Abstract
The human gut is host to almost 3000 microbial species, of which 90% are bacteria. Quorum sensing (QS) molecules generated by intestinal bacteria are important in establishing species- and strain-level structures within the gut microbiome but are also used to communicate with the host. Although we do not know which QS molecules have the most direct interaction with intestinal and sensory neurons, it is clear they affect our physiological and mental health. Signals produced by bacteria are diverse and include autoinducers (AIs), homoserine lactones (HSLs), quinolines, peptides, toxins and proteases. These signaling molecules activate specific receptors in the bacterial cell wall and trigger sensors in the cytoplasm that regulate gene expressions. A better understanding of the gene structures encoding the production of QS molecules is of importance when selecting strains with neurogenerative and other probiotic properties. Furthermore, QS molecules may be used as biomarkers in the diagnosis of inflammable bowel disease (IBD), irritable bowel syndrome (IBS) and colorectal cancer (CRC). In the future, it should be possible to use QS biomarkers to diagnose neurological and psychiatric diseases such as anxiety and depression, major depressive disorder (MDD), schizophrenia, bipolar disorder, autism and obsessive-compulsive disorder (OCD).
1. Introduction
In a highly competitive and ever-changing environment such as the gastrointestinal tract (GIT), microbiota have developed unique methods to communicate with each other. Quorum sensing (QS) molecules produced by the gut microbiota regulate a variety of cell functions such as expression of virulence genes, formation of biofilms, competence and sporulation and usually only initiate these processes when cell numbers reach a certain density [1,2,3]. These signals include autoinducers (AIs) such as AI-1, AI-2 and AI-3; 3-oxo-C12-homoserine lactone (3-oxo-C12-HSL); C4-homoserine lactone (C4-HSL); 2-heptyl-3-hydroxy-4(1H)-quinoline; 2-heptyl-4-hydroxyquinoline (HHQ); QS peptides (QSPs); peptide pheromone Agr; pore-forming toxins such as hemolysins, leucocidins and phenol-soluble modulins (psms) and proteases. Epinephrine (Epi), norepinephrine (NE), autoinducer 3 (AI-3), fucose, ethanolamine (EA) and vitamin B12 activate specific receptors in the bacterial cell wall and trigger sensors in the cytoplasm that regulate gene expressions. Conversation between microorganisms varies and ranges from interspecies communication, self-talk or intraspecies communication to cells from one genus responding to signals generated by another genus. Cells that are unable to produce their own communication signals are “listening” to signals generated by other cells, a phenomenon referred to as “eavesdropping” [4]. The gut microbiota uses certain metabolites as QS molecules to communicate with intestinal epithelial cells (IECs). Staphylococcus aureus, for instance, secretes a variety of virulence factors that manipulate the immune system of the host to safeguard its own survival [5]. The effect of these survival strategies on the host often manifests in the form of immune misfunctioning, neurological disorders, diarrhea and vast changes in the gut microbiome [6].
Microbial QS could be seen as a partnership or agreement amongst microbiota and, in the GIT, between the gut microbiota and the host. This requires microbiota and the host to develop specific adaptation strategies. In a complex environment such as the GIT with close to 3000 bacterial species, strains are in constant survival mode and produce an arsenal of inorganic and organic molecules participating in inter-microbial and inter-host communications. This review addresses changes brought about by QS amongst intestinal bacteria and the gut wall. The impact these communications may have on the central nervous system (CNS) and mental health is summarized.
2. Interbacterial Communication
2.1. Gram-Negative Bacteria
Gram-negative bacteria use small molecules as autoinducers (AIs) that either target transcription factors or transmembrane two-component histidine sensor kinases [7]. Amongst these, N-acyl-homoserine lactone (AHL), a small neutral lipid molecule with a homoserine lactone (HSL) moiety linked to a 4 to 18 carbon acyl side chain [8], is the best studied. AHL is synthesized from S-adenosylmethionine (SAM), catalyzed by either LuxI or LuxM synthetases (Figure 1, left image) [9]. Not all AHLs are the same. The length of the acyl side chain and substituents at the third position of the acyl chain differ and allow a LuxR-type receptor to discriminate between signals [10]. Furthermore, some species have a single AHL synthase enzyme and produce predominantly one type of AHL, whereas others have multiple AHL synthases and produce several forms of AHL [11]. The level at which AHLs are produced depends on the availability of substrates and is tightly controlled [11]. Bacteria lacking the LuxI-type synthase have orphan or “solo” LuxR-type receptors that respond to AHLs produced by other bacterial species in the same environment. SdiA (a LuxR homolog) in Escherichia coli and QscR in Pseudomonas aeruginosa are examples of such orphan LuxR-type receptors (Figure 1). These receptors are highly conserved with a 67–84% sequence identity [12] and are also found in Enterobacter, Citrobacter, Cronobacter, Klebsiella, Salmonella and Shigella [13].
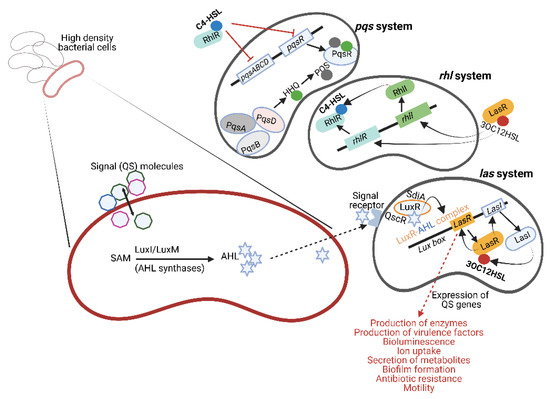
Figure 1.
Quorum sensing (QS) molecules produced by Gram-negative bacteria. Acyl-homoserine lactone (AHL) is produced from S-adenosylmethionine (SAM) by AHL synthase (shown on the left). LuxI-type synthases are major producers of AHLs. LuxM synthase, described for Vibrio harveyi, is important in intra-species communication. Signal receptors on the surface of the responding bacterial cell recognize AHL, form a LuxR–AHL complex and induce genes in the Lux box to produce 3-oxo-C12-homoserine lactone (3-oxo-C12-HSL, abbreviated as 3OC12HSL in this figure to conserve space). This is referred to as the las system. In the absence of LuxI synthases, LuxR proteins detect different AHL molecules produced by other bacterial species. Examples of these receptors are SdiA (LuxR homolog) in E. coli and QscR in P. aeruginosa. Pseudomonas aeruginosa has three major QS systems, namely las, rhl and pqs (shown on the right), that mediate cell-to-cell communication and control the synthesis and secretion of virulence factors, bioluminescence, biofilm formation, etc. Additionally, 3-oxo-C12-HSL may also induce the rhl system to produce C4-homoserine lactone (C4-HSL). The pqs system uses two signal molecules, i.e., 2-heptyl-3-hydroxy-4(1H)-quinoline (also referred to as Pseudomonas quinolone signal or PQS) and its biosynthetic precursor 2-heptyl-4-hydroxyquinoline (HHQ). This illustration was constructed using BioRender (https://biorender.com/, assessed on 27 September 2022).
Five QS systems have been described for pathogenic E. coli, i.e., AI-2 signaling produced by the enzyme LuxS, SdiA signaling that suppresses cell division, AI-3/Epi/NE signaling involved in host–bacteria communication, indole signaling and extracellular death factor (EDF) signaling that triggers the activation of toxin–antitoxin systems [14]. SdiA of E. coli (SdiAEC) is activated by AHLs produced by P. aeruginosa [15]. The SdiAEC/AHL complex increases the transcription of genes in the gad operon (gadW, gadE, yhiD and hdeA) of E. coli [16] that encodes an acid resistance system critical to the survival of enterohemorrhagic E. coli (EHEC) in a low-pH environment [17].
E. coli uses QS to regulate virulence genes, biofilm formation, mobility, the type III secretion system (T3SS), toxicity and the production of curli [18]. QS systems in Salmonella regulate the pathogenicity island SPI-1 (invasion), the expression of genes encoding flagella formation and the pefI-srgC plasmid operon regulating the genes rck (resistance to complement killing) and srgE (sdiA-regulated gene E) involved in the Zipper invasion mechanism [19,20,21].
Pseudomonas aeruginosa has three major QS systems, namely las, rhl and pqs (Figure 1, right image), involved in cell-to-cell communication, control over synthesis and secretion of virulence factors, bioluminescence, biofilm formation, etc. (listed in Figure 1). The las system regulates both the rhl and pqs systems by initiating the expression of the AI receptors RhlR and PqsR. These receptors also act as transcriptional activators when bound to their respective AIs. Freely diffusible AHL communicates with other bacterial cells or binds to LuxR-type receptors in the cytoplasm of producing cells to form stable LuxR–AHL complexes [9], as depicted in Figure 1. These LuxR–AHL complexes bind to the Lux box (las system, Figure 1) and regulate the expression of QS genes [22,23,24]. Mutants without the lasR gene (ΔlasR) are more motile, survive stationary growth much better and produce higher levels of β-lactamase and pyocyanin [25,26]. However, ΔlasR mutants produce fewer exoenzymes and elicit a higher immune response in host cells, as evident from an increase in the secretion of pro-inflammatory cytokines and neutrophil recruitment [27]. Complexes similar to the LasR/LasI system, e.g., RhlR/RhlI (rhl system, Figure 1) described for P. aeruginosa, produce and detect C4-homoserine lactone (C4-HSL) [28,29]. In the absence of LuxI synthases, LuxR proteins may bind to AHLs produced by other bacterial species and initiate interspecies communication [30,31,32]. The pqs system (Figure 1) employs two signal molecules, i.e., 2-heptyl-3-hydroxy-4(1H)-quinoline (also referred to as Pseudomonas quinolone signal or PQS) and its biosynthetic precursor 2-heptyl-4-hydroxyquinoline (HHQ).
2.2. Gram-Positive Bacteria
Gram-positive bacteria communicate using small linear or cyclized oligopeptides (QS peptides, QSPs) consisting of 5 to 17 amino acids [2,28,33,34]. The most studied QS systems are those produced by Bacillus, e.g., the competence sporulation factor (CSF), a pentapeptide, and the heptapeptide SDLPFEH (PapRIV). The heptapeptide forms after cleaving of the inactive 48-amino-acid pre-peptide by NprB protease [35,36].
QSPs are extracellularly secreted with the assistance of ATP-binding cassette transporters located in the cell membrane (Figure 2) and interact with either membrane-located receptors or cytoplasmic sensors such as the proteins Rap, NprR, PlcR and PrgX [33]. In the case of Staphylococcus aureus, the accessory gene regulator (agr), a four-gene operon encoding the peptide pheromone Agr, serves as the membrane-bound sensor. Agr regulates the expression of several genes, including virulence factors such as formylated peptides, proteases and pore-forming toxins (PFTs) such as hemolysins, leucocidins and phenol-soluble modulins (psms) [37]. Strains of S. aureus lacking the agr gene (Δagr) form biofilms and are more prone to causing chronic infections and bacteremia [38,39,40]. Enterococcus faecalis uses the Fsr-QS system, which is controlled by the four-gene locus fsrABDC [41]. Once cleaved, the activated peptide is intracellularly transported using a transmembrane kinase (Figure 2). A cascade of phosphorylation reactions excites the peptide and induces the expression of target genes (Figure 2). For more information on the chemical characteristics and microbial background of QSPs, the reader is referred to the Quorumpeps® database [42].
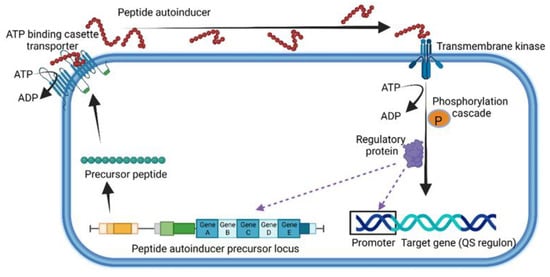
Figure 2.
General representation of quorum sensing used by Gram-positive bacteria. This illustration was constructed using BioRender (https://biorender.com/, assessed on 1 September 2022).
3. Interspecies Communication
Autoinducer-2 (AI-2), a furanosyl borate diester produced by Gram-negative and Gram-positive bacteria [43], plays a key role in interspecies communication and the altering of specific behavior such as virulence, luminescence and biofilm formation [44,45,46,47]. The AI-2 system is also used by the gut microbiota to overcome stressful conditions in the GIT [48,49]. Production of AI-2 is regulated by the luxS gene (Figure 3). S-adenosylhomocysteine (SAH) is converted to homocysteine by SAH hydrolase (SahH) in a one-step reaction but may also be produced from the cleavage of the thioether linkage of S-ribosylhomocysteine (SRH). This is a two-step reaction that requires SAH nucleosidase (Pfs) and LuxS. The intermediate, 4,5 dihydroxy-2,3-pentanedione (DPD), is rearranged to form AI-2 (Figure 3) [50].
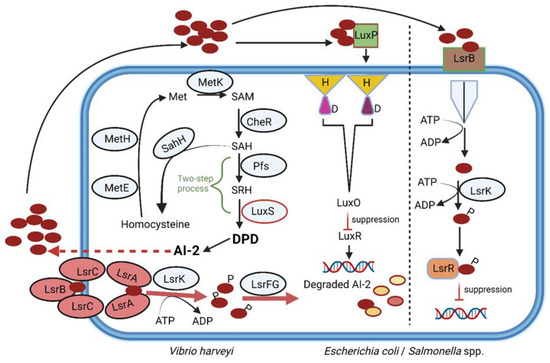
Figure 3.
