Computer-Based Identification of Potential Druggable Targets in Multidrug-Resistant Acinetobacter baumannii: A Combined In Silico, In Vitro and In Vivo Study
Abstract
1. Introduction
2. Materials and Methods
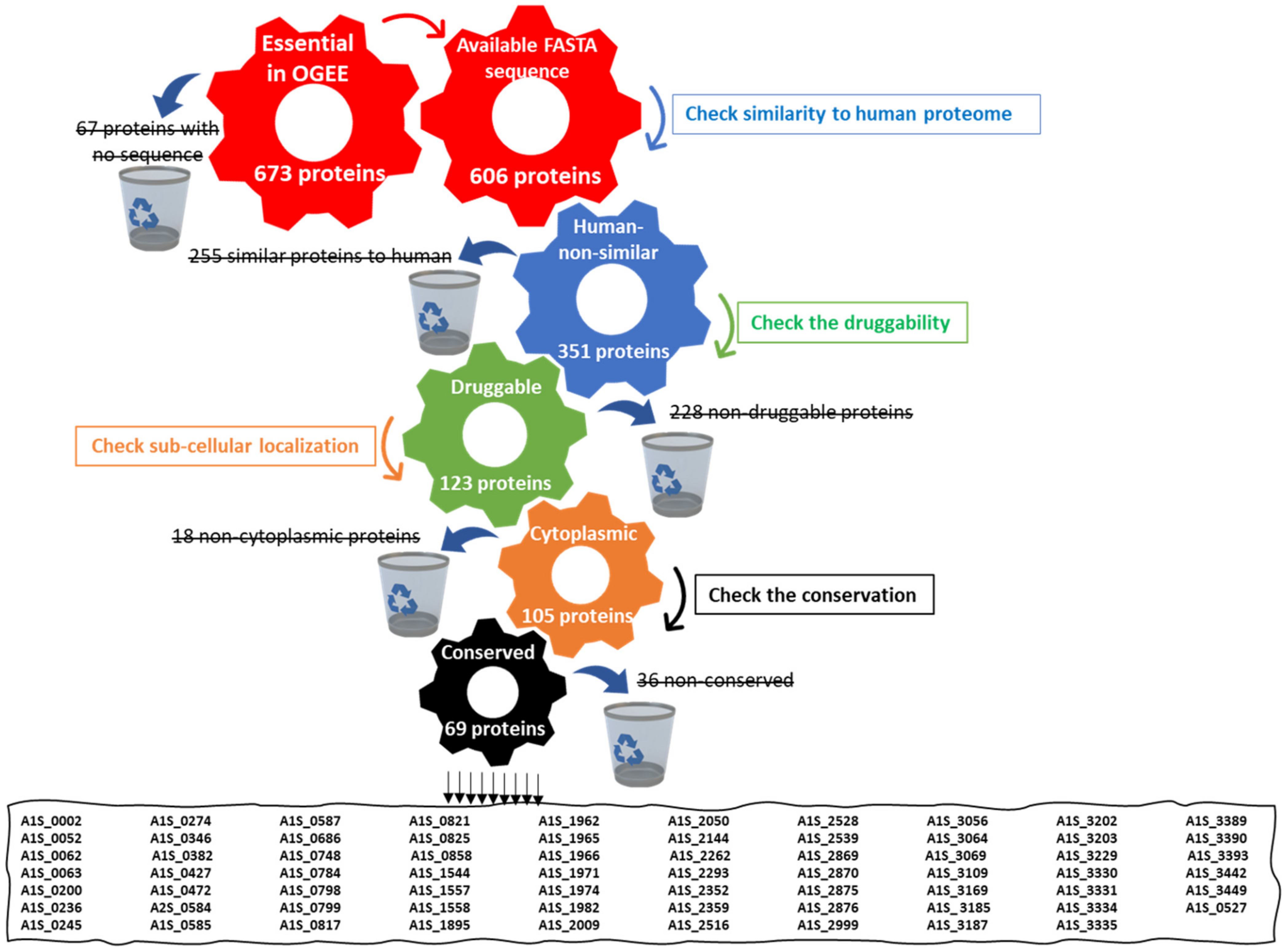
2.1. Retrieval of Essential Druggable Proteins Using the In Silico Approach
2.1.1. Retrieval of Essential Proteins
2.1.2. Identification of Human Non-Homologous Proteins
2.1.3. Druggability Analysis
2.1.4. Sub-Cellular Localization Prediction
2.1.5. Conservation of Potential Proteins in A. baumannii
2.1.6. Comparison of Shortlisted Proteins to the A. baumannii ATCC 19606 Proteome
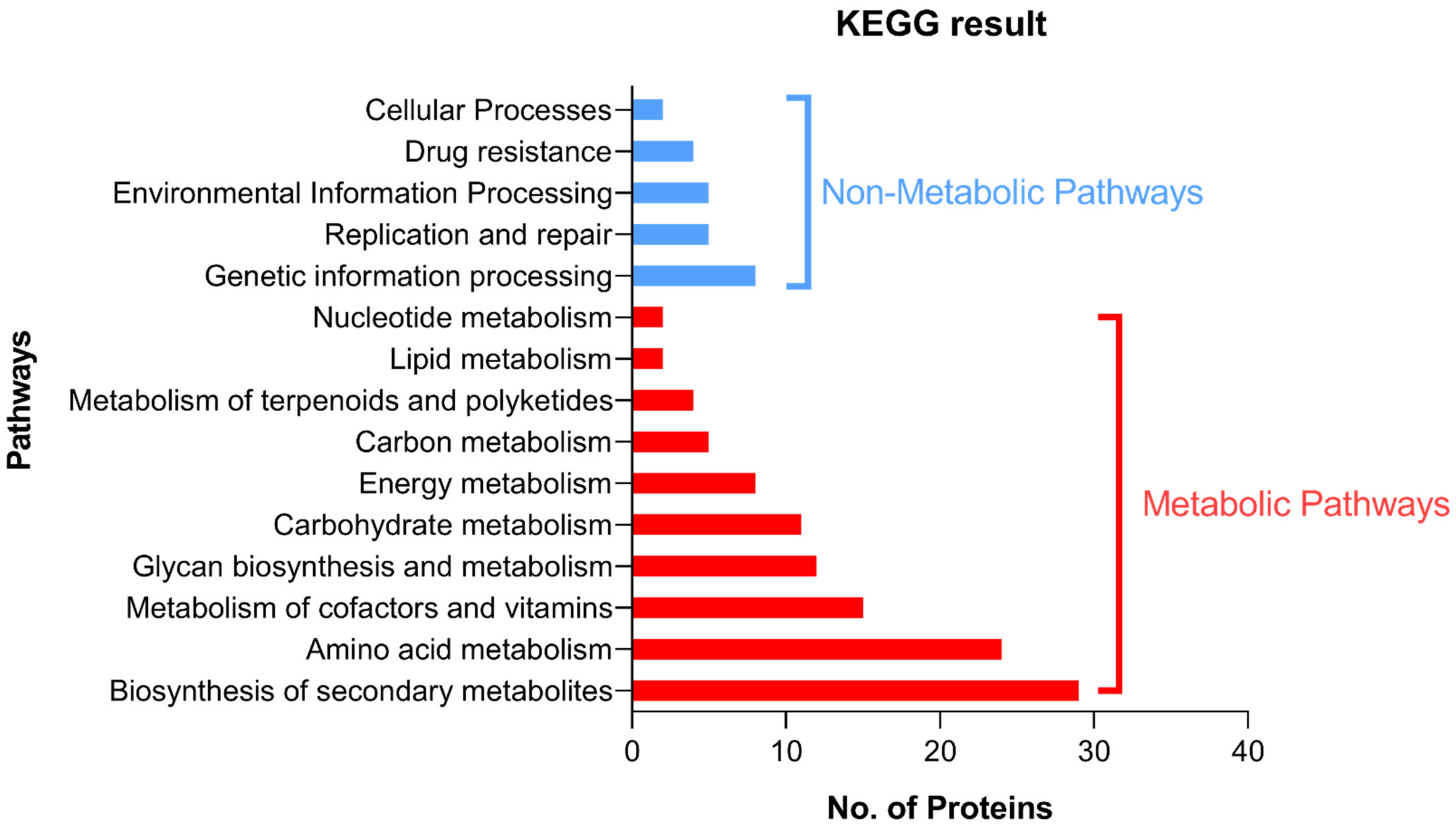
2.1.7. Detection of the Pathways Involving the Shortlisted Proteins
2.2. Testing of the In Vitro Antimicrobial Activity of Candidate Ligands against A. baumannii ATCC 19606
2.2.1. Preparation of the Ligand Solutions
