Abstract
Ultramicrobacteria (UMB) that can pass through a 0.22 µm filter are attractive because of their novelty and diversity. However, isolating UMB has been difficult because of their symbiotic or parasitic lifestyles in the environment. Some UMB have extracellular electron transfer (EET)-related genes, suggesting that these symbionts may grow on an electrode surface independently. Here, we attempted to culture from soil samples bacteria that passed through a 0.22 µm filter poised with +0.2 V vs. Ag/AgCl and isolated Cellulomonas sp. strain NTE-D12 from the electrochemical reactor. A phylogenetic analysis of the 16S rRNA showed 97.9% similarity to the closest related species, Cellulomonas algicola, indicating that the strain NTE-D12 is a novel species. Electrochemical and genomic analyses showed that the strain NTE-D12 generated the highest current density compared to that in the three related species, indicating the presence of a unique electron transfer system in the strain. Therefore, the present study provides a new isolation scheme for cultivating and isolating novel UMB potentially with a symbiotic relationship associated with interspecies electron transfer.
1. Introduction
The minimum size of prokaryotes such as bacteria and archaea is expected to be 0.2–0.3 μm, and 0.22 μm pore size filters have been used for microbial sterilization. However, Macdonell et al. reported that some Vibrio, Aeromonas, and Pseudomonas species were isolated after 0.22 μm filtration [1]. These bacteria are called ultramicrobacteria (UMB) and are reported to have unique functions and high phylogenetic novelty [2,3]. The phylogenetic diversity of UMB has reported to comprise more than 15% of the bacterial domain, caused a paradigm shift in evolutionary phylogenetics [4,5]. Many reasons for the ultra-micro cell size of bacteria have been proposed, including the lack of unnecessary genes and proteins for niche survival [6,7,8]. In fact, members of candidate phyla radiation (CPR), a highly diversified group of uncultivated bacteria dominant in the 0.22 μm filtrate, have a small genome (often <1 Mb) due to their symbiotic or parasitic lifestyle [4]. Since most of the UMB are in a viable but non-culturable state in conventional culture systems, information on UMB obtained from cultures (e.g., physiological and morphological characteristics) has been limited.
Interspecies electron transfer (IET) is a symbiotic form of metabolism in microorganisms [9,10,11]. In the case of direct IET nanowires that consist of outer-membrane cytochrome or conductive type IV pili (T4P) have highlighted the importance of transferring electrons to other species and driving their metabolism [12]. The genes encoding T4P are conserved in OD1 and WWE3 genomes, which are members of CPR, and the pili formed from ultra-microcells are bound to other cells [13]. Recently, several genes involved in iron reduction and oxidation in the extracellular membrane have been identified in CPR genomes [14]. In addition, several Vibrio and Aeromonas species possess extracellular electron transfer (EET)-related genes [15,16]. From these reports, we hypothesized that some UMB have EET capability enabling symbiosis associated with IET and can grow on an electrode independently. In the present study, we examined the capability of UMB from a soil sample filtered through a 0.22 μm membrane to grow in an electrochemical reactor. We isolated a bacterial strain and analyzed its molecular phylogenetic, electrochemical, and morphological characteristics under several culture conditions. Unlike previously reported electrochemical enrichment with a relatively large volume of at least 5 mL [17,18], we used a reactor with 200 µL volume that enabled us to compare different medium conditions at once. The present study provides a scheme for electrochemical enriching UMB for the first time.
2. Materials and Methods
2.1. Medium and Electrolyte Composition
Gifu anaerobic medium (GAM) broth (Nissui Pharmaceutical, Tokyo, Japan), purged with N2:CO2 (80:20 v/v) gas for 30 min, was used for the pre-enrichment of anaerobic bacteria in the soil. For this, 10-fold and 100-fold diluted BD DifcoTM LB Broth Miller (1/10, 1/100 LB; Becton, Dickinson and Company, Franklin Lakes, NJ, USA); 100-fold diluted BD DifcoTM Marine broth 2216 (1/100 MB; Becton, Dickinson and Company, Franklin Lakes, NJ, USA); and 10-fold and 100-fold R2A broth (1/10, 1/100 R2A; Nihon Seiyaku, Tokyo, Japan) were used for electrical enrichment. R2A agar (conc. 1.5%) was used for isolation.
The defined medium (DM), used as an electrolyte for electrochemical measurements as used in other electrogenic bacteria [19,20], had the following composition (L–1): NH4Cl: 1 g; MgCl2·6H2O: 0.2 g; CaCl2·2H2O: 0.08 g; NaHCO3: 2.5 g; NaCl: 10 g; and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid: 7.2 g (pH 8.1). Glucose: 10 g was used as an electron donor for electrochemical measurements of the DM (DMG). The DM and DMG were purged with N2 gas for 30 min.
2.2. Electrochemical Enrichment and Isolation of 0.22 μm Filter–Passable Bacteria
Soil was collected from Tomioka, Iwaki, Fukushima Prefecture, Japan (37°21′ N, 141°01′ E). The collected samples were suspended in an appropriate amount of phosphate-buffered saline (PBS), and GAM broth was added for pre-enrichment. After 24 h of incubation at 30 °C, the enrichment culture was filtered through a 0.22 μm PES filter membrane (Millipore Express, Bedford, OH, USA). Subsequently, 20 μL of the filtrate and 180 μL of five different media (1/10 LB, 1/100 LB, 1/100 MB, 1/10 R2A, and 1/100 R2A) were added to the reactor.