Production of AI-2, regulated by LuxS (an autoinducer synthase). S-adenosylhomocysteine (SAH) is converted to homocysteine by SAH hydrolase (SahH) in a one-step reaction but may also be produced from the cleavage of the thioether linkage of S-ribosylhomocysteine (SRH). This is a two-step reaction that requires SAH nucleosidase (Pfs) and LuxS. The intermediate 4,5 dihydroxy-2,3-pentanedione (DPD) is rearranged to form AI-2. Red circles = AI-2; LsrB-LsrC-LsrA = Lsr ABC-type transporter; LsrK = Lsr kinase; LsrFG = genes F and G, part of the Lsr operon; MetE = cobalamin-independent methionine synthetase; MetH = cobalamin-dependent methionine synthetase; MetK = adenosylmethionine synthetase; CheR = methyltransferase; P = phosphate group; LuxO = a central regulator; LuxR = repressor. This illustration was constructed using BioRender (https://biorender.com/, assessed on 5 September 2022).
Genes encoding homologues of luxS have been detected in more than a third of bacterial genomes, including Escherichia coli, Enterococcus faecalis, Campylobacter jejuni, S. aureus, Clostridium difficile, Bacillus spp., Streptococcus spp., Shigella flexneri, Helicobacter pylori, Salmonella enterica serotype Typhimurium, S. enterica serotype Typhi [43,51,52], Bifidobacterium [53], Lactobacillus [54,55], Eubacterium, Roseburia and Ruminococcus [56]. Strains of E. coli [57], Streptococcus pneumoniae [58], Streptococcus mutans [59,60] and Lactobacillus [61,62] use the luxS system to regulate genes encoding bacteriocin production. The same group of signaling factors is also used by Bifidobacterium to combat Salmonella infections [63]. An engineered strain of E. coli with increased production of AI-2 led to the reinstatement of streptomycin-repressed Firmicutes and suppressed the growth of Bacteroidetes [64]. It can be deduced from these findings that AI-2 may be used to restore balance in the gut microbiota after antibiotic treatment. Should this strategy be followed, it will have to be carefully controlled, as cytoplasmic levels of AI-2 are regulated by LsrK kinase (Figure 3). Mutants of E. coli with an inactive LsrK kinase were unable to phosphorylate AI-2 and lost their QS communication abilities [47,65]. However, when co-cultured with an AI-2-producing strain of Vibrio harveyi, the E. coli mutant responded to its own AI-2 and to that produced by V. harveyi. An increase in the adherence of Actinobacillus pleuropneumoniae to epithelial cells and an increase in the expression of motility genes in E. coli were observed in the presence of AI-2 [66,67]. In the case of Helicobacter pylori, AI-2 acted as a chemorepellent and prevented biofilm formation [68]. AI-2 may have the same anti-biofilm forming effect on Bacteroidetes, which would explain the decline in cell numbers as Firmicute numbers increase.
Changes in the populations of Firmicutes and Bacteroidetes alter the level and composition of SCFAs, which in turn affect gene expressions, cytokine secretion and regulatory T cell induction [69]. All these changes influence inflammatory responses. Increased levels of AI-2 could thus restore the balance between Firmicutes and Bacteroidetes and prevent, or revert, dysbiosis, IBD, obesity, autism and stress-related disorders. However positive this seems, the idea must be considered with care, as elevated levels of AI-2 may upregulate virulence, as shown with an increase in the release of bacteriophages from Enterococcus faecalis and an increase in gene transfer [70]. In mice, the administration of AI-2 had no effect on the expression of cytokines but aggravated P. aeruginosa lung infection by interfering with QS molecules produced by the pathogen [71]. These findings clearly show that implementing AI-2 aimed at maintaining gut homeostasis is far more complex and warrants further research.
Two classes of AI-2 receptors have been identified, namely LuxP, common among members of Vibrionales, and LsrB (Figure 3), widely distributed across Proteobacteria, Bacillus cereus and Bacillus anthracis [72,73,74]. The two receptors are structurally different and share a sequence similarity of only 11% [75]. Other members of Firmicutes, including gut microbiota, may respond to AI-2 using receptors similar to LuxP and LsrB. Streptococcus mutans and Staphylococcus epidermidis, however, respond to AI-2 in the absence of these receptors [43]. The LuxS/AI-2 QS system regulates the expression of several genes, including drug resistance [50]. The effect of AI-2 on the host’s immune system is less understood. Elevated levels of AI-2 were detected in tumors associated with colorectal cancer (CRC) [76]. This correlated with an increase in the expression of genes encoding TNFSF9 (tumor necrosis factor ligand superfamily member 9), as noted in tumor-associated macrophages [76]. AI-2 could thus be an important marker for CRC and warrants more research.
4. Interkingdom Communication
The autoinducer-3 (AI-3)/Epi/NE interkingdom signaling system [77] promotes the expression of virulence genes in pathogens such as S. typhimurium, Citrobacter rodentium and EHEC [78]. AI-3 controls the genes encoding the attachment of EHEC to epithelial cells in the colon (Figure 4), a process leading to the destruction of microvilli and the rearrangement of the cytoskeleton to form protective pedestal-like structures [79]. The direct effect of AI-3 on humans is unknown, apart from an increase in IL-8 production by THP-1 monocytes [80]. Epi and NE recognize the AI-3 receptor (Figure 4) [79] but do not activate or modulate adrenergic signaling [80]. Further research on microbial endocrinology is required to understand the effect of variations in AI-3 levels on the host.
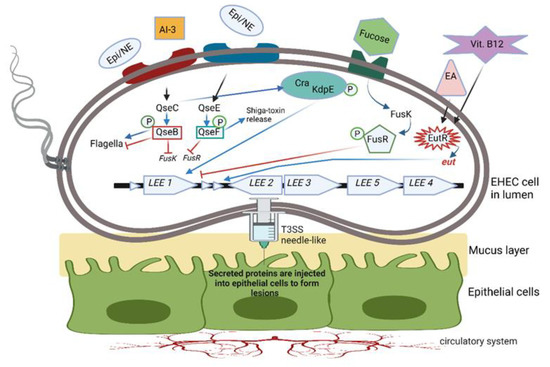
Figure 4.
Two-component QS system (TCS) used by enterohemorrhagic E. coli (EHEC). Bacterial cells sensing environmental signaling compounds such as epinephrine (Epi), norepinephrine (NE), autoinducer 3 (AI-3), fucose, ethanolamine (EA) and vitamin B12 activate transmembrane histidine kinase receptors (indicated here as blue and red rectangles). Response regulators either activate or repress the TCS. QseC histidine sensor activates QseB, and QseE activates QseF. QseB regulates the expression of flagella and, at the same time, represses the expression of fusK/-R, involved in fucose metabolism, and the expression of virulence genes. QseC controls the response regulator KdpE, which, together with Cra, stimulates genes in the LEE operon to form microscopic T3SS needle-like structures through which proteins are injected into the host cell. Microvilli on the surface of epithelial cells are eradicated, and lesions with actin-rich pedestals form, onto which EHEC cells attach. QseC also activates regulator QseF, which stimulates the production of Shiga toxin. FusK is activated by fucose and phosphorylates FusR to inhibit LEE expression. Genes in the eut operon encode the EutR transcription factor, which recognizes the presence of ethanolamine (EA) and vitamin B12. Both compounds are required to promote the transcription of genes in the LEE operon. This illustration was constructed using BioRender (https://biorender.com/, assessed on 3 September 2022).
Intestinal pathogens such as EHEC, Salmonella typhimurium and Citrobacter rodentium regulate virulence by using a two-component QS system (TCS). In the case of EHEC, the TCS consists of the quorum-sensing E. coli regulators QseBC and QseEF (Figure 4). QseB is a response regulator with a receiver domain and a helix-turn-helix (HTH) DNA binding domain, QseC functions as a bacterial adrenergic receptor, QseE is a sensor kinase and QseF is a response regulator [81]. Cells sensing environmental signaling compounds such as Epi, NE, AI-3, fucose and ethanolamine (EA) respond by activating transmembrane histidine kinase receptors (shown as blue and red rectangles in Figure 4). Response regulators either activate or repress the TCS. The QseC histidine sensor activates QseB, which regulates the expression of flagella and, at the same time, represses the expression of fusK/-R, encoding fucose metabolism and the expression of virulence genes. QseC can also phosphorylate the response regulator KdpE, which, together with Cra, stimulates genes in the LEE operon to encode the formation of microscopic “needles” through which proteins are injected into the host cell. At the same time, microvilli on the surface of epithelial cells are eradicated, and lesions with actin-rich pedestals form, onto which EHEC cells attach (not shown in Figure 4). QseC may also activate the regulator QseF, which stimulates the production of Shiga toxin. FusK is activated by fucose and phosphorylates FusR to inhibit LEE expression.
The 3-oxo-C12-HSL produced by P. aeruginosa (Figure 5) is actively transported across epithelial and immune cells [2,82] and destroys the permeability of the gut wall by repressing the expression of genes encoding tight junction proteins (TJs). This leads to the re-arranging (misplacing) of occludin, tricellulin, ZO-1, ZO-3, JAM-A, E-cadherin and β-catenin and prevents mucin production [2,83,84]. This not only exposes epithelial cells to infection but also activates the mucosal immune system, leading to an increase in leucocytes and the accumulation of pro-inflammatory cytokines [85]. Furthermore, 3-oxo-C12-HSL also inhibits tumor necrosis factor (TNF)-α and IL-12 production, causes malfunctioning of the T helper cell-1 (Th1) response and stimulates Th2 to produce immunoglobulin G1 [86]. Inhibition of Th1 and Th2 T lymphocyte differentiation increases cytokine production [87], intensifies oxidative stress, stimulates apoptosis and inactivates mitochondria [2].
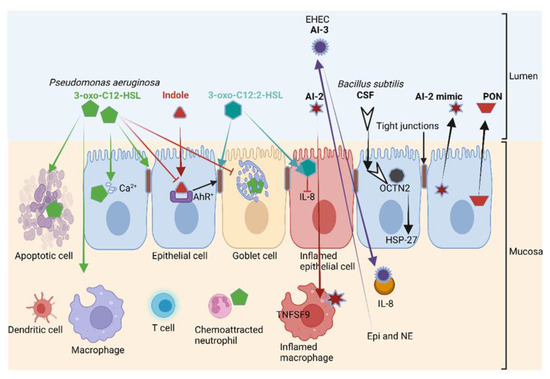
Figure 5.
The 3-oxo-C12-HSL produced by P. aeruginosa (also shown in Figure 1) induces apoptosis in various cell types, including epithelial cells; disrupts tight junctions and decreases mucin production. Intestinal acyl-homoserine lactone (3-oxo-C12:2-HSL) and indole (produced from tryptophan) protect tight junctions. The competence and sporulation factor (CSF), a pentapeptide produced by B. subtilis, binds to the cation transporter OCTN2 and activates heat shock protein 27 (HSP-27), which increases the strength of intestinal barriers. Meanwhile, 3-oxo-C12-HSL stimulates chemoattraction and phagocytosis in neutrophils and induces cell death. The autoinducers AI-2 and AI-3 induce the expression of the immune mediators TNSF9 and interleukin (IL)-8, respectively, leading to the inflammation of macrophages, while 3-oxo-C12:2-HSL reduces IL-8 production. Epinephrine (Epi) and norepinephrine (NE) bind to the AI-3 receptor in enterohemorrhagic E. coli (EHEC), thereby interfering with QS. Intestinal epithelial cells secrete a mimic form of AI-2 and paraoxonase (PON) to degrade HSLs. AhR: Aryl hydrocarbon receptor. This illustration was constructed using BioRender (https://biorender.com/, assessed on 6 September 2022).
A structurally similar form of 3-oxo-C12-HSL, 3-oxo-C12:2-HSL, has the opposite effect on the gut wall. Instead of destabilizing epithelial cells, 3-oxo-C12:2-HSL protects the tight junction proteins occludin and tricellulin and cytoplasmic ZO-1 from pro-inflammatory cytokines such as interferon-gamma (IFN-γ), TNF-α and IL-8 [88,89,90,91]. Apart from a few pioneering studies, the impact of 3-oxo-C12:2-HSL on immune cells in the human intestinal tract remains largely unknown. Landman et al. [88] reported much lower concentrations of 3-oxo-C12:2-HSL in patients diagnosed with IBD. This suggests that 3-oxo-C12:2 HSL plays a major role in the protection of epithelial cells exposed to an immune onslaught. Further research is required to determine if 3-oxo-C12:2-HSL could be used in the treatment of IBD. This also requires a better understanding of the processes involved in 3-oxo-C12:2-HSL quorum quenching, the cleaving of AHL and the hydrolysis of the homoserine lactone (HSL) ring. Thus far, three paraoxonases (PON1, PON2 and PON3) involved in the hydrolysis of the HSL ring have been identified in the GIT of humans and other mammals [92]. Of these, PON2 is the most active [93] and is predominantly expressed in the jejunum [94]. PON1 and PON3 are expressed at lower levels in patients diagnosed with Crohn’s disease and ulcerative colitis [95]. It is thus possible that these gastrointestinal disorders may be reversed by reinstating PON1 and PON3 levels. An in-depth study on the role paraoxonases play in different areas of the GIT, and their possible application in the treatment of gastric disorders, is required.
Other examples of intestinal receptors interacting with bacterial cells or bacterial compounds are pregnane X receptors (PXR1 and PXR2) and peroxisome proliferator-activated receptors (PPARα, PPARβ/δ and PPARγ). Pregnane X receptors (PXRs) are primarily expressed by intestinal epithelial cells, but also to a lesser extent by kidney cells, T cells, macrophages and dendritic cells [96], and regulate the expression of proteins involved in detoxification and the metabolism of glucose, lipid, cholesterol and bile acid [97]. Peroxisome proliferator-activated receptors (PPARs) are widely expressed in human cells, including intestinal cells [98], and regulate energy production, lipid metabolism and inflammation. PPARα represses nuclear factor kappa B (NF-κB) signaling, which decreases the production of inflammatory cytokines [99,100]. PPARγ inhibits the activation of macrophages and the production of inflammatory cytokines such as tumor necrosis factor-alpha (TNF-α), interleukin (IL)-6 and IL-1β. These anti-inflammatory responses may restore gut dysbiosis and alleviate IBDs such as ulcerative colitis and Crohn’s disease [101].