2.2.2. Culturing of the Bacterial Strain and Inoculum Preparation
2.2.3. Determination of the MIC of Candidate Ligands
2.3. Molecular Docking of Candidate Ligands to Their Target Proteins
2.4. In Vivo Effect of Candidate Ligands on Wound Infection in the Mouse Model
2.4.1. Ethical Statement
2.4.2. Induction of Infection
2.4.3. Treatment of A. baumannii-Infected Wounds by Candidate Ligands
- Group 1: Dulbecco’s Phosphate-buffered saline (PBS) (Biowest, France)
- Group 2: Cefepime solution (Sandoz, Egypt)
- Group 3: Citric acid solution
- Group 4: D-tartaric acid solution
- Group 5: Malonic acid solution
2.4.4. Mice Euthanization and Counting the Colonies on the Wounded Skin
2.5. In Vitro Cytotoxicity Assay of Candidate Ligands against Human Skin Fibroblast
2.6. In Vitro Confirmation of Aspartate 1-Decarboxylase Inhibition by Malonic Acid
2.7. Statistical Analysis
3. Results
3.1. Retrieval of Essential Druggable Proteins Using the In Silico Approach
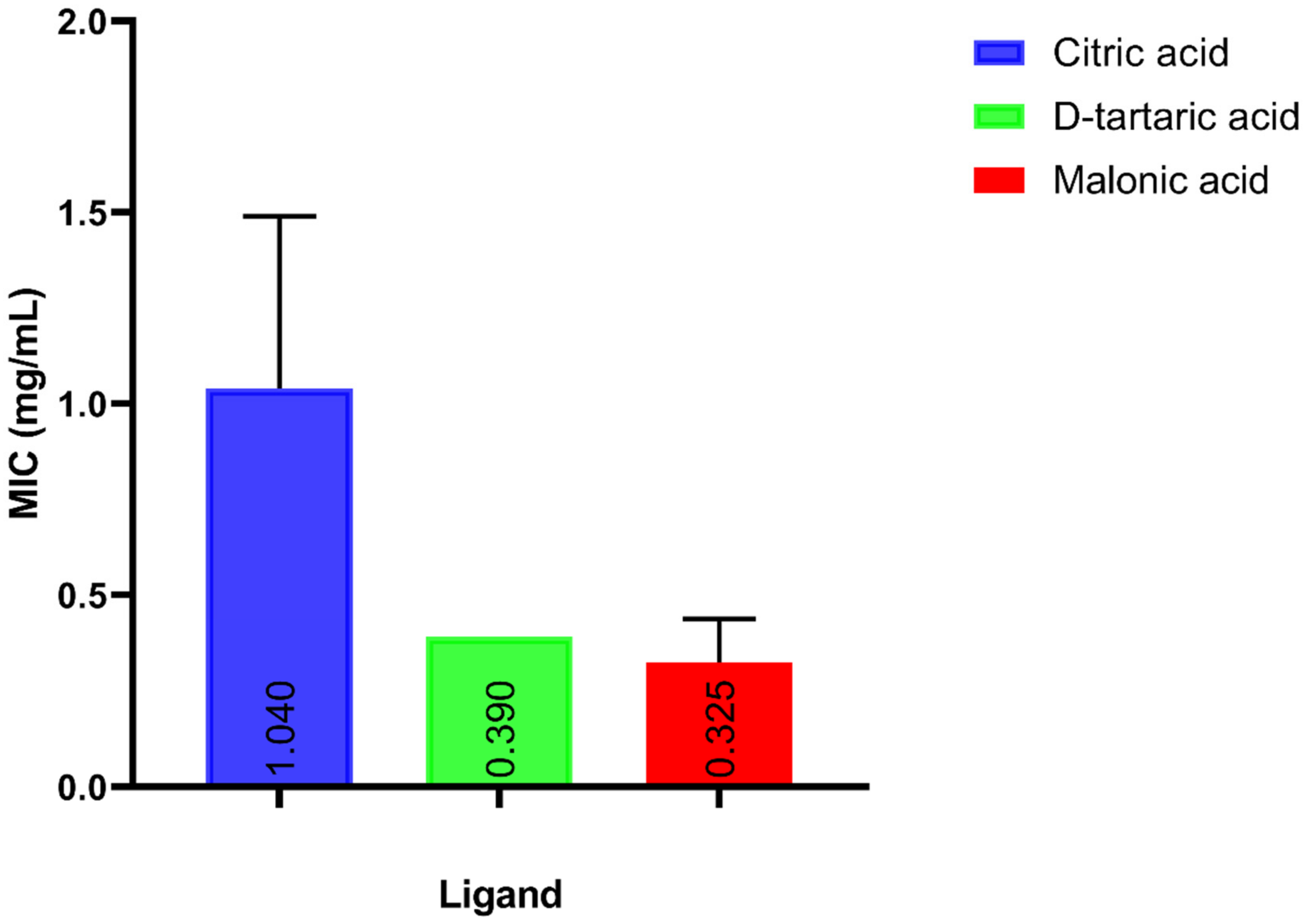
3.2. Testing the In Vitro Antimicrobial Activity of Candidate Ligands against A. baumannii
3.3. Molecular Docking of Candidate Ligands to Their Target Proteins
3.3.1. Docking of Citric Acid to Glutamine Synthetase
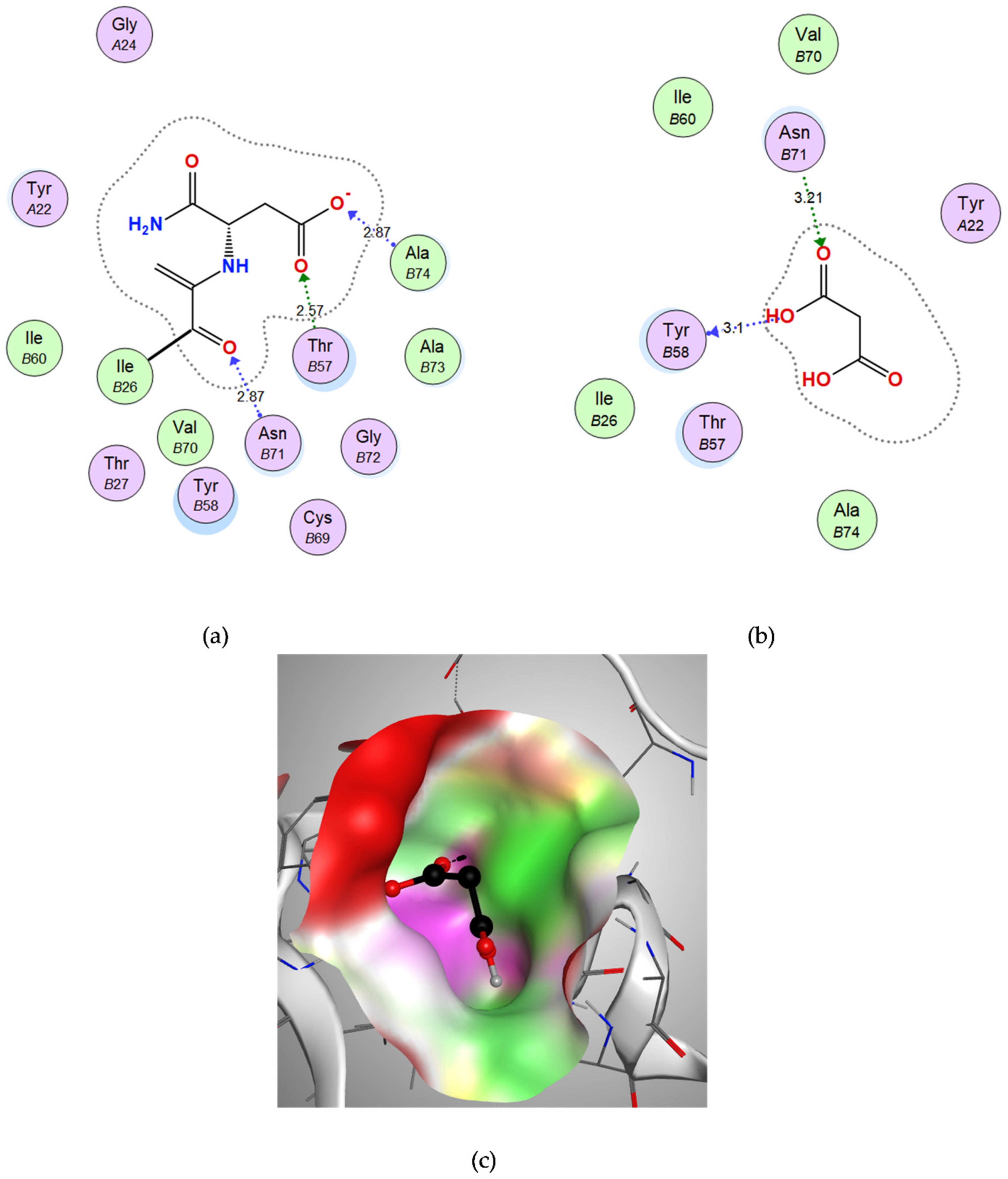
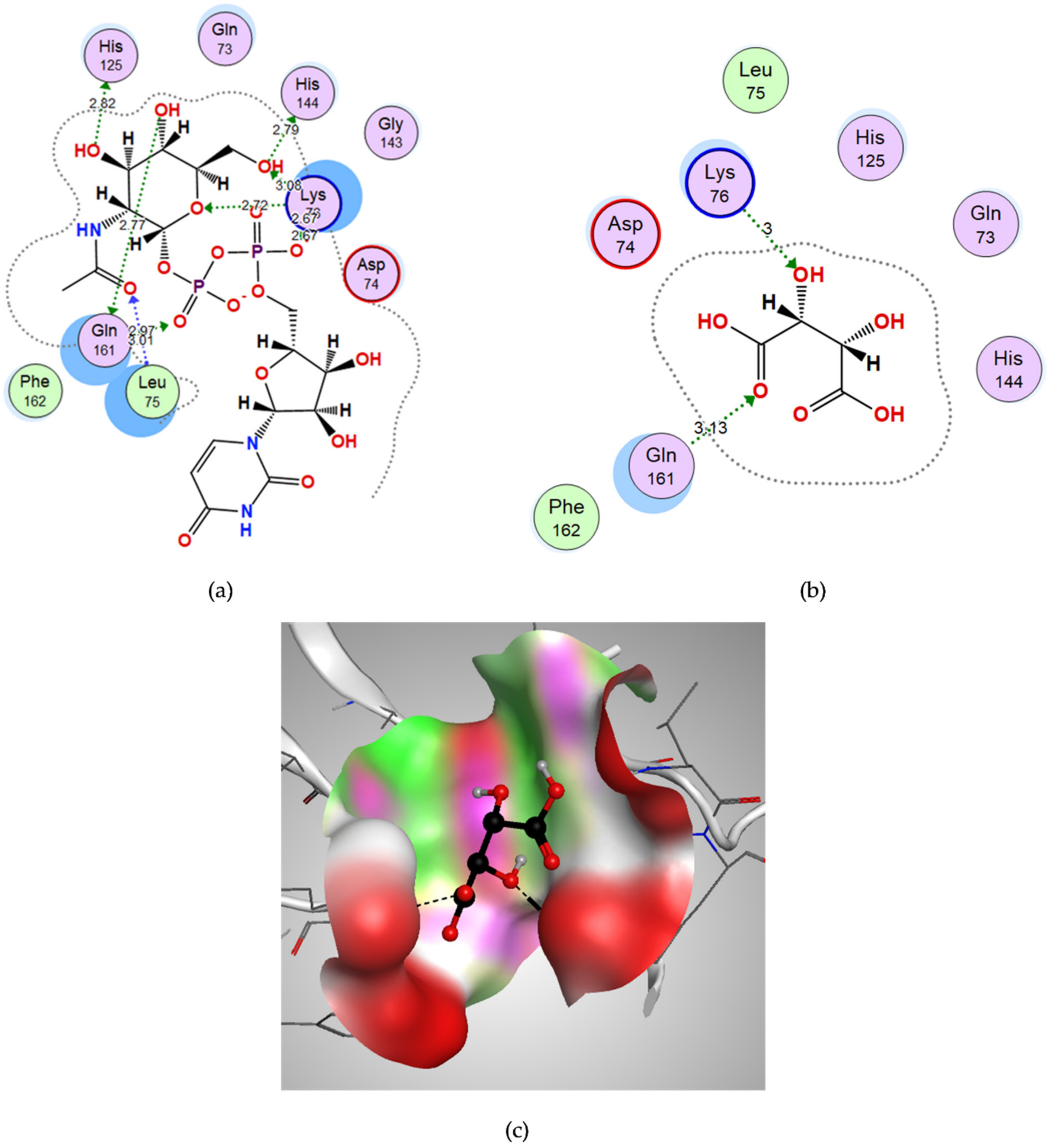
3.3.2. Docking of Malonic Acid to Aspartate 1-Decarboxylase
3.3.3. Docking of D-Tartaric Acid to UDP-N-acetylglucosamine Acyltransferase
3.4. In Vivo Effect of Candidate Ligands on Wound Infection in the Mouse Model
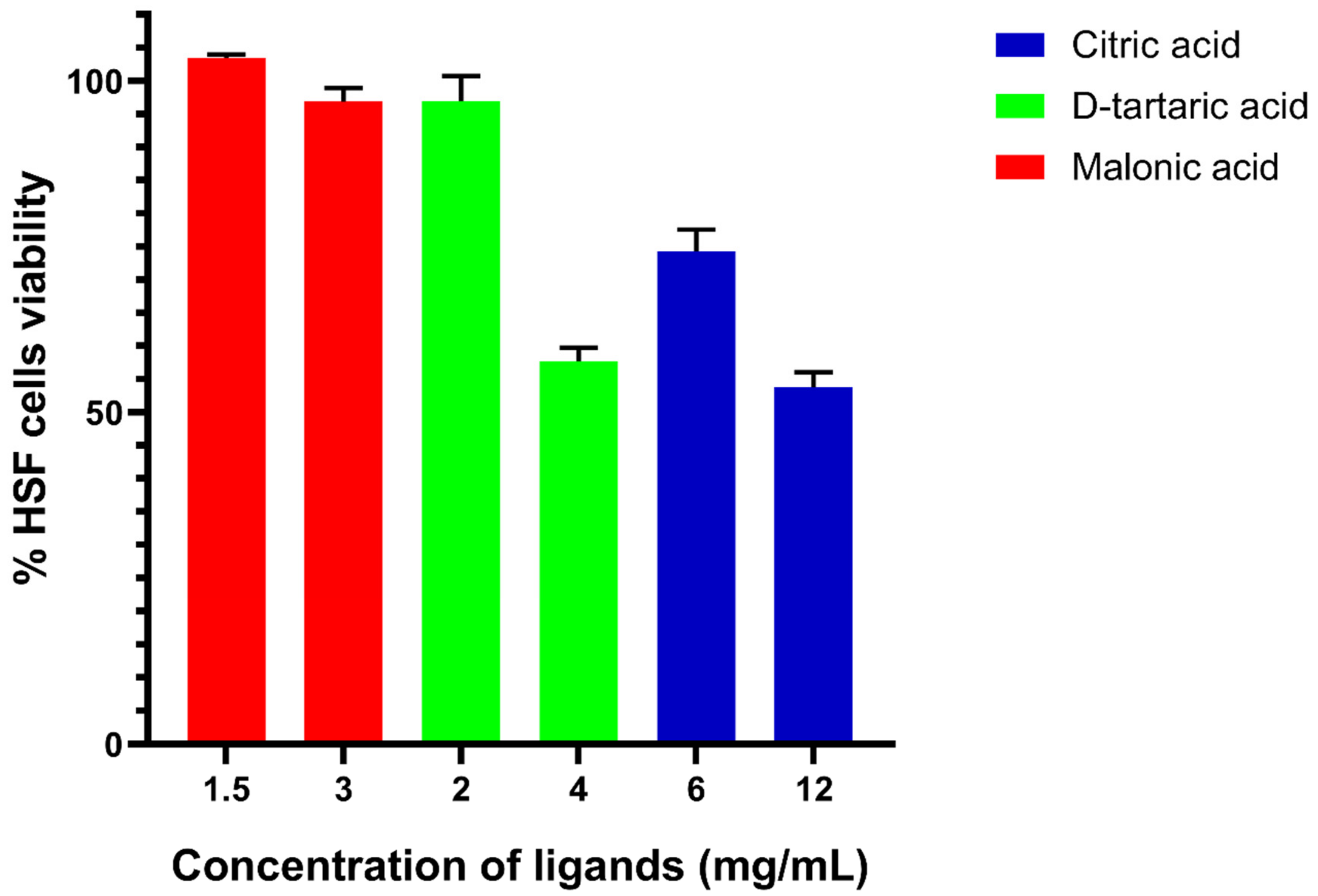
3.5. In Vitro Cytotoxicity Assay of Candidate Ligands against Human Skin Fibroblast
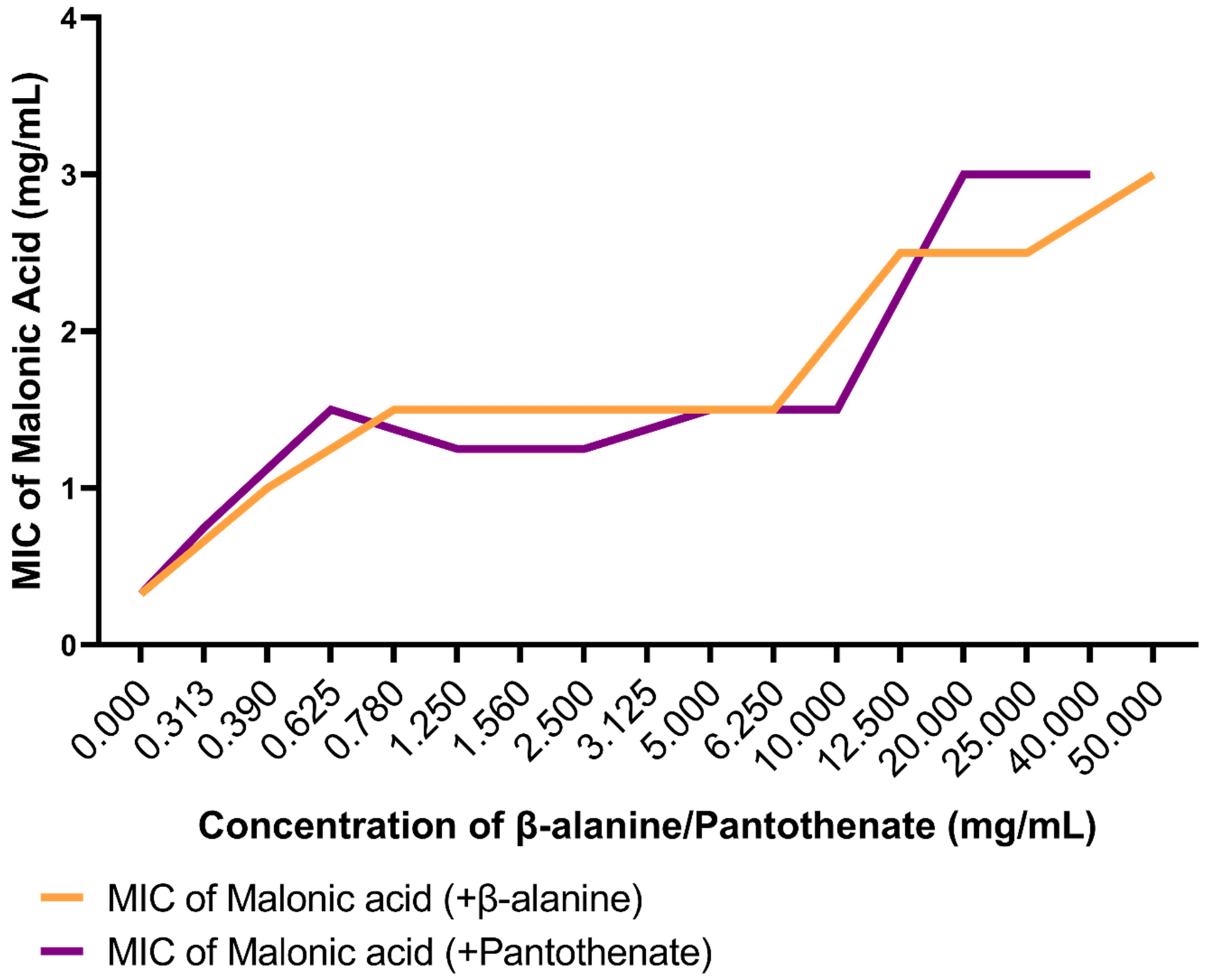
3.6. In Vitro Confirmation of Aspartate 1-Decarboxylase Inhibition by Malonic Acid
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a Successful Pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Eliopoulos, G.M.; Maragakis, L.L.; Perl, T.M. Acinetobacter baumannii: Epidemiology, Antimicrobial Resistance, and Treatment Options. Clin. Infect. Dis. 2008, 46, 1254–1263. [Google Scholar] [CrossRef]
- Brotfain, E.; Borer, A.; Koyfman, L.; Saidel-Odes, L.; Frenkel, A.; Gruenbaum, S.E.; Rosenzweig, V.; Zlotnik, A.; Klein, M. Multidrug Resistance Acinetobacter Bacteremia Secondary to Ventilator-Associated Pneumonia: Risk Factors and Outcome. J. Intensive Care Med. 2017, 32, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Perez, F.; Hujer, A.M.; Hujer, K.M.; Decker, B.K.; Rather, P.N.; Bonomo, R.A. Global Challenge of Multidrug-Resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007, 51, 3471–3484. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Priority Pathogens List for R&D of New Antibiotics. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 25 June 2022).
