Virol. 2022, 32, e2381. [Google Scholar] [CrossRef]
- Desingu, P.A.; Nagarajan, K. Omicron variant losing its critical mutations in the receptor-binding domain. J. Med. Virol. 2022, 94, 2365–2368. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, Y.; Iketani, S.; Nair, M.S.; Li, Z.; Mohri, H.; Wang, M.; Yu, J.; Bowen, A.D.; Chang, J.Y.; et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 2022, 608, 603–608. [Google Scholar] [CrossRef]
- Khan, K.; Karim, F.; Ganga, Y.; Bernstein, M.; Jule, Z.; Reedoy, K.; Cele, S.; Lustig, G.; Amoako, D.; Wolter, N.; et al. Omicron BA.4/BA.5 escape neutralizing immunity elicited by BA.1 infection. Nat. Commun. 2022, 13, 4686. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Yisimayi, A.; Jian, F.; Song, W.; Xiao, T.; Wang, L.; Du, S.; Wang, J.; Li, Q.; Chen, X.; et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022, 608, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; Delerce, J.; Beye, M.; Levasseur, A.; Boschi, C.; Houhamdi, L.; Tissot-Dupont, H.; Yahi, N.; Million, M.; La Scola, B.; et al. First cases of infection with the 21L/BA.2 Omicron variant in Marseille, France. J. Med. Virol. 2022, 94, 3421–3430. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Maggi, F. Recombination in Coronaviruses, with a Focus on SARS-CoV-2. Viruses 2022, 14, 1239. [Google Scholar] [CrossRef] [PubMed]
- Le Targa, L.; Wurtz, N.; Lacoste, A.; Penant, G.; Jardot, P.; Annessi, A.; Colson, P.; La Scola, B.; Aherfi, S. SARS-CoV-2 testing of aircraft wastewater shows that mandatory tests and vaccination pass before boarding did not prevent massive importation of Omicron variant in Europe. Viruses 2022, 14, 1511. [Google Scholar] [CrossRef]
- Hayer, J.; Urlaub, E. Evaluation of the Roche SARS-CoV-2 Rapid Antibody Test in Samples from Vaccinated Individuals. Microbiol. Spectr. 2022, 10, e02709-21. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.; Tiu, C.; Hu, Z.; Chen, V.C.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef]
- Zhu, F.; Althaus, T.; Tan, C.W.; Costantini, A.; Chia, W.N.; Van Vinh Chau, N.; Van Tan, L.; Mattiuzzo, G.; Rose, N.J.; Voiglio, E.; et al. WHO international standard for SARS-CoV-2 antibodies to determine markers of protection. Lancet Microbe 2022, 3, e81–e82. [Google Scholar] [CrossRef]
- Colson, P.; Delerce, J.; Marion-Paris, E.; Lagier, J.C.; Levasseur, A.; Fournier, P.E.; La Scola, B.; Raoult, D. A 21L/BA.2-21K/BA.1 “MixOmicron” SARS-CoV-2 hybrid undetected by qPCR that screen for variant in routine diagnosis. medRxiv 2022. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Rambaut, A.; Holmes, E.C.; O’Toole, A.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Alm, E.; Broberg, E.K.; Connor, T.; Hodcroft, E.B.; Komissarov, A.B.; Maurer-Stroh, S.; Melidou, A.; Neher, R.A.; O’Toole, A.; Pereyaslov, D.; et al. Geographical and temporal distribution of SARS-CoV-2 clades in the WHO European Region, January to June 2020. Eurosurveillance 2020, 25, 2001410. [Google Scholar] [CrossRef]
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N.; Colson, P.; Chahinian, H.; La Scola, B.; Raoult, D. The puzzling mutational landscape of the SARS-2-variant Omicron. J. Med. Virol. 2022, 94, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Benton, D.J.; Wrobel, A.G.; Roustan, C.; Borg, A.; Xu, P.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2021, 118, e2022586118. [Google Scholar] [CrossRef]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Pruitt, K.D.; Schoch, C.L.; Sherry, S.T.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2022, 50, D161–D164. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N.; Azzaz, F.; Chahinian, H. Structural dynamics of SARS-CoV-2 variants: A health monitoring strategy for anticipating Covid-19 outbreaks. J. Infect. 2021, 83, 197–206. [Google Scholar] [CrossRef]
- Xin, H.; Wong, J.Y.; Murphy, C.; Yeung, A.; Ali, S.T.; Wu, P.; Cowling, B.J. The Incubation Period Distribution of Coronavirus Disease 2019: A Systematic Review and Meta-analysis. Clin. Infect. Dis. 2021, 73, 2344–2352. [Google Scholar] [CrossRef]
- Colson, P.; Levasseur, A.; Gautret, P.; Fenollar, F.; Thuan Hoang, V.; Delerce, J.; Bitam, I.; Saile, R.; Maaloum, M.; Padane, A.; et al. Introduction into the Marseille geographical area of a mild SARS-CoV-2 variant originating from sub-Saharan Africa: An investigational study. Travel Med. Infect. Dis. 2021, 40, 101980. [Google Scholar] [CrossRef] [PubMed]
- Pascarella, S.; Bianchi, M.; Giovanetti, M.; Narzi, D.; Cauda, R.; Cassone, A.; Ciccozzi, M. The SARS-CoV-2 Mu variant should not be left aside: It warrants attention for its immuno-escaping ability. J. Med. Virol. 2022, 94, 2479–2486. [Google Scholar] [CrossRef]
- Yadav, P.D.; Nyayanit, D.A.; Gupta, N.; Shastri, J.; Sahay, R.R.; Patil, D.Y.; Shete, A.M.; Razdan, A.; Agrawal, S.; Kumar, A.; et al. Detection and isolation of SARS-CoV-2 Eta variant from the international travelers and local residents of India. J. Med. Virol. 2022, 94, 3404–3409. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Mohsin, M.; Mahmud, S. Omicron SARS-CoV-2 variant of concern: A review on its transmissibility, immune evasion, reinfection, and severity. Medicine 2022, 101, e29165. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Makatini, Z.; Moyo, S.; Amoako, D.G.; et al. Continued emergence and evolution of Omicron in South Africa: New BA.4 and BA.5 lineages. MedRxiv 2022. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colson, P.; Lavagna, C.; Delerce, J.; Groshenry, G.; Yahi, N.; Fantini, J.; La Scola, B.; Althaus, T. First Detection of the SARS-CoV-2 Omicron BA.5/22B in Monaco. Microorganisms 2022, 10, 1952. https://doi.org/10.3390/microorganisms10101952
Colson P, Lavagna C, Delerce J, Groshenry G, Yahi N, Fantini J, La Scola B, Althaus T. First Detection of the SARS-CoV-2 Omicron BA.5/22B in Monaco. Microorganisms. 2022; 10(10):1952. https://doi.org/10.3390/microorganisms10101952
Chicago/Turabian StyleColson, Philippe, Christian Lavagna, Jérémy Delerce, Guillaume Groshenry, Nouara Yahi, Jacques Fantini, Bernard La Scola, and Thomas Althaus. 2022. "First Detection of the SARS-CoV-2 Omicron BA.5/22B in Monaco" Microorganisms 10, no. 10: 1952. https://doi.org/10.3390/microorganisms10101952
APA StyleColson, P., Lavagna, C., Delerce, J., Groshenry, G., Yahi, N., Fantini, J., La Scola, B., & Althaus, T. (2022). First Detection of the SARS-CoV-2 Omicron BA.5/22B in Monaco. Microorganisms, 10(10), 1952. https://doi.org/10.3390/microorganisms10101952