PQS and HHQ produced by Pseudomonas (Figure 1) interact with lymphoid cells, dendritic cells and macrophages, leading to the suppression of innate and adaptive immune responses [102,103]. In response, the aryl hydrocarbon receptor (AhR) senses the PQS signals and alerts the immune system to activate the most beneficial immune response [104,105]. This involves the expression of IL-22 and IL-17 [106]. Activation of AhR also stimulates the p38 pathway, which initiates the apoptosis of epithelial cells [107]. This is an excellent example of “eavesdropping” on an interkingdom level.
CSF, produced by Bacillus subtilis, binds to the cation transporter OCTN2 (Figure 5). This activates HSP-27, mediates the uptake of organic cations and carnitine and promotes intestinal barrier integrity [108]. Once in the cell, CSF acts as a reporter monitoring changes in the behavior or composition of the gut microbiota [109]. HSP-27 acts as a protein chaperone and an antioxidant but also facilitates the refolding of damaged proteins, thus preventing apoptosis and actin cytoskeletal remodeling [2,110]. The autoinducers AI-2 and AI-3 promote the expression of the immune mediators TNSF9 and IL-8 (Figure 5), and 3-oxo-C12:2-HSL reduces the production of IL-8 by epithelial cells. Host cells retaliate against QS by the binding of Epi and NE to AI-3 receptors (Figure 5). Epithelial cells then secrete AI-2 and PON that degrade HSLs (Figure 5).
PapRIV, produced by Bacillus, crosses the gastrointestinal tract (GIT), albeit slowly, and enters the circulatory system from where most peptides (87%) cross the BBB in a one-directional way. It can be deduced from in vitro studies that PapRIV activates microglia and may thus play a role in gut–brain interactions [111]. According to Yorick Janssens et al. [111], the second (aspartic acid) and the fourth (proline) amino acids play a key role in the activation of microglia. PapRIV also induces the production of the pro-inflammatory cytokines IL-6 and TNFα, increases intracellular ROS and stimulates an increase in ameboid cells [112]. Autoinducer peptides (AIPs) produced by Clostridium acetobutylicum cross the BBB much more easily than AIPs produced by Streptococcus pneumonia [113]. AIPs produced by Gram-positive bacteria crossing the gut wall have been shown with in vivo studies on Caco-2 cells [102,103]. De Spiegeleer et al. [114] have shown that AIPs produced by Staphylococcus, Streptococcus, Lactobacillus and Bacillus in the GIT have pro- and anti-inflammatory effects on muscle cells. The crossing of these barriers seems to depend on the structure and size of the peptide. Small diffusible molecules produced during the degradation of signal peptides, referred to as diffusible signal factors (DSFs), may also act as autoinducers [115,116].
Signals generated by gut bacteria are recorded by specialized cells in the gut wall (Figure 6), resulting in temporary or long-lasting changes in physical or mental health. These cells differentiate between signals produced by autochthonous (endemic) and foreign, potentially pathogenic, microbiota by using pattern recognition receptors (PRRs). The pro-inflammatory properties of AHLs, associated with an increase in neutrophil activity and the differentiation of fibroblasts into myofibroblasts, are crucial for tissue regeneration [117,118]. These onslaughts on the immune system are driven by systems independent of pathogen-associated molecular pattern (PAMP) recognition pathways, TLRs and the nucleotide-binding oligomerization domain proteins Nod1 and Nod2 [99]. Toll-like receptors (TLRs) and Nod-like receptors (NLRs) specialize in the recognition of microbial cell wall components, and G protein-coupled receptors (GPRs) activate G proteins involved in hormonal regulation [119,120] (Figure 6). GPRs are localized on the surface of intestinal immune cells, leukocytes, regulatory T cells, monocytes, macrophages and colonic epithelial and mesenchymal cells, thus playing a role in anti-inflammatory and pro-inflammatory processes and barrier control [121] (Figure 6).
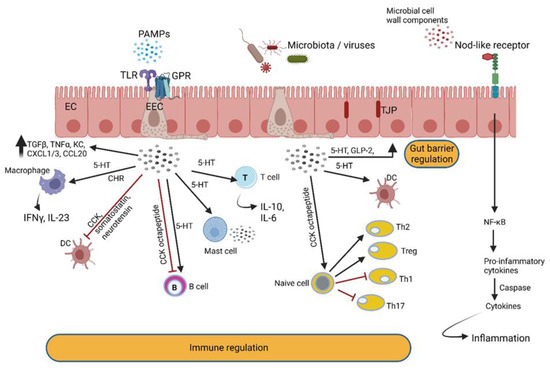
Figure 6.
Enteroendocrine cells (EECs) in the gut wall detect intestinal bacteria and microbial metabolites and react by secreting peptide hormones and cytokines that react with immune cells. Hormones produced by EECs modulate intestinal barrier functions and react with enteric nerves. The latter communicate with the central nervous system via the vagus nerve. PAMPs = pathogen-associated molecular patterns; TLR = Toll-like receptor; GPR = G protein-coupled receptor; TGF = tumor growth factor; TNF = tumor necrosis factor; KC = neutrophil chemokine; CXLC = chemokine (C-X-C motif) ligand; CCL = chemokine ligand; IFN = interferon; IL = interleukin; CCK = cholecystokinin; DC = dendritic cell; 5-HT = serotonin; Th = T helper cell; Treg = regulatory T cell. This illustration was constructed using BioRender (https://biorender.com/, assessed on 6 September 2022).
AhRs regulate immune responses and pathogenesis (Figure 7) [122]. Large quantities of AhR are expressed by intestinal epithelial cells and immune cells such as innate lymphoid cells, intraepithelial lymphocytes, TH17 cells and Treg cells but are also present in the liver, lung, bladder and placenta [123,124,125]. One of the key functions of AhR is restoration of barrier homeostasis, a phenomenon eminent in IBD [7].
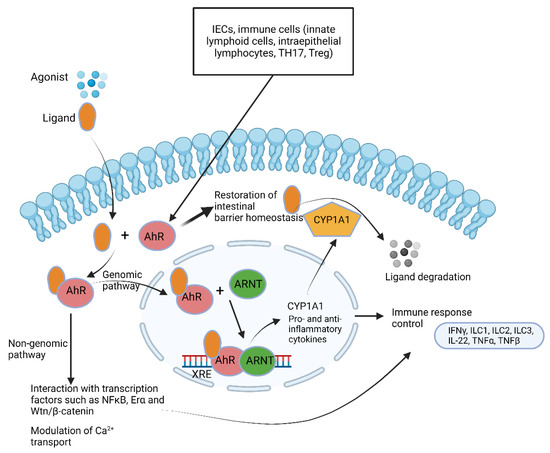
Figure 7.
Summary of aryl hydrocarbon receptor (AhR) pathways. AhR ligands such as 6-formylindolo [3,2-b]carbazole; 2,3,7,8-tetrachlorodibenzo-p-dioxin and polycyclic aromatic hydrocarbons cross the cytoplasmic membrane and bind to AhR. The ligand–AhR complex crosses into the nucleus and forms a heterodimer with the AhR nuclear translocator (ARNT). Binding of the ligand–AhR–ARNT complex with xenobiotic response elements (XREs) leads to the expression of cytochrome P450, family 1, subfamily A, polypeptide 1 (CYP1A1). CYP1A1 degrades AhR ligands. This leads to the induction of several genes encoding pro-inflammatory and anti-inflammatory cytokines. The non-genomic pathway involves interaction between AhR and other transcription factors and modulates Ca2+ transport. This illustration was constructed using BioRender (https://biorender.com/, assessed on 6 September 2022).
AhR is activated by metabolic compounds produced from the bacterial degradation of tryptophan [126] but also by 2,4-dihydroxyquinoline (2,4-DHQ) and quinolone derivatives [127], pyocyanin, 1-hydroxyphenazine, phenazine-1-carboxylic acid and phenazine-1-carboxamide [128]. Tryptophan degradation products such as indole, indolo [3,2-b]carbazole, indole acetic acid (IAA), 3-methylindole and tryptamine react with AhR and stimulate the production of interleukin-22 [126,129,130]. Individuals with inflammatory bowel disease (IBD) [131], metabolic syndrome [132] or celiac disease [133] have decreased fecal concentrations of AhR ligands and reduced AhR activity. Higher indole concentrations were detected in patients suffering from Clostridioides difficile (Clostridium difficile) infection (CDI) compared to healthy individuals [134]. Elevated levels of indole produced by enterotoxigenic E. coli stimulated the colonization of C. difficile whilst other intestinal bacteria were repressed [134]. When indole was administered to GF mice, the expression of genes encoding tight junction proteins increased and improved the resistance of epithelial cells to colitis [135]. Since Trp is not synthesized by gut microbiota or the host, indole levels are directly linked to diet. Roasted cashew nuts, sunflower seeds, cheddar cheese, chicken breast and boiled eggs are rich in Trp [136].
The importance of AhR cannot be overemphasized as it plays a crucial role in barrier integrity; intestinal and immune homeostasis, mostly due to the regulation of tight junction proteins; the generation and survival of intraepithelial lymphocytes (IELs); the production of IL-22 and IL-10; the regulation of peristalsis and microbiota density; the regulation of goblet cell differentiation in the colon, specifically preventing goblet cell depletion in the elderly [2,122,127,137]; and the stimulation of antimicrobial peptide production via IL-22 [138,139,140,141,142]. Individuals with IBD and celiac disease have low levels of AhR in their feces [133]. Similarly, patients with Crohn’s disease have decreased levels of AhR, especially in the ileum, and have the tendency to convert ILC3 to type 1 innate lymphoid cells (ILC1) [143,144]. ILC1 elicits the production of IFNγ and TNFα, which are both known to induce apoptosis of epithelial cells in vitro, and most likely also in vivo, disrupting the intestinal barrier and increasing microbial-driven inflammation [145]. LC1 also induces TGFβ, which is associated with tumor formation and colorectal cancer (CRC) [145]. Further research is required to have a better understanding of the cross-talk between host AhR and bacterial QS molecules.
5. Can QS Be Used to Control Microbial Infections?
Five years ago, the World Health Organization (WHO) published a list of pathogenic bacteria most resistant to currently used antimicrobials. Acinetobacter baumannii, P. aeruginosa and enterobacteria resistant to carbapenems and species producing extended spectrum beta-lactamases (ESBLs) were amongst the top ones on the list [146]. This urged many scientists to investigate the possibility of using anti-QS therapy, referred to as quorum quenching (QQ), to prevent or control bacterial infections [147]. In recent years, many published articles have reported promising results indicating the possibility of reducing the pathogenicity of microorganisms and easier eradication when co-treated with antibiotics. Wu et al. [148] suggested that AHL-based QS may be used to control infections caused by Gram-negative bacteria. This approach is only possible where disrupting QS has a major effect on the expression of virulence genes [149]. Limited successes were reported using QQ in the treatment of infections caused by P. aeruginosa and S. aureus [150], notably concerning biofilm-associated infections [151]. The rationale behind this concept is to interrupt receptor proteins, degrade autoinducing signals or inhibit the synthesis of QS signaling molecules [152,153,154,155]. Another approach is using synthetic compounds analogous to QS signaling molecules [156].
Van den Abbeele et al. [157] reported a decline of approximately 60% of mucosa-associated pathogens, mostly Clostridium spp., when QQ was applied. Although promising from a perspective of infection management, such drastic changes may lead to the development of pro-inflammatory diseases such as cystic fibrosis [158], sclerosis [159] and IBD [160,161] and an increase in Enterococcus and C. difficile cell numbers [162]. Perhaps the most alarming of all is evidence of increased cell aggregation and biofilm formation in bacteria with a dysfunctional or absent luxS QS system, as reported for Helicobacter pylori [163], Vibrio cholerae [164], Aggregatibacter actinomycetemcomitans [165], Actinobacillus pleuropneumoniae [166], Haemophilus parasuis [167], S. aureus [168], S. epidermidis [169], Streptococcus mutans [170], Enterococcus faecalis [171] and Bacillus cereus [172]. Meropenem and levofloxacin stimulated the expression of an efflux pump in A. baumannii that promoted the release of an AHL, resulting in an increase in QS-mediated biofilm formation [173]. Ciprofloxacin, ceftazidime and azithromycin prevented QS when used at sub-inhibitory concentrations [174]. Despite these challenges and limitations, QQ may still be the answer to controlling bacterial infections [147,153,154,155,175,176,177], especially those caused by multi-drug-resistant (MDR) and extensively resistant microorganisms (XDR) [178]. However, with the variable reactions of pathogens to QQ therapy combined with antibiotics, the treatment of infections will have to be evaluated on a case-to-case basis.
Strains may develop resistance to QS inhibitors, as shown for P. aeruginosa exposed to brominated furanones. Resistant cells had mutations in genes encoding efflux pumps [179]. Strains of P. aeruginosa resistant to carbapenems and azithromycin lost antibiotic-associated QS inhibition [180]. Streptococcus pyogenes and S. aureus mutants without LuxS (ΔluxS) were more resistant to macrophages [181,182]. This may lead to the development of persistent pathogens difficult to eradicate. For further information on resistance to QQ, the reader is referred to Defoirdt et al. [183], García-Contreras et al. [179], Kalia et al. [184] and Liu et al. [185].
In the case of S. aureus, the accessory gene regulator (agr) regulates the expression of several genes, including virulence factors such as formylated peptides, proteases and pore-forming toxins (PFTs) such as hemolysins, leucocidins and phenol-soluble modulins (psms) [38]. Strains of S. aureus lacking the agr gene (Δagr) are more prone to causing chronic infections and bacteremia [38,39]. These findings suggest that treatment of S. aureus infections with QQ is not an option.