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States; US Department of Health and Human Services: Washington, DC, USA, 2019.
- Fields, F.R.; Lee, S.W.; McConnell, M.J. Using bacterial genomes and essential genes for the development of new antibiotics. Biochem. Pharmacol. 2017, 134, 74–86. [Google Scholar] [CrossRef]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007, 6, 29–40. [Google Scholar] [CrossRef]
- Zhang, X.; Acencio, M.L.; Lemke, N. Predicting Essential Genes and Proteins Based on Machine Learning and Network Topological Features: A Comprehensive Review. Front. Physiol. 2016, 7, 75. [Google Scholar] [CrossRef]
- Serral, F.; Castello, F.A.; Sosa, E.J.; Pardo, A.M.; Palumbo, M.C.; Modenutti, C.; Palomino, M.M.; Lazarowski, A.; Auzmendi, J.; Ramos, P.I.P.; et al. From Genome to Drugs: New Approaches in Antimicrobial Discovery. Front. Pharmacol. 2021, 12, 647060. [Google Scholar] [CrossRef]
- Mondal, S.I.; Ferdous, S.; Jewel, N.A.; Akter, A.; Mahmud, Z.; Islam, M.M.; Afrin, T.; Karim, N. Identification of potential drug targets by subtractive genome analysis of Escherichia coli O157:H7: An in silico approach. Adv. Appl. Bioinform. Chem. 2015, 8, 49–63. [Google Scholar] [CrossRef]
- Ibrahim, K.A.; Helmy, O.M.; Kashef, M.T.; Elkhamissy, T.R.; Ramadan, M.A. Identification of Potential Drug Targets in Helicobacter pylori Using In Silico Subtractive Proteomics Approaches and Their Possible Inhibition through Drug Repurposing. Pathogens 2020, 9, 747. [Google Scholar] [CrossRef]
- Uddin, R.; Siraj, B.; Rashid, M.; Khan, A.; Ahsan Halim, S.; Al-Harrasi, A. Genome Subtraction and Comparison for the Identification of Novel Drug Targets against Mycobacterium avium subsp. hominissuis. Pathogens 2020, 9, 368. [Google Scholar] [CrossRef] [PubMed]
- Uddin, R.; Jamil, F. Prioritization of potential drug targets against P. aeruginosa by core proteomic analysis using computational subtractive genomics and Protein-Protein interaction network. Comput. Biol. Chem. 2018, 74, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.U.; Khan, M.A.; Hashem, A.; Islam, M.M.; Morshed, M.N.; Keya, C.A.; Salimullah, M. Finding Potential Therapeutic Targets against Shigella flexneri through Proteome Exploration. Front. Microbiol. 2016, 7, 1817. [Google Scholar] [CrossRef] [PubMed]
- Shahid, F.; Ashfaq, U.A.; Saeed, S.; Munir, S.; Almatroudi, A.; Khurshid, M. In Silico Subtractive Proteomics Approach for Identification of Potential Drug Targets in Staphylococcus saprophyticus. Int. J. Environ. Res. Public Health 2020, 17, 3644. [Google Scholar] [CrossRef] [PubMed]
- Saleem, H.; Ashfaq, U.A.; Nadeem, H.; Zubair, M.; Siddique, M.H.; Rasul, I. Subtractive genomics and molecular docking approach to identify drug targets against Stenotrophomonas maltophilia. PLoS ONE 2021, 16, e0261111. [Google Scholar] [CrossRef]
- Khan, K.; Jalal, K.; Khan, A.; Al-Harrasi, A.; Uddin, R. Comparative Metabolic Pathways Analysis and Subtractive Genomics Profiling to Prioritize Potential Drug Targets Against Streptococcus pneumoniae. Front. Microbiol. 2022, 12, 796363. [Google Scholar] [CrossRef]
- Naorem, R.S.; Pangabam, B.D.; Bora, S.S.; Goswami, G.; Barooah, M.; Hazarika, D.J.; Fekete, C. Identification of Putative Vaccine and Drug Targets against the Methicillin-Resistant Staphylococcus aureus by Reverse Vaccinology and Subtractive Genomics Approaches. Molecules 2022, 27, 2083. [Google Scholar] [CrossRef]
- Kaur, H.; Kalia, M.; Taneja, N. Identification of novel non-homologous drug targets against Acinetobacter baumannii using subtractive genomics and comparative metabolic pathway analysis. Microb. Pathog. 2021, 152, 104608. [Google Scholar] [CrossRef]
- Solanki, V.; Tiwari, V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci. Rep. 2018, 8, 9044. [Google Scholar] [CrossRef]
- Kaur, N.; Khokhar, M.; Jain, V.; Bharatam, P.V.; Sandhir, R.; Tewari, R. Identification of druggable targets for Acinetobacter baumannii via subtractive genomics and plausible inhibitors for MurA and MurB. Appl. Biochem. Biotechnol. 2013, 171, 417–436. [Google Scholar] [CrossRef]
- Chen, W.-H.; Minguez, P.; Lercher, M.J.; Bork, P. OGEE: An online gene essentiality database. Nucleic Acids Res. 2011, 40, D901–D906. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2017, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-S.; Chen, Y.-C.; Lu, C.-H.; Hwang, J.-K. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2020, 49, D545–D551. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- CLSI M07-A9; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012; Volume 32.