Pseudomonas aeruginosa uses psms to alter cell membrane properties [186] and activate the immune system [187]. These amphipathic peptides lyse neutrophils, erythrocytes and T cells [186]. By using QS inhibitors, the number of microbial enzymes degrading mediators of inflammatory responses may be limited. Since oxidative stress selects for cells with an active QS system [188], success in treatment using QQ techniques depends on the exposure of the infected area to oxygen and the overall immune status of the individual [189].
An interesting advance in QS research in the last decade is the discovery that QSPs may promote tumor cell invasion and angiogenesis (at least in vitro), suggesting that these peptides may stimulate stem cell differentiation and the migration of cancer stem cells [190,191]. The influence microbiota have over colon cancer stem cells, “instructing” them to become treatable or non-treatable, was raised by Trosko and Lenz [192].
6. Effect of QS on the CNS and Mental Health
The effect of QS molecules on the CNS is ill-researched. Several QSPs can diffuse through the intestinal mucosa and enter the circulatory system, from where they may penetrate the blood–brain barrier (BBB) [113]. Based on these findings, QSPs may play a key role in communication between the gut microbiome and the brain. If this is the case, QSPs may affect neurodevelopment and initiate neurodegenerative diseases. Further research is needed to confirm these findings.
Exotoxins produced by S. aureus activate the transcription factor accessory gene regulator (Agr)A, which regulates the expression of several genes, including virulence factors, pore-forming toxins (PFTs) and bacterial proteases [37]. These toxins increase intracellular calcium levels, leading to the activation of sensory neurons [193]. This is especially true for psms attached to formyl peptide receptor-like proteins (FPRs) [194]. The structural similarity of FPRs to b-defensins and ligands of mas-related G protein-coupled receptor (MRGPR) X2 [195] suggests that MRGPRs are involved in psm-mediated effects such as skin allergies [196]. The expression of FPRs in sensory and dorsal root ganglia of the colon has been well documented and linked with QS-dependent pathways involved in the gut–brain axis (GBA) [197,198]. The pore-forming toxin alpha-hemolysin (Hla) produced by S. aureus excites neurons by increasing the transfer of calcium [199]. According to Uhlig et al. [198], Hla produces smaller, less disruptive pores in cell membranes compared to psms [200]. The authors have also observed the expression of Adam10, a membrane-bound metalloprotease produced in sensory neurons, to which Hla binds [201]. The importance of exotoxins in GBA communication is unknown. However, since S. aureus is associated with irritable bowel syndrome and food [202,203], these QS molecules have the potential to directly modulate gut–brain communication and intestinal reflex.
Janssens et al. [204] screened 85 quorum sensing peptides on six different neuronal cell lines and found 22 peptides with a possible effect on the GBA. Of these, four peptides induced neurite outgrowth, two peptides inhibited nerve growth factor (NGF)-induced neurite outgrowth and eight peptides induced neurite outgrowth in human SH-SY5Y neuroblastoma cells. Two peptides killed SH-SY5Y cells and six peptides induced either IL-6 expression or nitric oxide (NO) production.
Several reports have been published on the role that cell wall components such as lipopolysaccharides, polysaccharides and peptidoglycans play in neuron activation and the GBA [37,205,206,207,208]. Cell wall components also induce the release of neuropeptides, ATP and cytokines [209]. Short-chain fatty acids, tryptophan, trace amines [142,210] and exotoxins [182] also have neuromodulator properties. Serotonin and histamine excite mast cells in the proximity of nerve endings [210,211].
Neuronal conditions such as Alzheimer’s disease (AD), autism spectrum disorder (ASD), multiple sclerosis (MS), Parkinson’s disease (PD) and amyotrophic lateral sclerosis (ALS) are associated with dysfunctional microglia [212]. Fecal transplants from humans with attention deficit hyperactivity disorder (ADHD), AD and PD to mice activated microglia in the brain and aggravated cognitive and physical impairments [213,214,215]. These findings along with more evidence of a clear link between microbial dysbiosis and neurodevelopmental, neurodegenerative and psychiatric disorders such as ASD, schizophrenia, AD, major depressive disorder (MDD) and PD [216,217,218,219] prompted researchers to have a closer look at the GBA. For more information on gut bacteria and neurotransmitters, the reader is referred to a recent review by Dicks [220]. The role that gut bacteria play in neuropsychiatric disorders has recently been reviewed by Dicks et al. [221].
7. Conclusions
With bacteria representing the largest fraction of almost 3000 microbial species in the GIT, it is no surprise that they have developed mechanisms to communicate with host cells. Some QS molecules are genus-specific, but a few are used by Gram-negative and Gram-positive bacteria. Hormones such as Epi and NE and certain carbohydrates (e.g., fucose and EA) activate specific receptors in bacteria that, in turn, trigger sensors in the cytoplasm to regulate gene expressions. In a healthy GIT, these signaling molecules are important in maintaining a homeostatic status. Some QS molecules, such as 3-oxo-C12:2-HSL, protect tight junction proteins and may be important in the treatment of leaky gut syndrome. Some QS molecules stimulate tumor growth and are closely associated with the development of specific cancers, whilst others are linked to neurological disorders. QSPs that penetrate the blood–brain barrier (BBB) constitutes an area that warrants more research, especially since the gut microbiome is increasingly recognized as a key player in neuropsychiatry. This warrants a better understanding of the underlying mechanisms involved in the regulation of gut bacterial QS systems. We also need to have a better understanding of bacterially produced QS molecules in adrenal pathways. With more in-depth knowledge of the different QS systems produced by intestinal bacteria, we may be able to develop biomarkers that can be used to diagnose neurological and psychiatric diseases such as anxiety and depression, MDD, schizophrenia, bipolar disorder, autism and OCD. More research is required to understand the integral communications between autoinducer-based signaling molecules and neurons in the human CNS. Research on using QS inhibitors (QQ therapy) to control microbial infections is gaining interest. Although a microbial infection may not be fully controlled using this strategy, it may provide the host’s immune system a better chance to overcome the infection. Before this approach can be implemented, we need to investigate the effect that QS inhibitors may have on non-pathogenic, beneficial, commensal gut microbiota.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The author has no conflict of interest.
References
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Coquant, G.; Aguanno, D.; Pham, S.; Grellier, N.; Thenet, S.; Carrière, V.; Grill, J.-P.; Seksik, P. Gossip in the gut: Quorum sensing, a new player in the host microbiota interactions. World J. Gastroenterol. 2021, 27, 7247–7270. [Google Scholar] [CrossRef] [PubMed]
- Yashiroda, Y.; Yoshida, M. Intraspecies cell–cell communication in yeast. FEMS Yeast Res. 2019, 19, foz071. [Google Scholar] [CrossRef]
- Prescott, R.; Decho, A.W. Flexibility and Adaptability of Quorum Sensing in Nature. Trends Microbiol. 2020, 28, 436–444. [Google Scholar] [CrossRef]
- Tam, K.; Torres, V.J. Staphylococcus aureus Secreted Toxins and Extracellular Enzymes. Microbiol. Spectr. 2018, 7, GPP3-0039-2018. [Google Scholar] [CrossRef] [PubMed]
- Fülling, C.; Dinan, T.G.; Cryan, J.F. Gut Microbe to Brain Signaling: What Happens in Vagus…. Neuron 2019, 101, 998–1002. [Google Scholar] [CrossRef] [PubMed]
- Krasulova, K.; Illes, P. Intestinal interplay of quorum sensing molecules and human receptors. Biochimie 2021, 189, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Galloway, W.R.J.D.; Hodgkinson, J.T.; Bowden, S.D.; Welch, M.; Spring, D.R. Quorum Sensing in Gram-Negative Bacteria: Small-Molecule Modulation of AHL and AI-2 Quorum Sensing Pathways. Chem. Rev. 2010, 111, 28–67. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Luo, Y. Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Front. Microbiol. 2021, 12, 611413. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, C.; Eberhard, A. Signal generation in autoinduction systems: Synthesis of acylated homoserine lactones by LuxI-type proteins. In Cell-Cell Signaling in Bacteria; Dunny, G., Winans, S.C., Eds.; ASM Press: Washington, DC, USA, 1999; pp. 211–230. [Google Scholar]
- Churchill, M.E.A.; Herman, J.P. Acyl-homoserinelactone biosynthesis: Structure and mechanism. In Chemical Communication among Bacteria; Stephen, W., Bonnie, B., Eds.; ASM Press: Washington, DC, USA, 2008; Chapter 17. [Google Scholar]
- Styles, M.J.; Blackwell, H.E. Non-native autoinducer analogs capable of modulating the SdiA quorum sensing receptor in Salmonella enterica serovar Typhimurium. Beilstein J. Org. Chem. 2018, 14, 2651–2664. [Google Scholar] [CrossRef] [PubMed]
- Sabag-Daigle, A.; Ahmer, B.M.M. ExpI and PhzI Are Descendants of the Long Lost Cognate Signal Synthase for SdiA. PLoS ONE 2012, 7, e47720. [Google Scholar] [CrossRef] [PubMed]
- Zohar, B.A.; Kolodkin-Gal, I. Quorum sensing in Escherichia coli: Interkingdom, inter- and intraspecies dialogues, and a suicide inducing peptide. In Quorum Sensing vs. Quorum Quenching: A Battle with No End in Sight; Springer: New Delhi, India, 2015; pp. 85–99. [Google Scholar]
- Hughes, D.T.; Terekhova, D.A.; Liou, L.; Hovde, C.J.; Sahl, J.W.; Patankar, A.V.; Gonzalez, J.E.; Edrington, T.S.; Rasko, D.A.; Sperandio, V. Chemical sensing in mammalian host–bacterial commensal associations. Proc. Natl. Acad. Sci. USA 2010, 107, 9831–9836. [Google Scholar] [CrossRef] [PubMed]
- Dyszel, J.L.; Soares, J.A.; Swearingen, M.C.; Lindsay, A.; Smith, J.N.; Ahmer, B.M.M. E. coli K-12 and EHEC Genes Regulated by SdiA. PLoS ONE 2010, 5, e8946. [Google Scholar] [CrossRef] [PubMed]
- Price, S.B.; Wright, J.C.; DeGraves, F.J.; Castanie-Cornet, M.-P.; Foster, J.W. Acid Resistance Systems Required for Survival of Escherichia coli O157:H7 in the Bovine Gastrointestinal Tract and in Apple Cider Are Different. Appl. Environ. Microbiol. 2004, 70, 4792–4799. [Google Scholar] [CrossRef] [PubMed]
- Witsø, I.L.; Rukke, H.V.; Benneche, T.; Scheie, A.A. Thiophenone Attenuates Enteropathogenic Escherichia coli O103:H2 Virulence by Interfering with AI-2 Signaling. PLoS ONE 2016, 11, e0157334. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Shin, D.; Kim, M.; Park, J.; Lim, S.; Ryu, S. LsrR-mediated quorum sensing controls invasiveness of Salmonella typhimurium by regulating SPI-1 and flagella genes. PLoS ONE 2012, 7, e37059. [Google Scholar] [CrossRef] [PubMed]
- Abed, N.; Grépinet, O.; Canepa, S.; Hurtado-Escobar, G.A.; Guichard, N.; Wiedemann, A.; Velge, P.; Virlogeux-Payant, I. Direct regulation of the pefI-srgC operon encoding the Rck invasin by the quorum-sensing regulator SdiA in Salmonella typhimurium. Mol. Microbiol. 2014, 94, 254–271. [Google Scholar] [CrossRef]
- Habyarimana, F.; Sabag-Daigle, A.; Ahmer, B.M. The SdiA-regulated gene srgE encodes a type III secreted effector. J. Bacteriol. 2014, 96, 2301–2312. [Google Scholar] [CrossRef]
- Coquant, G.; Grill, J.-P.; Seksik, P. Impact of N-Acyl-Homoserine Lactones, Quorum Sensing Molecules, on Gut Immunity. Front. Immunol. 2020, 11, 1827. [Google Scholar] [CrossRef]
- Zhu, J.; Winans, S.C. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. USA 2001, 98, 1507–1512. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, D.A.; Wu, M.; Hoffman, L.R.; Kulasekara, H.D.; Déziel, E.; Smith, E.E.; Nguyen, H.; Ernst, R.K.; Freeman, T.J.L.; Spencer, D.H.; et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 2007, 64, 512–533. [Google Scholar] [CrossRef] [PubMed]
- Luján, A.M.; Moyano, A.J.; Segura, I.; Argaraña, C.E.; Smania, A.M. Quorum-sensing-deficient (lasR) mutants emerge at high frequency from a Pseudomonas aeruginosa mutS strain. Microbiology 2007, 153, 225–237. [Google Scholar] [CrossRef] [PubMed]
- LaFayette, S.L.; Houle, D.; Beaudoin, T.; Wojewodka, G.; Radzioch, D.; Hoffman; Burns, J.L.; Dandekar, A.A.; Smalley, N.E.; Chandler, J.R.; et al. Cystic fibrosis–adapted Pseudomonas aeruginosa quorum sensing lasR mutants cause hyperinflammatory responses. Sci. Adv. 2015, 1, e1500199. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Mukherjee, S.; Moustafa, D.; Smith, C.D.; Goldberg, J.B.; Bassler, B.L. The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. PLOS Pathog. 2017, 13, e1006504. [Google Scholar] [CrossRef] [PubMed]
- Agodi, A.; Barchitta, M.; Cipresso, R.; Giaquinta, L.; Romeo, M.A.; Denaro, C. Pseudomonas aeruginosa carriage, colonization, and infection in ICU patients. Intensive Care Med. 2007, 33, 1155–1161. [Google Scholar] [CrossRef]
- Bertrand, X.; Thouverez, M.; Talon, D.; Boillot, A.; Capellier, G.; Floriot, C.; Hélias, J.P. Endemicity, molecular diversity and colonisation routes of Pseudomonas aeruginosa in intensive care units. Intensive Care Med. 2001, 27, 1263–1268. [Google Scholar] [CrossRef]
- Swearingen, M.C.; Sabag-Daigle, A.; Ahmer, B.M. Are there acyl-homoserine lactones within mammalian intestines? J. Bacteriol. 2013, 195, 173–179. [Google Scholar] [CrossRef]
- Debunne, N.; Verbeke, F.; Janssens, Y.; Wynendaele, E.; De Spiegeleer, B. Chromatography of Quorum Sensing Peptides: An Important Functional Class of the Bacterial Peptidome. Chromatographia 2017, 81, 25–40. [Google Scholar] [CrossRef]
- Verbeke, F.; De Craemer, S.; Debunne, N.; Janssens, Y.; Wynendaele, E.; Van de Wiele, C.; De Spiegeleer, B. Peptides as Quorum Sensing Molecules: Measurement Techniques and Obtained Levels In Vitro and In Vivo. Front. Neurosci. 2017, 11, 183. [Google Scholar] [CrossRef]
- Bouillaut, L.; Perchat, S.; Arold, S.T.; Zorrilla, S.; Slamti, L.; Henry, C.; Gohar, M.; Declerck, N.; Lereclus, D. Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucleic Acids Res. 2008, 36, 3791–3801. [Google Scholar] [CrossRef] [PubMed]
- Pomerantsev, A.P.; Pomerantseva, O.M.; Camp, A.S.; Mukkamala, R.; Goldman, S.; Leppla, S.H. PapR peptide maturation: Role of the NprB protease in Bacillus cereus 569 PlcR/PapR global gene regulation. FEMS Immunol. Med Microbiol. 2009, 55, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Braughton, K.R.; Kretschmer, D.; Bach, T.-H.L.; Queck, S.Y.; Li, M.; Kennedy, A.D.; Dorward, D.W.; Klebanoff, S.J.; Peschel, A.; et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 2007, 13, 1510–1514. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Chong, Y.P.; Park, H.J.; Park, K.-H.; Moon, S.M.; Jeong, J.-Y.; Kim, M.-N.; Kim, S.-H.; Lee, S.-O.; Choi, S.-H.; et al. agr dysfunction and persistent methicillin-resistant Staphylococcus aureus bacteremia in patients with removed eradicable foci. Infection 2012, 41, 111–119. [Google Scholar] [CrossRef]
- Kang, C.K.; the Korea INfectious Diseases (KIND) study group; Kim, Y.K.; Jung, S.-I.; Park, W.B.; Song, K.-H.; Park, K.-H.; Choe, P.G.; Jang, H.-C.; Lee, S.; et al. Agr functionality affects clinical outcomes in patients with persistent methicillin-resistant Staphylococcus aureus bacteraemia. Eur. J. Clin. Microbiol. 2017, 36, 2187–2191. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Le, K.Y.; Khan, B.A.; Nguyen, T.H.; Hunt, R.; Bae, J.; Kabat, J.; Zheng, Y.; Cheung, G.Y.C.; Li, M.; et al. Resistance to leukocytes ties benefits of quorum sensing dysfunctionality to biofilm infection. Nat. Microbiol. 2019, 4, 1114–1119. [Google Scholar] [CrossRef] [PubMed]
- Ali, L.; Goraya, M.U.; Arafat, Y.; Ajmal, M.; Chen, J.-L.; Yu, D. Molecular Mechanism of Quorum-Sensing in Enterococcus faecalis: Its Role in Virulence and Therapeutic Approaches. Int. J. Mol. Sci. 2017, 18, 960. [Google Scholar] [CrossRef]
- Wynendaele, E.; Bronselaer, A.; Nielandt, J.; D’Hondt, M.; Stalmans, S.; Bracke, N.; Verbeke, F.; Van De Wiele, C.; De Tré, G.; De Spiegeleer, B. Quorumpeps database: Chemical space, microbial origin and functionality of quorum sensing peptides. Nucleic Acids Res. 2012, 41, D655–D659. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.S.; Thompson, J.A.; Xavier, K.B. AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 2013, 37, 156–181. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, C.E.; Hong, W.; Pang, B.; Weimer, K.E.D.; Juneau, R.A.; Turner, J.; Swords, W.E. Indirect Pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in Polymicrobial Otitis Media Occurs via Interspecies Quorum Signaling. mBio 2010, 1, e00102-10. [Google Scholar] [CrossRef] [PubMed]
- Cuadra-Saenz, G.; Rao, D.L.; Underwood, A.J.; Belapure, S.A.; Campagna, S.R.; Sun, Z.; Tammariello, S.; Rickard, A.H. Autoinducer-2 influences interactions amongst pioneer colonizing streptococci in oral biofilms. Microbiology 2012, 158, 1783–1795. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.S.; McAuley, J.R.; Taga, M.E.; Xavier, K.B.; Miller, S.T. Sinorhizobium meliloti, a bacterium lacking the autoinducer-2 (AI-2) synthase, responds to AI-2 supplied by other bacteria. Mol. Microbiol. 2008, 70, 1223–1235. [Google Scholar] [CrossRef]
- Xavier, K.; Bassler, B.L. Interference with AI-2-mediated bacterial cell–cell communication. Nature 2005, 437, 750–753. [Google Scholar] [CrossRef] [PubMed]
- Yeo, S.; Park, H.; Ji, Y.; Park, S.; Yang, J.; Lee, J.; Mathara, J.M.; Shin, H.; Holzapfel, W. Influence of gastrointestinal stress on autoinducer-2 activity of two Lactobacillus species. FEMS Microbiol. Ecol. 2015, 91, fiv065. [Google Scholar] [CrossRef]
- Liu, L.; Wu, R.; Zhang, J.; Li, P. Overexpression of luxS Promotes Stress Resistance and Biofilm Formation of Lactobacillus paraplantarum L-ZS9 by Regulating the Expression of Multiple Genes. Front. Microbiol. 2018, 9, 2628. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, B.; Grenier, D.; Yi, L. Regulatory Mechanisms of the LuxS/AI-2 System and Bacterial Resistance. Antimicrob. Agents Chemother. 2019, 63, e01186-19. [Google Scholar] [CrossRef] [PubMed]
- Schauder, S.; Shokat, K.; Surette, M.G.; Bassler, B.L. The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 2001, 41, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Khan, C.M.A. The dynamic interactions between Salmonella and the microbiota, within the challenging niche of the gastroin-testinal tract. Int. Sch. Res. Not. 2014, 2014, 846049. Available online: https://www.hindawi.com/journals/isrn/2014/846049/ (accessed on 14 July 2022).
- Sun, Z.; He, X.; Brancaccio, V.F.; Yuan, J.; Riedel, C.U. Bifidobacteria Exhibit LuxS-Dependent Autoinducer 2 Activity and Biofilm Formation. PLoS ONE 2014, 9, e88260. [Google Scholar] [CrossRef]
- Lebeer, S.; De Keersmaecker, S.C.J.; Verhoeven, T.L.A.; Fadda, A.A.; Marchal, K.; Vanderleyden, J. Functional Analysis of luxS in the Probiotic Strain Lactobacillus rhamnosus GG Reveals a Central Metabolic Role Important for Growth and Biofilm Formation. J. Bacteriol. 2007, 189, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wu, R.; Zhang, J.; Shang, N.; Li, P. D-Ribose Interferes with Quorum Sensing to Inhibit Biofilm Formation of Lactobacillus paraplantarum L-ZS9. Front. Microbiol. 2017, 8, 1860. [Google Scholar] [CrossRef] [PubMed]
- Lukáš, F.; Gorenc, G.; Kopečný, J. Detection of possible AI-2-mediated quorum sensing system in commensal intestinal bacteria. Folia Microbiol. 2008, 53, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.-Y.; Zhao, Z.; Avillan, J.J.; Liu, J.; Call, D.R. Autoinducer-2 Quorum Sensing Contributes to Regulation of Microcin PDI in Escherichia coli. Front. Microbiol. 2017, 8, 2570. [Google Scholar] [CrossRef]
- Miller, E.L.; Kjos, M.; Abrudan, M.I.; Roberts, I.S.; Veening, J.-W.; Rozen, D.E. Eavesdropping and crosstalk between secreted quorum sensing peptide signals that regulate bacteriocin production in Streptococcus pneumoniae. ISME J. 2018, 12, 2363–2375. [Google Scholar] [CrossRef]
- Merritt, J.; Kreth, J.; Shi, W.; Qi, F. LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Mol. Microbiol. 2005, 57, 960–969. [Google Scholar] [CrossRef]
- Sztajer, H.; Lemme, A.; Vilchez, R.; Schulz, S.; Geffers, R.; Yip, C.Y.Y.; Levesque, C.M.; Cvitkovitch, D.G.; Wagner-Döbler, I. Autoinducer-2-Regulated Genes in Streptococcus mutans UA159 and Global Metabolic Effect of the luxS Mutation. J. Bacteriol. 2008, 190, 401–415. [Google Scholar] [CrossRef]
- Jia, F.-F.; Pang, X.-H.; Zhu, D.-Q.; Zhu, Z.-T.; Sun, S.-R.; Meng, X.-C. Role of the luxS gene in bacteriocin biosynthesis by Lactobacillus plantarum KLDS1.0391: A proteomic analysis. Sci. Rep. 2017, 7, 13871. [Google Scholar] [CrossRef]
- Li, J.; Yang, X.; Shi, G.; Chang, J.; Liu, Z.; Zeng, M. Cooperation of lactic acid bacteria regulated by the AI-2/LuxS system involve in the biopreservation of refrigerated shrimp. Food Res. Int. 2019, 120, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Christiaen, S.E.A.; O’Connell Motherway, M.; Bottacini, F.; Lanigan, N.; Casey, P.G.; Huys, G.; Nelis, H.J.; van Sinderen, D.; Coenye, T. Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PLoS ONE 2014, 9, e98111. [Google Scholar] [CrossRef]
- Thompson, J.A.; Oliveira, R.A.; Djukovic, A.; Ubeda, C.; Xavier, K.B. Manipulation of the Quorum Sensing Signal AI-2 Affects the Antibiotic-Treated Gut Microbiota. Cell Rep. 2015, 10, 1861–1871. [Google Scholar] [CrossRef]
- Xavier, K.B.; Bassler, B.L. Regulation of Uptake and Processing of the Quorum-Sensing Autoinducer AI-2 in Escherichia coli. J. Bacteriol. 2005, 187, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Hegde, M.; Englert, D.L.; Schrock, S.; Cohn, W.B.; Vogt, C.; Wood, T.K.; Manson, M.D.; Jayaraman, A. Chemotaxis to the Quorum-Sensing Signal AI-2 Requires the Tsr Chemoreceptor and the Periplasmic LsrB AI-2-Binding Protein. J. Bacteriol. 2011, 193, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Barrios, A.F.G.; Zuo, R.; Hashimoto, Y.; Yang, L.; Bentley, W.E.; Wood, T.K. Autoinducer 2 Controls Biofilm Formation in Escherichia coli through a Novel Motility Quorum-Sensing Regulator (MqsR, B3022). J. Bacteriol. 2006, 188, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.K.; Huang, J.Y.; Wreden, C.; Sweeney, E.G.; Goers, J.; Remington, S.J.; Guillemin, K. Chemorepulsion from the Quorum Signal Autoinducer-2 Promotes Helicobacter pylori Biofilm Dispersal. mBio 2015, 6, e00379-15. [Google Scholar] [CrossRef]
- Watt, R.; Parkin, K.; Martino, D. The Potential Effects of Short-Chain Fatty Acids on the Epigenetic Regulation of Innate Immune Memory. Challenges 2020, 11, 25. [Google Scholar] [CrossRef]
- Rossmann, F.S.; Raček, T.; Wobser, D.; Puchalka, J.; Rabener, E.M.; Reiger, M.; Hendrickx, A.P.A.; Diederich, A.-K.; Jung, K.; Klein, C.; et al. Phage-mediated Dispersal of Biofilm and Distribution of Bacterial Virulence Genes Is Induced by Quorum Sensing. PLOS Pathog. 2015, 11, e1004653. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, X.; Song, C.; Zhang, Y.; Wang, Z.; Liu, Z.; Wei, H.; Yu, J. Autoinducer-2 Facilitates Pseudomonas aeruginosa PAO1 Pathogenicity in Vitro and in Vivo. Front. Microbiol. 2017, 8, 1944. [Google Scholar] [CrossRef]
- Pereira, C.S.; de Regt, A.K.; Brito, P.H.; Miller, S.T.; Xavier, K.B. Identification of functional LsrB-like autoinducer-2 receptors. J. Bacteriol. 2009, 191, 6975–6987. [Google Scholar] [CrossRef]
- Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.L.; Hughson, F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002, 415, 545–549. [Google Scholar] [CrossRef]