- Ismail, M.M.; Samir, R.; Saber, F.R.; Ahmed, S.R.; Farag, M.A. Pimenta Oil as a Potential Treatment for Acinetobacter baumannii Wound Infection: In Vitro and In Vivo Bioassays in Relation to Its Chemical Composition. Antibiotics 2020, 9, 679. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Iregui, A.; Landman, D.; Quale, J. Activity of cefepime/zidebactam (WCK 5222) against Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter baumannii endemic to New York City medical centres. J. Antimicrob. Chemother. 2019, 74, 2938–2942. [Google Scholar] [CrossRef]
- Zhu, Y.; Lu, J.; Zhao, J.; Zhang, X.; Yu, H.H.; Velkov, T.; Li, J. Complete genome sequence and genome-scale metabolic modelling of Acinetobacter baumannii type strain ATCC 19606. Int. J. Med. Microbiol. IJMM 2020, 310, 151412. [Google Scholar] [CrossRef] [PubMed]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. JNCI J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Allam, R.M.; Al-Abd, A.M.; Khedr, A.; Sharaf, O.A.; Nofal, S.M.; Khalifa, A.E.; Mosli, H.A.; Abdel-Naim, A.B. Fingolimod interrupts the cross talk between estrogen metabolism and sphingolipid metabolism within prostate cancer cells. Toxicol. Lett. 2018, 291, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Kotb, A.; Abutaleb, N.S.; Seleem, M.A.; Hagras, M.; Mohammad, H.; Bayoumi, A.; Ghiaty, A.; Seleem, M.N.; Mayhoub, A.S. Phenylthiazoles with tert-Butyl side chain: Metabolically stable with anti-biofilm activity. Eur. J. Med. Chem. 2018, 151, 110–120. [Google Scholar] [CrossRef]
- Shi, W.; Chen, J.; Feng, J.; Cui, P.; Zhang, S.; Weng, X.; Zhang, W.; Zhang, Y. Aspartate decarboxylase (PanD) as a new target of pyrazinamide in Mycobacterium tuberculosis. Emerg. Microbes Infect. 2014, 3, e58. [Google Scholar] [CrossRef] [PubMed]
- Miyasaki, Y.; Rabenstein, J.D.; Rhea, J.; Crouch, M.-L.; Mocek, U.M.; Kittell, P.E.; Morgan, M.A.; Nichols, W.S.; Van Benschoten, M.M.; Hardy, W.D.; et al. Isolation and Characterization of Antimicrobial Compounds in Plant Extracts against Multidrug-Resistant Acinetobacter baumannii. PLoS ONE 2013, 8, e61594. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.; Roy, R.; Tiwari, V. Screening of Herbal-Based Bioactive Extract Against Carbapenem-Resistant Strain of Acinetobacter baumannii. Microb. Drug Resist. 2016, 22, 364–371. [Google Scholar] [CrossRef]
- Tsai, C.-C.; Lin, C.-S.; Hsu, C.-R.; Chang, C.-M.; Chang, I.W.; Lin, L.-W.; Hung, C.-H.; Wang, J.-L. Using the Chinese herb Scutellaria barbata against extensively drug-resistant Acinetobacter baumannii infections: In vitro and in vivo studies. BMC Complementary Altern. Med. 2018, 18, 96. [Google Scholar] [CrossRef]
- Kusradze, I.; Karumidze, N.; Rigvava, S.; Dvalidze, T.; Katsitadze, M.; Amiranashvili, I.; Goderdzishvili, M. Characterization and Testing the Efficiency of Acinetobacter baumannii Phage vB-GEC_Ab-M-G7 as an Antibacterial Agent. Front. Microbiol. 2016, 7, 1590. [Google Scholar] [CrossRef]
- Jeon, J.; Ryu, C.-M.; Lee, J.-Y.; Park, J.-H.; Yong, D.; Lee, K.; Besser, T.E. In Vivo Application of Bacteriophage as a Potential Therapeutic Agent To Control OXA-66-Like Carbapenemase-Producing Acinetobacter baumannii Strains Belonging to Sequence Type 357. Appl. Environ. Microbiol. 2016, 82, 4200–4208. [Google Scholar] [CrossRef]
- Tiwari, V.; Raghav, R.; Tiwari, M. Comparative anti-bacterial activity of differently capped silver nanomaterial on the carbapenem sensitive and resistant strains of Acinetobacter baumannii. J. Nanomed. Nanotechnol. 2015, 6, 1000314. [Google Scholar] [CrossRef]
- Tiwari, V.; Tiwari, M.; Solanki, V. Polyvinylpyrrolidone-Capped Silver Nanoparticle Inhibits Infection of Carbapenem-Resistant Strain of Acinetobacter baumannii in the Human Pulmonary Epithelial Cell. Front. Immunol. 2017, 8, 973. [Google Scholar] [CrossRef] [PubMed]
- Navid, A.; Ahmad, S.; Sajjad, R.; Raza, S.; Azam, S.S. Structure Based in Silico Screening Revealed a Potent Acinetobacter baumannii Ftsz Inhibitor from Asinex Antibacterial Library. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 19, 3008–3018. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Tiwari, M.; Tiwari, V. In silico high-throughput virtual screening and molecular dynamics simulation study to identify inhibitor for AdeABC efflux pump of Acinetobacter baumannii. J. Biomol. Struct. Dyn. 2018, 36, 1182–1194. [Google Scholar] [CrossRef] [PubMed]
- Barh, D.; Tiwari, S.; Jain, N.; Ali, A.; Santos, A.R.; Misra, A.N.; Azevedo, V.; Kumar, A. In silico subtractive genomics for target identification in human bacterial pathogens. Drug Dev. Res. 2011, 72, 162–177. [Google Scholar] [CrossRef]
- Uddin, R.; Masood, F.; Azam, S.S.; Wadood, A. Identification of putative non-host essential genes and novel drug targets against Acinetobacter baumannii by in silico comparative genome analysis. Microb. Pathog. 2019, 128, 28–35. [Google Scholar] [CrossRef]
- Ball, H.S.; Girma, M.B.; Zainab, M.; Soojhawon, I.; Couch, R.D.; Noble, S.M. Characterization and Inhibition of 1-Deoxy-d-Xylulose 5-Phosphate Reductoisomerase: A Promising Drug Target in Acinetobacter baumannii and Klebsiella pneumoniae. ACS Infect. Dis. 2021, 7, 2987–2998. [Google Scholar] [CrossRef]
- Read, B.J.; Fisher, G.; Wissett, O.L.R.; Machado, T.F.G.; Nicholson, J.; Mitchell, J.B.O.; da Silva, R.G. Allosteric Inhibition of Acinetobacter baumannii ATP Phosphoribosyltransferase by Protein:Dipeptide and Protein:Protein Interactions. ACS Infect. Dis. 2022, 8, 197–209. [Google Scholar] [CrossRef]
- Jobelius, H.; Bianchino, G.I.; Borel, F.; Chaignon, P.; Seemann, M. The Reductive Dehydroxylation Catalyzed by IspH, a Source of Inspiration for the Development of Novel Anti-Infectives. Molecules 2022, 27, 708. [Google Scholar] [CrossRef]
- Amera, G.M.; Khan, R.J.; Pathak, A.; Kumar, A.; Singh, A.K. Structure based in-silico study on UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase (MurE) from Acinetobacter baumannii as a drug target against nosocomial infections. Inform. Med. Unlocked 2019, 16, 100216. [Google Scholar] [CrossRef]
- Hutton, C.A.; Perugini, M.A.; Gerrard, J.A. Inhibition of lysine biosynthesis: An evolving antibiotic strategy. Mol. BioSyst. 2007, 3, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.D.; Parkes, A.L.; Corbett, D.; Dickie, A.P.; Southey, M.; Andersen, O.A.; Stein, D.B.; Barbeau, O.R.; Sanzone, A.; Thommes, P.; et al. Discovery of Novel UDP-N-Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa. J. Med. Chem. 2021, 64, 14377–14425. [Google Scholar] [CrossRef] [PubMed]
- Dangkulwanich, M.; Raetz, C.R.H.; Williams, A.H. Structure guided design of an antibacterial peptide that targets UDP-N-acetylglucosamine acyltransferase. Sci. Rep. 2019, 9, 3947. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.H.; Immormino, R.M.; Gewirth, D.T.; Raetz, C.R.H. Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide. Proc. Natl. Acad. Sci. USA 2006, 103, 10877–10882. [Google Scholar] [CrossRef] [PubMed]
- Gopal, P.; Sarathy, J.P.; Yee, M.; Ragunathan, P.; Shin, J.; Bhushan, S.; Zhu, J.; Akopian, T.; Kandror, O.; Lim, T.K.; et al. Pyrazinamide triggers degradation of its target aspartate decarboxylase. Nat. Commun. 2020, 11, 1661. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.P.; Blanchard, J.E.; Murphy, C.; Roderick, S.L.; Brown, E.D. High-Throughput Screening Identifies Novel Inhibitors of the Acetyltransferase Activity of Escherichia coli GlmU. Antimicrob. Agents Chemother. 2009, 53, 2306–2311. [Google Scholar] [CrossRef] [PubMed]
- Buurman, E.T.; Andrews, B.; Gao, N.; Hu, J.; Keating, T.A.; Lahiri, S.; Otterbein, L.R.; Patten, A.D.; Stokes, S.S.; Shapiro, A.B. In vitro validation of acetyltransferase activity of GlmU as an antibacterial target in Haemophilus influenzae. J. Biol. Chem. 2011, 286, 40734–40742. [Google Scholar] [CrossRef] [PubMed]
- Laudy, A.E.; Mrowka, A.; Krajewska, J.; Tyski, S. The Influence of Efflux Pump Inhibitors on the Activity of Non-Antibiotic NSAIDS against Gram-Negative Rods. PLoS ONE 2016, 11, e0147131. [Google Scholar] [CrossRef]
- Parr, A.M.; Zoutman, D.E.; Davidson, J.S. Antimicrobial activity of lidocaine against bacteria associated with nosocomial wound infection. Ann. Plast. Surg. 1999, 43, 239–245. [Google Scholar] [CrossRef]
- Bohnert, J.A.; Szymaniak-Vits, M.; Schuster, S.; Kern, W.V. Efflux inhibition by selective serotonin reuptake inhibitors in Escherichia coli. J. Antimicrob. Chemother. 2011, 66, 2057–2060. [Google Scholar] [CrossRef]
- Lancellotti, P.; Musumeci, L.; Jacques, N.; Servais, L.; Goffin, E.; Pirotte, B.; Oury, C. Antibacterial Activity of Ticagrelor in Conventional Antiplatelet Dosages Against Antibiotic-Resistant Gram-Positive Bacteria. JAMA Cardiol. 2019, 4, 596–599. [Google Scholar] [CrossRef] [PubMed]
- El-Nakeeb, M.A.; Abou-Shleib, H.M.; Khalil, A.M.; Omar, H.G.; El-Halfawy, O.M. In vitro antibacterial activity of some antihistaminics belonging to different groups against multi-drug resistant clinical isolates. Braz. J. Microbiol. 2011, 42, 980–991. [Google Scholar] [CrossRef] [PubMed]
- Masadeh, M.; Mhaidat, N.; Alzoubi, K.; Al-azzam, S.; Alnasser, Z. Antibacterial activity of statins: A comparative study of Atorvastatin, Simvastatin, and Rosuvastatin. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Kovanda, L.; Zhang, W.; Wei, X.; Luo, J.; Wu, X.; Atwill, E.R.; Vaessen, S.; Li, X.; Liu, Y. In Vitro Antimicrobial Activities of Organic Acids and Their Derivatives on Several Species of Gram-Negative and Gram-Positive Bacteria. Molecules 2019, 24, 3770. [Google Scholar] [CrossRef]
- Zazgornik, J.; Mittermayer, H. Citric acid inhibits growth of Helicobacter pylori in vitro: A new strategy for eradication. Wien Klin. Wochenschr. 2011, 123, 38–40. [Google Scholar] [CrossRef] [PubMed]
- Al-Rousan, W.M.; Olaimat, A.N.; Osaili, T.M.; Al-Nabulsi, A.A.; Ajo, R.Y.; Holley, R.A. Use of acetic and citric acids to inhibit Escherichia coli O157:H7, Salmonella Typhimurium and Staphylococcus aureus in tabbouleh salad. Food Microbiol. 2018, 73, 61–66. [Google Scholar] [CrossRef]
- Nagoba, B.S.; Gandhi, R.C.; Wadher, B.J.; Deshmukh, S.R.; Gandhi, S.P. Citric acid treatment of severe electric burns complicated by multiple antibiotic resistant Pseudomonas aeruginosa. Burns 1998, 24, 481–483. [Google Scholar] [CrossRef]
- Over, K.F.; Hettiarachchy, N.; Johnson, M.G.; Davis, B. Effect of Organic Acids and Plant Extracts on Escherichia coli O157:H7, Listeria monocytogenes, and Salmonella Typhimurium in Broth Culture Model and Chicken Meat Systems. J. Food Sci. 2009, 74, M515–M521. [Google Scholar] [CrossRef]