- Miller, S.T.; Xavier, K.B.; Campagna, S.R.; Taga, M.E.; Semmelhack, M.F.; Bassler, B.L.; Hughson, F.M. Salmonella typhimurium Recognizes a Chemically Distinct Form of the Bacterial Quorum-Sensing Signal AI-2. Mol. Cell 2004, 15, 677–687. [Google Scholar] [CrossRef]
- Thompson, J.A.; Oliveira, R.; Xavier, K. Chemical conversations in the gut microbiota. Gut Microbes 2016, 7, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Peng, W.; Wu, J.; Wang, X.; Ren, Y.; Li, H.; Peng, Y.; Tang, X.; Fu, X. Autoinducer-2 of gut microbiota, a potential novel marker for human colorectal cancer, is associated with the activation of TNFSF9 signaling in macrophages. OncoImmunology 2019, 8, e1626192. [Google Scholar] [CrossRef] [PubMed]
- Moreira, C.G.; Weinshenker, D.; Sperandio, V. QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo. Infect. Immun. 2010, 78, 914–926. [Google Scholar] [CrossRef] [PubMed]
- Moreira, C.G.; Sperandio, V. The Epinephrine/Norepinephrine/Autoinducer-3 Interkingdom Signaling System in Escherichia coli O157:H7. Microb. Endocrinol. 2016, 874, 247–261. [Google Scholar] [CrossRef]
- Sperandio, V.; Torres, A.G.; Jarvis, B.; Nataro, J.P.; Kaper, J.B. Bacteria–host communication: The language of hormones. Proc. Natl. Acad. Sci. USA 2003, 100, 8951–8956. [Google Scholar] [CrossRef]
- Kim, C.S.; Gatsios, A.; Cuesta, S.; Lam, Y.C.; Wei, Z.; Chen, H.; Russell, R.M.; Shine, E.E.; Wang, R.; Wyche, T.P.; et al. Characterization of autoinducer-3 structure and bio-synthesis in E. coli. ACS Cent. Sci. 2020, 6, 197–206. [Google Scholar] [CrossRef]
- Lustri, B.C.; Sperandio, V.; Moreira, C.G. Bacterial chat: Intestinal metabolites and signals in host-microbiota-pathogen inter-actions. Infect. Immun. 2017, 85, e00476-17. [Google Scholar] [CrossRef]
- Holm, A.; Vikström, E. Quorum sensing communication between bacteria and human cells: Signals, targets, and functions. Front. Plant Sci. 2014, 5, 309. [Google Scholar] [CrossRef]
- Vikström, E.; Bui, L.; Konradsson, P.; Magnusson, K.-E. Role of calcium signalling and phosphorylations in disruption of the epithelial junctions by Pseudomonas aeruginosa quorum sensing molecule. Eur. J. Cell Biol. 2010, 89, 584–597. [Google Scholar] [CrossRef]
- Vikström, E.; Bui, L.; Konradsson, P.; Magnusson, K.-E. The junctional integrity of epithelial cells is modulated by Pseudomonas aeruginosa quorum sensing molecule through phosphorylation-dependent mechanisms. Exp. Cell Res. 2009, 315, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Neurath, M.F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 2014, 14, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Telford, G.; Wheeler, D.; Williams, P.; Tomkins, P.T.; Appleby, P.; Sewell, H.; Stewart, G.S.A.B.; Bycroft, B.W.; Pritchard, D.I. The Pseudomonas aeruginosa Quorum-Sensing Signal Molecule N-(3-Oxododecanoyl)-l-Homoserine Lactone Has Immunomodulatory Activity. Infect. Immun. 1998, 66, 36–42. [Google Scholar] [CrossRef]
- Ritchie, A.J.; Jansson, A.; Stallberg, J.; Nilsson, P.; Lysaght, P.; Cooley, M.A. The Pseudomonas aeruginosa Quorum-Sensing Molecule N-3-(Oxododecanoyl)-l-Homoserine Lactone Inhibits T-Cell Differentiation and Cytokine Production by a Mechanism Involving an Early Step in T-Cell Activation. Infect. Immun. 2005, 73, 1648–1655. [Google Scholar] [CrossRef]
- Landman, C.; Grill, J.P.; Mallet, J.M.; Marteau, P.; Humbert, L.; Le Balch, E.; Maubert, M.A.; Perez, K.; Chaara, W.; Brot, L. Inter-kingdom effect on epithelial cells of the N-Acyl homoserine lactone 3-oxo-C12:2, a major quorum-sensing molecule from gut microbiota. PLoS ONE 2018, 13, e0202587. [Google Scholar] [CrossRef] [PubMed]
- Aguanno, D.; Coquant, G.; Postal, B.G.; Osinski, C.; Wieckowski, M.; Stockholm, D.; Grill, J.-P.; Carrière, V.; Seksik, P.; Thenet, S. The intestinal quorum sensing 3-oxo-C12:2 Acyl homoserine lactone limits cytokine-induced tight junction disruption. Tissue Barriers 2020, 8, 1832877. [Google Scholar] [CrossRef] [PubMed]
- Beck, P.L.; A Cotton, J.; Platnich, J.M.; A Muruve, D.; Buret, A.G.; Jijon, H.B. Interleukin-8 in gastrointestinal inflammation and malignancy: Induction and clinical consequences. Int. J. Interf. Cytokine Mediat. Res. 2016, 8, 13–34. [Google Scholar] [CrossRef]
- Ghezzal, S.; Postal, B.G.; Quevrain, E.; Brot, L.; Seksik, P.; Leturque, A.; Thenet, S.; Carrière, V. Palmitic acid damages gut epithelium integrity and initiates inflammatory cytokine production. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2019, 1865, 158530. [Google Scholar] [CrossRef]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. Rev. 2016, 40, 86–116. [Google Scholar] [CrossRef]
- Teiber, J.F.; Horke, S.; Haines, D.C.; Chowdhary, P.K.; Xiao, J.; Kramer, G.L.; Haley, R.W.; Draganov, D.I. Dominant Role of Paraoxonases in Inactivation of the Pseudomonas aeruginosa Quorum-Sensing Signal N-(3-Oxododecanoyl)-l-Homoserine Lactone. Infect. Immun. 2008, 76, 2512–2519. [Google Scholar] [CrossRef]
- Levy, E.; Trudel, K.; Bendayan, M.; Seidman, E.; Delvin, E.; Elchebly, M.; Lavoie, J.-C.; Precourt, L.-P.; Amre, D.; Sinnett, D. Biological role, protein expression, subcellular localization, and oxidative stress response of paraoxonase 2 in the intestine of humans and rats. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G1252–G1261. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rothem, L.; Hartman, C.; Dahan, A.; Lachter, J.; Eliakim, R.; Shamir, R. Paraoxonases are associated with intestinal inflammatory diseases and intracellularly localized to the endoplasmic reticulum. Free Radic. Biol. Med. 2007, 43, 730–739. [Google Scholar] [CrossRef] [PubMed]
- Ranhotra, H.S.; Flannigan, K.L.; Brave, M.; Mukherjee, S.; Lukin, D.J.; Hirota, S.A.; Mani, S. Xenobiotic Receptor-Mediated Regulation of Intestinal Barrier Function and Innate Immunity. Nucl. Recept. Res. 2016, 3, 101199. [Google Scholar] [CrossRef] [PubMed]
- Pavek, P. Pregnane X Receptor (PXR)-Mediated Gene Repression and Cross-Talk of PXR with Other Nuclear Receptors via Coactivator Interactions. Front. Pharmacol. 2016, 7, 456. [Google Scholar] [CrossRef] [PubMed]
- Cooley, M.A.; Whittall, C.; Rolph, M.S. Pseudomonas signal molecule 3-oxo-C12-homoserine lactone interferes with binding of rosiglitazone to human PPARgamma. Microb. Infect. 2020, 12, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Kravchenko, V.V.; Kaufmann, G.F.; Mathison, J.C.; Scott, D.A.; Katz, A.Z.; Wood, M.R.; Brogan, A.P.; Lehmann, M.; Mee, J.M.; Iwata, K.; et al. N-(3-Oxo-acyl)homoserine Lactones Signal Cell Activation through a Mechanism distinct from the Canonical Pathogen-associated Molecular Pattern Recognition Receptor Pathways. J. Biol. Chem. 2006, 281, 28822–28830. [Google Scholar] [CrossRef]
- Kravchenko, V.V.; Kaufmann, G.F.; Mathison, J.C.; Scott, D.A.; Katz, A.Z.; Grauer, D.C.; Lehmann, M.; Meijler, M.M.; Janda, K.D.; Ulevitch, R.J. Modulation of gene expression via disruption of NFB signaling by a bacterial small molecule. Science 2008, 321, 259–263. [Google Scholar] [CrossRef]
- Decara, J.; Rivera, P.; López-Gambero, A.J.; Serrano, A.; Pavón, F.J.; Baixeras, E.; De Fonseca, F.R.; Suárez, J. Peroxisome Proliferator-Activated Receptors: Experimental Targeting for the Treatment of Inflammatory Bowel Diseases. Front. Pharmacol. 2020, 11, 730. [Google Scholar] [CrossRef]
- Kim, K.; Kim, S.-H.; Lépine, F.; Cho, Y.-H.; Lee, G.R. Global gene expression analysis on the target genes of PQS and HHQ in J774A.1 monocyte/macrophage cells. Microb. Pathog. 2010, 49, 174–180. [Google Scholar] [CrossRef]
- Lin, J.; Cheng, J.; Wang, Y.; Shen, X. The Pseudomonas Quinolone Signal (PQS): Not Just for Quorum Sensing Anymore. Front. Cell. Infect. Microbiol. 2018, 8, 230. [Google Scholar] [CrossRef]
- Moura-Alves, P.; Puyskens, A.; Stinn, A.; Klemm, M.; Guhlich-Bornhof, U.; Dorhoi, A.; Furkert, J.; Kreuchwig, A.; Protze, J.; Lozza, L.; et al. Host monitoring of quorum sensing during Pseudomonas aeruginosa infection. Science 2019, 366, eaaw1629. [Google Scholar] [CrossRef] [PubMed]
- Trikha, P.; Lee, D.A. The role of AhR in transcriptional regulation of immune cell development and function. Biochim. Biophys. Acta (BBA) Rev. Cancer 2019, 1873, 188335. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Ma, N.; He, T.; Johnston, L.J.; Ma, X. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit. Rev. Food Sci. Nutr. 2019, 60, 1760–1768. [Google Scholar] [CrossRef] [PubMed]
- Kurata, K.; Kawahara, H.; Nishimura, K.; Jisaka, M.; Yokota, K.; Shimizu, H. Skatole regulates intestinal epithelial cellular functions through activating aryl hydrocarbon receptors and p38. Biochem. Biophys. Res. Commun. 2019, 510, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, J.-D.; Bibbs, L.; Ulevitch, R.J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 1994, 265, 808–811. [Google Scholar] [CrossRef] [PubMed]
- Fujiya, M.; Musch, M.W.; Nakagawa, Y.; Hu, S.; Alverdy, J.; Kohgo, Y.; Schneewind, O.; Jabri, B.; Chang, E.B. The Bacillus subtilis Quorum-Sensing Molecule CSF Contributes to Intestinal Homeostasis via OCTN2, a Host Cell Membrane Transporter. Cell Host Microbe 2007, 1, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Vidyasagar, A.; A Wilson, N.; Djamali, A. Heat shock protein 27 (HSP27): Biomarker of disease and therapeutic target. Fibrogenesis Tissue Repair 2012, 5, 7. [Google Scholar] [CrossRef]
- Janssens, Y.; Debunne, N.; De Spiegeleer, A.; Wynendaele, E.; Planas, M.; Feliu, L.; Quarta, A.; Claes, C.; Van Dam, D.; De Deyn, P.P.; et al. PapRIV, a BV-2 microglial cell acti-vating quorum sensing peptide. Sci. Rep. 2021, 11, 10723. [Google Scholar] [CrossRef]
- Yang, D.; Elner, S.G.; Bian, Z.-M.; Till, G.O.; Petty, H.R.; Elner, V.M. Pro-inflammatory cytokines increase reactive oxygen species through mitochondria and NADPH oxidase in cultured RPE cells. Exp. Eye Res. 2007, 85, 462–472. [Google Scholar] [CrossRef]
- Wynendaele, E.; Verbeke, F.; Stalmans, S.; Gevaert, B.; Janssens, Y.; Van De Wiele, C.; Peremans, K.; Burvenich, C.; De Spiegeleer, B. Quorum Sensing Peptides Selectively Penetrate the Blood-Brain Barrier. PLoS ONE 2015, 10, e0142071. [Google Scholar] [CrossRef]
- De Spiegeleer, A.; Elewaut, D.; Noortgate, N.V.D.; Janssens, Y.; Debunne, N.; Van Langenhove, S.; Govindarajan, S.; De Spiegeleer, B.; Wynendaele, E. Quorum sensing molecules as a novel microbial factor impacting muscle cells. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2019, 1866, 165646. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pascual, D.; Monnet, V.; Gardan, R. Bacterial Cell–Cell Communication in the Host via RRNPP Peptide-Binding Regulators. Front. Microbiol. 2016, 7, 706. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.-L.; Liu, Y.-N.; Barber, C.E.; Dow, J.M.; Wootton, J.C.; Daniels, M.J. Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris. Mol. Gen. Genet. MGG 1991, 226, 409–417. [Google Scholar] [CrossRef]
- Kanno, E.; Kawakami, K.; Miyairi, S.; Tanno, H.; Otomaru, H.; Hatanaka, A.; Sato, S.; Ishii, K.; Hayashi, D.; Shibuya, N.; et al. Neutrophil-derived tumor necrosis factor-α contributes to acute wound healing promoted by N-(3-oxododecanoyl)-l-homoserine lactone from Pseudomonas aeruginosa. J. Dermatol. Sci. 2013, 70, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Kanno, E.; Kawakami, K.; Miyairi, S.; Tanno, H.; Suzuki, A.; Kamimatsuno, R.; Takagi, N.; Miyasaka, T.; Ishii, K.; Gotoh, N.; et al. Promotion of acute-phase skin wound healing by Pseudomonas aeruginosa C4-HSL. Int. Wound J. 2016, 13, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Krupnick, J.G.; Benovic, J.L. The role of receptor kinases and arrestins in g protein–coupled receptor regulation. Annu. Rev. Pharmacol. Toxicol. 1998, 38, 289–319. [Google Scholar] [CrossRef] [PubMed]