- Peh, E.; Kittler, S.; Reich, F.; Kehrenberg, C. Antimicrobial activity of organic acids against Campylobacter spp. and development of combinations—A synergistic effect? PLoS ONE 2020, 15, e0239312. [Google Scholar] [CrossRef]
- Feng, S.; Zeng, W.; Luo, F.; Zhao, J.; Yang, Z.; Sun, Q. Antibacterial activity of organic acids in aqueous extracts from pine needles (Pinus massoniana Lamb.). Food Sci. Biotechnol. 2010, 19, 35–41. [Google Scholar] [CrossRef]
- El Baaboua, A.; El Maadoudi, M.; Bouyahya, A.; Belmehdi, O.; Kounnoun, A.; Zahli, R.; Abrini, J. Evaluation of Antimicrobial Activity of Four Organic Acids Used in Chicks Feed to Control Salmonella typhimurium: Suggestion of Amendment in the Search Standard. Int. J. Microbiol. 2018, 2018, 7352593. [Google Scholar] [CrossRef] [PubMed]
- Gómez-García, M.; Sol, C.; de Nova, P.J.G.; Puyalto, M.; Mesas, L.; Puente, H.; Mencía-Ares, Ó.; Miranda, R.; Argüello, H.; Rubio, P.; et al. Antimicrobial activity of a selection of organic acids, their salts and essential oils against swine enteropathogenic bacteria. Porc. Health Manag. 2019, 5, 32. [Google Scholar] [CrossRef] [PubMed]
- Dittoe, D.K.; Ricke, S.C.; Kiess, A.S. Organic Acids and Potential for Modifying the Avian Gastrointestinal Tract and Reducing Pathogens and Disease. Front. Vet. Sci. 2018, 5, 216. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, C.E.V.; Stamford, T.L.M.; Neto, N.J.G.; de Souza, E.L. Inhibition of Staphylococcus aureus in broth and meat broth using synergies of phenolics and organic acids. Int. J. Food Microbiol. 2010, 137, 312–316. [Google Scholar] [CrossRef]
- Ben Braïek, O.; Smaoui, S. Chemistry, Safety, and Challenges of the Use of Organic Acids and Their Derivative Salts in Meat Preservation. J. Food Qual. 2021, 2021, 6653190. [Google Scholar] [CrossRef]
- Cisneros, J.M.; Rodríguez-Baño, J. Nosocomial bacteremia due to Acinetobacter baumannii: Epidemiology, clinical features and treatment. Clin. Microbiol. Infect. 2002, 8, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Mihu, M.R.; Sandkovsky, U.; Han, G.; Friedman, J.M.; Nosanchuk, J.D.; Martinez, L.R. The use of nitric oxide releasing nanoparticles as a treatment against Acinetobacter baumannii in wound infections. Virulence 2010, 1, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Farshadzadeh, Z.; Pourhajibagher, M.; Taheri, B.; Ekrami, A.; Modarressi, M.H.; Azimzadeh, M.; Bahador, A. Antimicrobial and anti-biofilm potencies of dermcidin-derived peptide DCD-1L against Acinetobacter baumannii: An in vivo wound healing model. BMC Microbiol. 2022, 22, 25. [Google Scholar] [CrossRef] [PubMed]
- Karumathil, D.P.; Surendran-Nair, M.; Venkitanarayanan, K. Efficacy of Trans-cinnamaldehyde and Eugenol in Reducing Acinetobacter baumannii Adhesion to and Invasion of Human Keratinocytes and Controlling Wound Infection In Vitro. Phytother. Res. 2016, 30, 2053–2059. [Google Scholar] [CrossRef]
- Cunha, B.A. Optimal therapy for multidrug-resistant Acinetobacter baumannii. Emerg. Infect. Dis. 2010, 16, 170. [Google Scholar] [CrossRef]
- Dillon, N.A.; Peterson, N.D.; Rosen, B.C.; Baughn, A.D. Pantothenate and Pantetheine Antagonize the Antitubercular Activity of Pyrazinamide. Antimicrob. Agents Chemother. 2014, 58, 7258–7263. [Google Scholar] [CrossRef] [PubMed]
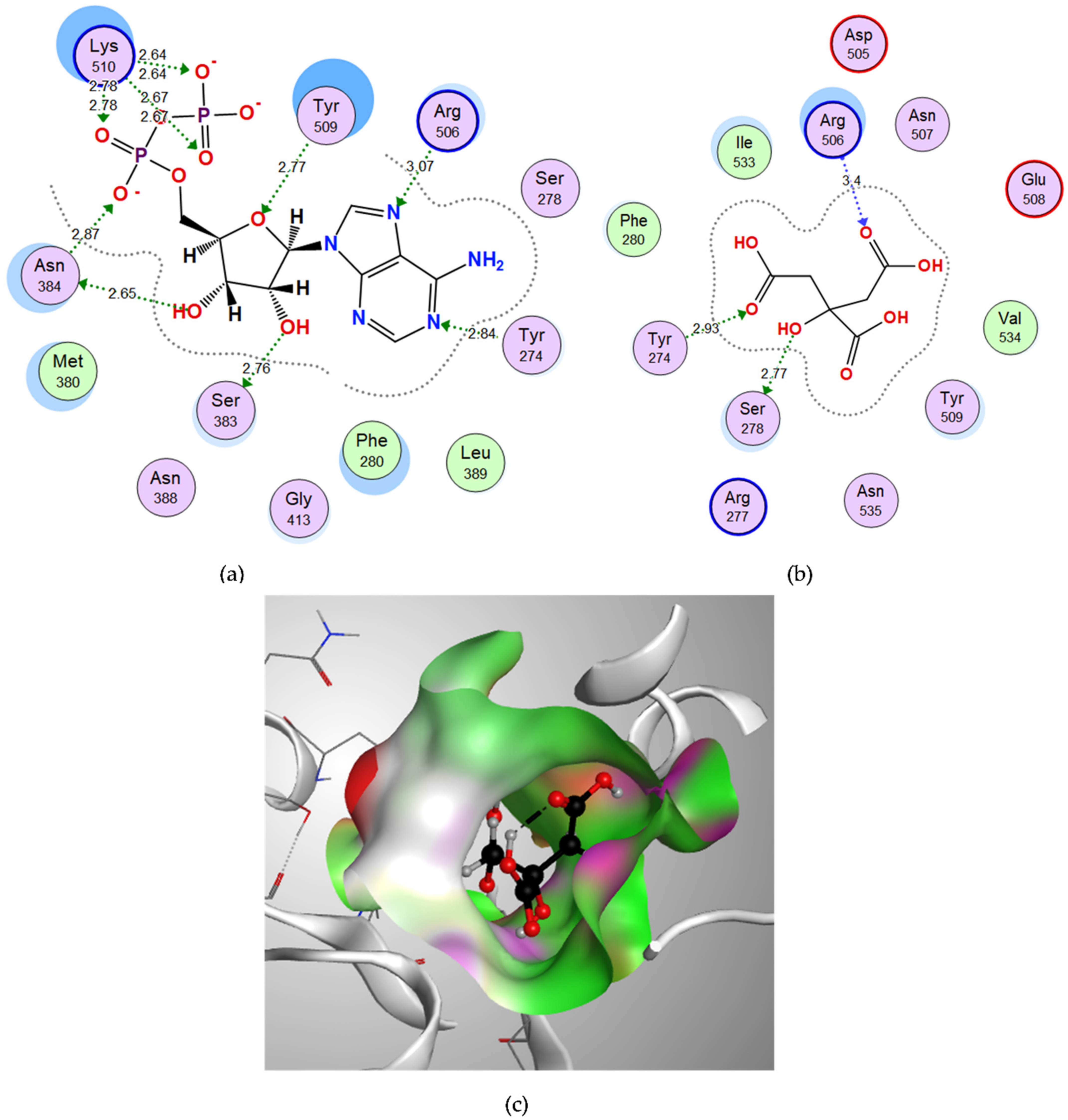


| Compound | Binding Score (kcal/mol) | Amino Acids at Active Site | Interacting Groups of Ligand | Type of Interaction | Bond Length |
|---|---|---|---|---|---|
| Citric acid | −13.0519 | Tyr274 | O (C=O) | H-bond acceptor | 2.93 |
| Ser278 | OH | H-bond donor | 2.77 | ||
| Arg506 | O (C=O) | H-bond acceptor | 3.40 | ||
| Malonic acid | −8.5187 | Tyr-B58 | OH | H-bond donor | 3.10 |
| Asn-B71 | O (C=O) | H-bond acceptor | 3.21 | ||
| D-tartaric acid | −9.2053 | Lys76 | OH | H-bond acceptor | 3.00 |
| Gln161 | O (C=O) | H-bond acceptor | 3.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badie, O.H.; Basyony, A.F.; Samir, R. Computer-Based Identification of Potential Druggable Targets in Multidrug-Resistant Acinetobacter baumannii: A Combined In Silico, In Vitro and In Vivo Study. Microorganisms 2022, 10, 1973. https://doi.org/10.3390/microorganisms10101973
Badie OH, Basyony AF, Samir R. Computer-Based Identification of Potential Druggable Targets in Multidrug-Resistant Acinetobacter baumannii: A Combined In Silico, In Vitro and In Vivo Study. Microorganisms. 2022; 10(10):1973. https://doi.org/10.3390/microorganisms10101973
Chicago/Turabian StyleBadie, Omar H., Ahmed F. Basyony, and Reham Samir. 2022. "Computer-Based Identification of Potential Druggable Targets in Multidrug-Resistant Acinetobacter baumannii: A Combined In Silico, In Vitro and In Vivo Study" Microorganisms 10, no. 10: 1973. https://doi.org/10.3390/microorganisms10101973
APA StyleBadie, O. H., Basyony, A. F., & Samir, R. (2022). Computer-Based Identification of Potential Druggable Targets in Multidrug-Resistant Acinetobacter baumannii: A Combined In Silico, In Vitro and In Vivo Study. Microorganisms, 10(10), 1973. https://doi.org/10.3390/microorganisms10101973