- Plouffe, B.; Thomsen, A.R.B.; Irannejad, R. Emerging Role of Compartmentalized G Protein-Coupled Receptor Signaling in the Cardiovascular Field. ACS Pharmacol. Transl. Sci. 2020, 3, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Veldhoen, M.; Brucklacher-Waldert, V. Dietary influences on intestinal immunity. Nat. Rev. Immunol. 2012, 12, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Lamorte, S.; Shinde, R.; McGaha, T.L. Nuclear receptors, the aryl hydrocarbon receptor, and macrophage function. Mol. Asp. Med. 2021, 78, 100942. [Google Scholar] [CrossRef]
- Quintana, F.J.; Basso, A.S.; Iglesias, A.H.; Korn, T.; Farez, M.F.; Bettelli, E.; Caccamo, M.; Oukka, M.; Weiner, H.L. Control of Treg and TH17 cell differentiation by the aryl hydrocarbon receptor. Nature 2008, 453, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Kiss, E.A.; Vonarbourg, C.; Kopfmann, S.; Hobeika, E.; Finke, D.; Esser, C.; Diefenbach, A. Natural Aryl Hydrocarbon Receptor Ligands Control Organogenesis of Intestinal Lymphoid Follicles. Science 2011, 334, 1561–1565. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Wang, Q.; Ma, Y.; Li, L.; Yu, K.; Zhang, Z.; Chen, G.; Li, X.; Xiao, W.; Xu, P.; et al. Aryl Hydrocarbon Receptor Activation Modulates Intestinal Epithelial Barrier Function by Maintaining Tight Junction Integrity. Int. J. Biol. Sci. 2018, 14, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Vyhlídalová, B.; Krasulová, K.; Pečinková, P.; Marcalíková, A.; Vrzal, R.; Zemánková, L.; Vančo, J.; Trávníček, Z.; Vondráček, J.; Karasová, M.; et al. Gut Microbial Catabolites of Tryptophan Are Ligands and Agonists of the Aryl Hydrocarbon Receptor: A Detailed Characterization. Int. J. Mol. Sci. 2020, 21, 2614. [Google Scholar] [CrossRef] [PubMed]
- Heeb, S.; Fletcher, M.P.; Chhabra, S.R.; Diggle, S.P.; Williams, P.; Cámara, M. Quinolones: From antibiotics to autoinducers. FEMS Microbiol. Rev. 2011, 35, 247–274. [Google Scholar] [CrossRef] [PubMed]
- Moura-Alves, P.; Faé, K.; Houthuys, E.; Dorhoi, A.; Kreuchwig, A.; Furkert, J.; Barison, N.; Diehl, A.; Munder, A.; Constant, P.; et al. AhR sensing of bacterial pigments regulates antibacterial defence. Nature 2014, 512, 387–392. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, T.D.; Murray, I.A.; Bisson, W.H.; Lahoti, T.S.; Gowda, K.; Amin, S.G.; Patterson, A.D.; Perdew, G.H. Adaptation of the human aryl hydro-carbon receptor to sense microbiota-derived indoles. Sci. Rep. 2015, 5, 2689. [Google Scholar] [CrossRef] [PubMed]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.-P.; Michel, M.-L.; DA Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Natividad, J.M.; Agus, A.; Planchais, J.; Lamas, B.; Jarry, A.C.; Martin, R.; Michel, M.-L.; Chong-Nguyen, C.; Roussel, R.; Straube, M.; et al. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab. 2018, 28, 737–749.e4. [Google Scholar] [CrossRef] [PubMed]
- Lamas, B.; Hernandez-Galan, L.; Galipeau, H.J.; Constante, M.; Clarizio, A.; Jury, J.; Breyner, N.M.; Caminero, A.; Rueda, G.; Hayes, C.L.; et al. Aryl hydrocarbon receptor ligand production by the gut microbiota is decreased in celiac disease leading to intestinal inflammation. Sci. Transl. Med. 2020, 12, eaba0624. [Google Scholar] [CrossRef] [PubMed]
- Darkoh, C.; Plants-Paris, K.; Bishoff, D.; DuPont, H.L. Clostridium difficile modulates the gut microbiota by inducing the production of indole, an interkingdom signaling and antimicrobial molecule. mSystems 2019, 4, e00346-18. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.; Kinoshita, M.; Harada, K.; Mizutani, M.; Masahata, K.; Kayama, H.; Takeda, K. Commensal Bacteria-Dependent Indole Production Enhances Epithelial Barrier Function in the Colon. PLoS ONE 2013, 8, e80604. [Google Scholar] [CrossRef] [PubMed]
- Soh, N.L.; Walter, G. Tryptophan and depression: Can diet alone be the answer? Acta Neuropsychiatr. 2011, 23, 3–11. [Google Scholar] [CrossRef]
- Tudela, H.; Claus, S.P.; Saleh, M. Next Generation Microbiome Research: Identification of Keystone Species in the Metabolic Regulation of Host-Gut Microbiota Interplay. Front. Cell Dev. Biol. 2021, 9, 719072. [Google Scholar] [CrossRef] [PubMed]
- Taleb, S. Tryptophan Dietary Impacts Gut Barrier and Metabolic Diseases. Front. Immunol. 2019, 10, 2113. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, P.; Tian, H.; Tian, F.; Zhang, Y.; Zhang, L.; Gao, X.; Wang, X. Aryl hydrocarbon receptor/IL-22/Stat3 signaling pathway is involved in the modulation of intestinal mucosa antimicrobial molecules by commensal microbiota in mice. Innate Immun. 2018, 24, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Barragan, L.; Chai, J.N.; Tianero, M.D.; Di Luccia, B.; Ahern, P.P.; Merriman, J.; Cortez, V.S.; Caparon, M.G.; Donia, M.S.; Gilfillan, S.; et al. Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells. Science 2017, 357, 806–810. [Google Scholar] [CrossRef]
- Powell, D.N.; Swimm, A.; Sonowal, R.; Bretin, A.; Gewirtz, A.T.; Jones, R.M.; Kalman, D. Indoles from the commensal microbiota act via the AHR and IL-10 to tune the cellular composition of the colonic epithelium during aging. Proc. Natl. Acad. Sci. USA 2020, 117, 21519–21526. [Google Scholar] [CrossRef]
- Obata, Y.; Castaño, Á.; Boeing, S.; Bon-Frauches, A.C.; Fung, C.; Fallesen, T.; de Agüero, M.G.; Yilmaz, B.; Lopes, R.; Huseynova, A.; et al. Neuronal programming by microbiota regulates intestinal physiology. Nature 2020, 578, 284–289. [Google Scholar] [CrossRef]
- Gomez de Agüero, M.; Ganal-Vonarburg, S.C.; Fuhrer, T.; Rupp, S.; Uchimura, Y.; Li, H.; Steinert, A.; Heikenwalder, M.; Hapfelmeier, S.; Sauer, U.; et al. The maternal microbiota drives early postnatal innate immune development. Science 2016, 351, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Doty, A.; Glover, S.C. Aryl hydrocarbon receptor signaling involves in the human intestinal ILC3/ILC1 conversion in the inflamed terminal ileum of Crohn’s disease patients. Inflamm. Cell Signal. 2016, 3, e1404. [Google Scholar] [CrossRef]
- Schroeder, J.-H.; Howard, J.K.; Lord, G.M. Transcription factor-driven regulation of ILC1 and ILC3. Trends Immunol. 2022, 43, 564–579. [Google Scholar] [CrossRef]
- Willyard, C. The drug-resistant bacteria that pose the greatest health threats. Nature 2017, 543, 15. [Google Scholar] [CrossRef] [PubMed]
- Krzyżek, P. Challenges and Limitations of Anti-quorum Sensing Therapies. Front. Microbiol. 2019, 10, 2473. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, J.; Liu, C.; Yang, A.; Qiao, J. Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell. Mol. Life Sci. 2019, 77, 1319–1343. [Google Scholar] [CrossRef] [PubMed]
- Maura, D.; E Ballok, A.; Rahme, L.G. Considerations and caveats in anti-virulence drug development. Curr. Opin. Microbiol. 2016, 33, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Sommer, R.; Hinsberger, S.; Lu, C.; Hartmann, R.W.; Empting, M.; Titz, A. Novel Strategies for the Treatment of Pseudomonas aeruginosa Infections. J. Med. Chem. 2016, 59, 5929–5969. [Google Scholar] [CrossRef] [PubMed]
- Munir, S.; Shah, A.A.; Shahid, M.; Manzoor, I.; Aslam, B.; Rasool, M.H.; Saeed, M.; Ayaz, S.; Khurshid, M. Quorum Sensing Interfering Strategies and Their Implications in the Management of Biofilm-Associated Bacterial Infections. Braz. Arch. Biol. Technol. 2020, 63, e20190555. [Google Scholar] [CrossRef]
- García-Contreras, R. Is quorum sensing interference a viable alternative to treat Pseudomonas aeruginosa infections? Front. Microbiol. 2016, 7, 1454. [Google Scholar] [CrossRef]
- Mellini, M.; Di Muzio, E.; D’Angelo, F.; Baldelli, V.; Ferrillo, S.; Visca, P.; Leoni, L.; Polticelli, F.; Rampioni, G. In silico selection and experimental validation of FDA-approved drugs as anti-quorum sensing agents. Front. Microbiol. 2019, 10, 2355. [Google Scholar] [CrossRef] [PubMed]
- LaSarre, B.; Federle, M.J. Exploiting Quorum Sensing To Confuse Bacterial Pathogens. Microbiol. Mol. Biol. Rev. 2013, 77, 73–111. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, L.; Zhang, M.; Liu, H.; Lu, P.; Lin, K. Quorum sensing inhibitors: A patent review (2014–2018). Expert Opin. Ther. Patents 2018, 28, 849–865. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.C. Quorum sensing inhibitors: An overview. Biotechnol. Adv. 2013, 31, 224–245. [Google Scholar] [CrossRef] [PubMed]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.M.; Van De Wiele, T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Duytschaever, G.; Huys, G.; Bekaert, M.; Boulanger, L.; De Boeck, K.; Vandamme, P. Dysbiosis of bifidobacteria and Clostridium cluster XIVa in the cystic fibrosis fecal microbiota. J. Cyst. Fibros. 2012, 12, 206–215. [Google Scholar] [CrossRef]
- Miyake, S.; Kim, S.; Suda, W.; Oshima, K.; Nakamura, M.; Matsuoka, T.; Chihara, N.; Tomita, A.; Sato, W.; Kim, S.-W.; et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS ONE 2015, 10, e0137429. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Seksik, P.; Rigottier-Gois, L.; Lay, C.; Lepage, P.; Podglajen, I.; Marteau, P.; Doré, J. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm. Bowel Dis. 2006, 12, 106–111. [Google Scholar] [CrossRef]
- Imhann, F.; Vila, A.V.; Bonder, M.J.; Fu, J.; Gevers, D.; Visschedijk, M.C.; Spekhorst, L.M.; Alberts, R.; Franke, L.; Van Dullemen, H.M.; et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2016, 67, 108–119. [Google Scholar] [CrossRef]
- Livanos, A.; Snider, E.J.; Whittier, S.; Chong, D.H.; Wang, T.C.; Abrams, J.A.; Freedberg, D.E. Rapid gastrointestinal loss of Clostridial Clusters IV and XIVa in the ICU associates with an expansion of gut pathogens. PLoS ONE 2018, 13, e0200322. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, E.G.; Nishida, A.; Weston, A.; Bañuelos, M.S.; Potter, K.; Conery, J.; Guillemin, K. Agent-Based Modeling Demonstrates How Local Chemotactic Behavior Can Shape Biofilm Architecture. mSphere 2019, 4, e00285-19. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.A.; Benitez, J.A. Differential response of Vibrio cholerae planktonic and biofilm cells to autoinducer 2 deficiency. Microbiol. Immunol. 2009, 53, 582–586. [Google Scholar] [CrossRef] [PubMed]
- Velusamy, S.K.; Sampathkumar, V.; Godboley, D.; Fine, D.H. Survival of an Aggregatibacter actinomycetemcomitans quorum sensing luxS mutant in the mouths of Rhesus monkeys: Insights into ecological adaptation. Mol. Oral Microbiol. 2017, 32, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhou, R.; Li, T.; Kang, M.; Wan, Y.; Xu, Z.; Chen, H. Enhanced biofilm formation and reduced virulence of Actinobacillus pleuropneumoniae luxS mutant. Microb. Pathog. 2008, 45, 192–200. [Google Scholar] [CrossRef]
- Zhang, B.; Ku, X.; Zhang, X.; Zhang, Y.; Chen, G.; Chen, F.; Zeng, W.; Li, J.; Zhu, L.; He, Q. The AI-2/luxS quorum sensing system affects the growth characteristics, biofilm formation, and virulence of Haemophilus parasuis. Front. Cell. Infect. Microbiol. 2019, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Qiu, S.; Jiang, Q.; Sun, H.; Xue, T.; Cai, G.; Sun, B. AI-2 quorum sensing negatively regulates rbf expression and biofilm formation in Staphylococcus aureus. Int. J. Med Microbiol. 2017, 307, 257–267. [Google Scholar] [CrossRef]
- Xue, T.; Ni, J.; Shang, F.; Chen, X.; Zhang, M. Autoinducer-2 increases biofilm formation via an ica- and bhp-dependent manner in Staphylococcus epidermidis RP62A. Microbes Infect. 2015, 17, 345–352. [Google Scholar] [CrossRef]
- He, Z.; Liang, J.; Tang, Z.; Ma, R.; Peng, H.; Huang, Z. Role of the luxS Gene in Initial Biofilm Formation by Streptococcus mutans. Microb. Physiol. 2015, 25, 60–68. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Liang, J.; Zhou, W.; Xie, Q.; Tang, Z.; Ma, R.; Huang, Z. Effect of the quorum-sensing luxS gene on biofilm formation by Enterococcus faecalis. Eur. J. Oral Sci. 2016, 124, 234–240. [Google Scholar] [CrossRef]
- Auger, S.; Krin, E.; Aymerich, S.; Gohar, M. Autoinducer 2 Affects Biofilm Formation by Bacillus cereus. Appl. Environ. Microbiol. 2006, 72, 937–941. [Google Scholar] [CrossRef]
- He, X.; Lu, F.; Yuan, F.; Jiang, D.; Zhao, P.; Zhu, J.; Cheng, H.; Cao, J.; Lu, G. Biofilm Formation Caused by Clinical Acinetobacter baumannii Isolates Is Associated with Overexpression of the AdeFGH Efflux Pump. Antimicrob. Agents Chemother. 2015, 59, 4817–4825. [Google Scholar] [CrossRef] [PubMed]
- Starkey, M.; Lepine, F.; Maura, D.; Bandyopadhaya, A.; Lesic, B.; He, J.; Kitao, T.; Righi, V.; Milot, S.; Tzika, A.; et al. Identification of Anti-virulence Compounds That Disrupt Quorum-Sensing Regulated Acute and Persistent Pathogenicity. PLOS Pathog. 2014, 10, e1004321. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T. Quorum-Sensing Systems as Targets for Antivirulence Therapy. Trends Microbiol. 2018, 26, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Rémy, B.; Mion, S.; Plener, L.; Elias, M.; Chabrière, E.; Daudé, D. Interference in Bacterial Quorum Sensing: A Biopharmaceutical Perspective. Front. Pharmacol. 2018, 9, 203. [Google Scholar] [CrossRef]
- Fleitas Martínez, O.; Rigueiras, P.O.; Pires, Á.D.S.; Porto, W.F.; Silva, O.N.; de la Fuente-Nunez, C.; Franco, O.L. Interference with quorum-sensing signal biosynthesis as a promising therapeutic strategy against multidrug-resistant pathogens. Front. Cell. Infect. Microbiol. 2019, 8, 444. [Google Scholar] [CrossRef]
- Gohil, N.; Ramírez-García, R.; Panchasara, H.; Patel, S.; Bhattacharjee, G.; Singh, V. Book Review: Quorum Sensing vs. Quorum Quenching: A Battle With No End in Sight. Front. Cell. Infect. Microbiol. 2018, 8, 106. [Google Scholar] [CrossRef]
- García-Contreras, R.; Maeda, T.; Wood, T. Resistance to Quorum-Quenching Compounds. Appl. Environ. Microbiol. 2013, 79, 6840–6846. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Teo, J.W.P.; Drautz-Moses, D.I.; Schuster, S.C.; Givskov, M.; Yang, L. Acquisition of resistance to carbapenem and macrolide-mediated quorum sensing inhibition by Pseudomonas aeruginosa via ICETn43716385. Commun. Biol. 2018, 1, 57. [Google Scholar] [CrossRef]
- Siller, M.; Janapatla, R.P.; A Pirzada, Z.; Hassler, C.; Zinkl, D.; Charpentier, E. Functional analysis of the group A streptococcal luxS/AI-2 system in metabolism, adaptation to stress and interaction with host cells. BMC Microbiol. 2008, 8, 188. [Google Scholar] [CrossRef]
- Zhao, L.; Xue, T.; Shang, F.; Sun, H.; Sun, B. Staphylococcus aureus AI-2 Quorum Sensing Associates with the KdpDE Two-Component System To Regulate Capsular Polysaccharide Synthesis and Virulence. Infect. Immun. 2010, 78, 3506–3515. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T.; Boon, N.; Bossier, P. Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 2010, 6, e1000989. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.C.; Wood, T.; Kumar, P. Evolution of Resistance to Quorum-Sensing Inhibitors. Microb. Ecol. 2013, 68, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qin, Q.; Defoirdt, T. Does quorum sensing interference affect the fitness of bacterial pathogens in the real world? Environ. Microbiol. 2018, 20, 3918–3926. [Google Scholar] [CrossRef]
- Laabei, M.; Jamieson, W.D.; Yang, Y.; van den Elsen, J.; Jenkins, A.T.A. Investigating the lytic activity and structural properties of Staphylococcus aureus phenol soluble modulin (PSM) peptide toxins. Biochim. Biophys. Acta (BBA) Biomembr. 2014, 1838, 3153–3161. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Meng, J.; Cheng, J.; Fan, Z.; Chen, P.; Ruan, H.; Tu, Z.; Kang, N.; Li, N.; Xu, Y.; et al. Pseudomonas aeruginosa quorum-sensing metabolite induces host immune cell death through cell surface lipid domain dissolution. Nat. Microbiol. 2018, 4, 97–111. [Google Scholar] [CrossRef]
- García-Contreras, R.; Nuñez-López, L.; Jasso-Chávez, R.; Kwan, B.W.; A Belmont, J.; Rangel-Vega, A.; Maeda, T.; Wood, T. Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J. 2014, 9, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Kostylev, M.; Kim, D.Y.; Smalley, N.E.; Salukhe, I.; Greenberg, E.P.; Dandekar, A.A. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc. Natl. Acad. Sci. USA 2019, 116, 7027–7032. [Google Scholar] [CrossRef] [PubMed]
- De Spiegeleer, B.; Verbeke, F.; D’Hondt, M.; Hendrix, A.; Van De Wiele, C.; Burvenich, C.; Peremans, K.; De Wever, O.; Bracke, M.; Wynendaele, E. The Quorum Sensing Peptides PhrG, CSP and EDF Promote Angiogenesis and Invasion of Breast Cancer Cells In Vitro. PLoS ONE 2015, 10, e0119471. [Google Scholar] [CrossRef] [PubMed]
- Wynendaele, E.; Verbeke, F.; D’Hondt, M.; Hendrix, A.; Van De Wiele, C.; Burvenich, C.; Peremans, K.; De Wever, O.; Bracke, M.; De Spiegeleer, B. Crosstalk between the micro-biome and cancer cells by quorum sensing peptides. Peptides 2015, 64, 40–48. [Google Scholar] [PubMed]
- Trosko, J.E.; Lenz, H.J. What roles do colon stem cells and gap junctions play in the left and right location of origin of colorectal cancers? J. Cell. Commun. Signal. 2017, 11, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Blake, K.J.; Baral, P.; Voisin, T.; Lubkin, A.; Pinho-Ribeiro, F.A.; Adams, K.L.; Roberson, D.P.; Ma, Y.C.; Otto, M.; Woolf, C.J.; et al. Staphylococcus aureus produces pain through pore-forming toxins and neuronal TRPV1 that is silenced by QX-314. Nat. Commun. 2018, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Rautenberg, M.; Joo, H.; Otto, M.; Peschel, A. Neutrophil responses to staphylococcal pathogens and commensals via the formyl peptide receptor 2 relates to phenol-soluble modulin release and virulence. FASEB J. 2010, 25, 1254–1263. [Google Scholar] [CrossRef]
- Bader, M.; Alenina, N.; Andrade-Navarro, M.A.; Santos, R.A. Mas and its related G protein-coupled receptors, Mrgprs. Pharmacol. Rev. 2014, 66, 1080–1105. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Oscherwitz, J.; Cease, K.B.; Chan, S.M.; Munoz-Planillo, R.; Hasegawa, M.; Villaruz, A.E.; Cheung, G.Y.; McGavin, M.J.; Travers, J.B.; et al. Staphylococcus d-toxin induces allergic skin disease by activating mast cells. Nature 2013, 503, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Hockley, J.; Taylor, T.S.; Callejo, G.; Wilbrey, A.L.; Gutteridge, A.; Bach, K.; Winchester, W.J.; Bulmer, D.C.; McMurray, G.; Smith, E.S.J. Single-cell RNAseq reveals seven classes of colonic sensory neuron. Gut 2018, 68, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Uhlig, F.; Grundy, L.; Garcia-Caraballo, S.; Brierley, S.M.; Foster, S.J.; Grundy, D. Identification of a Quorum Sensing-Dependent Communication Pathway Mediating Bacteria-Gut-Brain Cross Talk. iScience 2020, 23, 101695. [Google Scholar] [CrossRef]
- Chiu, I.M.; Heesters, B.A.; Ghasemlou, N.; Von Hehn, C.A.; Zhao, F.; Tran, J.; Wainger, B.; Strominger, A.; Muralidharan, S.; Horswill, A.R.; et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature 2013, 501, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Vandenesch, F.; Lina, G.; Henry, T. Staphylococcus aureus hemolysins, bicomponent leukocidins, and cytolytic peptides: A redundant arsenal of membrane-damaging virulence factors? Front. Cell. Infect. Microbiol. 2012, 2, 12. [Google Scholar] [CrossRef]
- Wilke, G.A.; Wardenburg, J.B. Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus-hemolysin-mediated cel-lular injury. Proc. Natl. Acad. Sci. USA 2010, 107, 13473–13478. [Google Scholar] [CrossRef]
- Rinttilä, T.; Lyra, A.; Krogius-Kurikka, L.; Palva, A. Real-time PCR analysis of enteric pathogens from fecal samples of irritable bowel syndrome subjects. Gut Pathog. 2011, 3, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Denayer, S.; Delbrassinne, L.; Nia, Y.; Botteldoorn, N. Food-Borne Outbreak Investigation and Molecular Typing: High Diversity of Staphylococcus aureus Strains and Importance of Toxin Detection. Toxins 2017, 9, 407. [Google Scholar] [CrossRef]
- Janssens, Y.; Wynendaele, E.; Verbeke, F.; Debunne, N.; Gevaert, B.; Audenaert, K.; Van DeWiele, C.; De Spiegeleer, B. Screening of quorum sensing peptides for biological effects in neuronal cells. Peptides 2018, 101, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.K.; Kasper, D.L.; Wang, B.; Forsythe, P.; Bienenstock, J.; Kunze, W.A. Bacteroides fragilis polysaccharide A is necessary and sufficient for acute activation of intestinal sensory neurons. Nat. Commun. 2013, 4, 1465. [Google Scholar] [CrossRef]
- Pokusaeva, K.; Johnson, C.; Luk, B.; Uribe, G.; Fu, Y.; Oezguen, N.; Matsunami, R.K.; Lugo, M.; Major, A.; Mori-Akiyama, Y.; et al. GABA producing Bifidobacterium dentium modulates visceral sensitivity in the intestine. Neurogastroenterol. Motil. 2017, 29, e12904. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, Y.; Williams, B.B.; Battaglioli, E.J.; Whitaker, W.R.; Till, L.; Grover, M.; Linden, D.R.; Akiba, Y.; Kandimalla, K.K.; Zachos, N.C.; et al. Gut Microbiota-Produced Tryptamine Activates an Epithelial G-Protein-Coupled Receptor to Increase Colonic Secretion. Cell Host Microbe 2018, 23, 775–785.e5. [Google Scholar] [CrossRef] [PubMed]
- Strandwitz, P.; Kim, K.H.; Terekhova, D.; Liu, J.K.; Sharma, A.; Levering, J.; McDonald, D.; Dietrich, D.; Ramadhar, T.R.; Lekbua, A.; et al. GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 2019, 4, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Gourbeyre, P.; Berri, M.; Lippi, Y.; Meurens, F.; Vincent-Naulleau, S.; Laffitte, J.; Rogel-Gaillard, C.; Pinton, P.; Oswald, I.P. Pattern recognition receptors in the gut: Analysis of their expression along the intestinal tract and the crypt/villus axis. Physiol. Rep. 2015, 3, e12225. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Harvima, I.T. Mast cell-neural interactions contribute to pain and itch. Immunol. Rev. 2018, 282, 168–187. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhang, L.; Bai, T.; Qian, W.; Li, R.; Hou, X. Mast Cell-dependent Mesenteric Afferent Activation by Mucosal Supernatant From Different Bowel Segments of Guinea Pigs With Post-infectious Irritable Bowel Syndrome. J. Neurogastroenterol. Motil. 2015, 21, 236–246. [Google Scholar] [CrossRef]
- Abdel-Haq, R.; Schlachetzki, J.C.M.; Glass, C.K.; Mazmanian, S.K. Microbiome-microglia connections via the gut-brain axis. J. Exp. Med. 2019, 216, 41–59. [Google Scholar] [CrossRef]
- Sampson, T.R.; Debelius, J.W.; Thron, T.; Janssen, S.; Shastri, G.G.; Ilhan, Z.E.; Challis, C.; Schretter, C.E.; Rocha, S.; Gradinaru, V.; et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016, 167, 1469–1480.e12. [Google Scholar] [CrossRef]
- Shen, H.; Guan, Q.; Zhang, X.; Yuan, C.; Tan, Z.; Zhai, L.; Hao, Y.; Gu, Y.; Han, C. New mechanism of neuroinflammation in Alzheimer’s disease: The activation of NLRP3 inflammasome mediated by gut microbiota. Prog. Neuropsychopharmacol. Biol. Psychiatry 2020, 100, 109884. [Google Scholar] [CrossRef] [PubMed]
- Tengeler, A.C.; Dam, S.A.; Wiesmann, M.; Naaijen, J.; Van Bodegom, M.; Belzer, C.; Dederen, P.J.; Verweij, V.; Franke, B.; Kozicz, T.; et al. Gut microbiota from persons with attention-deficit/hyperactivity disorder affects the brain in mice. Microbiome 2020, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wu, B.; Liang, J.; He, F.; Gu, W.; Li, K.; Luo, Y.; Chen, J.; Gao, Y.; Wu, Z.; et al. Altered gut microbiota and mucosal immunity in patients with schizo-phrenia. Brain Behav. Immun. 2020, 85, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-J.; He, S.; Fang, L.; Wang, B.; Bai, S.-J.; Xie, J.; Zhou, C.-J.; Wang, W.; Xie, P. Age-specific differential changes on gut microbiota composition in patients with major depressive disorder. Aging 2020, 12, 2764–2776. [Google Scholar] [CrossRef] [PubMed]
- Panza, F.; Lozupone, M.; Solfrizzi, V.; Watling, M.; Imbimbo, B. Time to test antibacterial therapy in Alzheimer’s disease. Brain 2019, 142, 2905–2929. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef]
- Dicks, L.M.T. Gut Bacteria and Neurotransmitters. Microorganisms 2022, 10, 1838. [Google Scholar] [CrossRef]
- Dicks, L.M.T.; Hurn, D.; Hermanus, D. Gut Bacteria and Neuropsychiatric Disorders. Microorganisms 2021, 9, 2583. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).