Three screen-printed electrode systems were used for electrochemical enrichment (Metrohm DropSens, Oviedo, Spain). A carbon electrode, formed from carbon nanotubes, was used as a working electrode (surface area, 7.07 mm2) and counter electrode. Printed Ag/AgCl was used as a reference electrode. All the electrodes were placed at the bottom of the reactor. The working electrode was poised at +0.2 V (vs. Ag/AgCl) throughout electrochemical enrichment at 30 °C for 5 days. Enriched samples were spread on R2A agar under aerobic conditions. A single isolated strain, NTE-D12, was streaked onto fresh agar plates twice to ensure reliable colony purification.
2.3. DNA Extraction and 16S rRNA Gene Phylogenetic Analysis
Bacterial cells were lysed according to a previously described protocol [21] and centrifuged at 9100× g for 15 min to remove intact cells. The supernatant containing extracted genomic DNA was used for polymerase chain reaction (PCR). The 16S rRNA gene was amplified using TaKaRa Ex Taq (Takara Bio, Kusatsu, Shiga, Japan) DNA polymerase. The primers for PCR were 10 F (5′-GTTTGATCCTGGCTCA-3′) and 1492 R (5′-GGTTACCTTGTTACGACTT-3′). The amplified DNA was purified using the NucleoSpin® Gel and PCR Clean-up (Takara Bio, Kusatsu, Shiga, Japan), followed by sequencing. The sequence data obtained were compiled using BioEdit to construct the full-length 16S rRNA gene sequence [22]. The 16S rRNA gene sequences of type species were collected from the GenBank database; multiple sequence alignment was performed with MUSCLE [23], and a phylogenetic tree was constructed using the neighbor-joining method [24]. Tree topology was evaluated by bootstrapping with 1000 resamplings [25].
2.4. Comparison of the Current Production of the Isolated Strain with Related Species and Differential Pulse Voltammetry (DPV) Analysis
Related Cellulomonas type strains, including Cellulomonas algicola strain NBRC112905T, Cellulomonas fimi strain NBRC15513T, and Cellulomonas biazotea strain NBRC12680T, were purchased from the NITE Biological Resource Center’s (NBRC) (Kisaradsu, Chiba, Japan) online catalog. The isolated strain NTE-D12 and the three related species were precultured in R2A broth under aerobic conditions. Cultured cells were harvested during the mid-log phase for each species. The cells were centrifuged at 7150× g at 4 °C for 10 min. The harvested cells were washed twice with the DM by resuspending, centrifuging the cells, and removing the supernatant. Washed cells were resuspended with DMG for the electrochemical experiment, adjusted to OD600 (optical density measured at a wavelength of 600 nm) = 0.5, and purged with N2 gas for 30 min. Resuspended cells were added to three screen-printed electrode systems, and chronoamperometry (CA) was measured poised at +200 mV vs. Ag/AgCl for 12 h.
For DPV measurements, we used a three-electrode reactor with a tin-doped In2O3 (ITO) electrode as the working electrode (surface area, 3.1 cm2) previously used for characterizing S. oneidensis MR-1 [26,27]. The working electrode was placed at the bottom of the reactor. A platinum wire and Ag/AgCl saturated KCl were used as the counter and reference electrodes, respectively. DPV was measured under the following conditions: pulse increment, 5.0 mV; pulse amplitude, 50 mV; pulse width, 300 mV; and pulse period, 5.0 s [26,27]. DPV measurement of NTE-D12 was performed before adding cells and 12 and 24 h after adding cells during CA measurement. All the electrochemical measurements were conducted in an anaerobic chamber filled with N2 gas and maintained at 30 °C.
2.5. Evaluation of Cell Size under Various Pure Culture Conditions Using Scanning Electron Microscopy (SEM) and Filtration
After culturing the cells in R2A for 24 h, aerobically in DMG for 3 days, anaerobically in GAM for 5 days, and electrically in DMG for 24 h, SEM observations were performed. To promote cell adsorption, poly-l-lysine was applied to the ITO electrode, incubated for 40 min, and washed off with sterilized water. The culture medium was smeared onto the coated surface of ITO and incubated for 50 min. Cells were fixed with 2.5% glutaraldehyde for 30 min at 25 °C, followed by washing three times in 0.1 M phosphate buffer for 10 min. This sample was dehydrated with 25%, 50%, 75%, and 99.5% ethanol gradients for 10 min each. The dehydrated samples were exchanged with 100% t-butanol three times and freeze-dried under vacuum. These samples were platinum-coated under vacuum and observed using a Keyence VE-9800 microscope (Keyence, Osaka-shi, Osaka, Japan). To compare cell size, the cells were counted, and the major and minor axes were calculated using ImageJ ver. 1.53 k [28]. Under the same culture conditions as for SEM, the filter-passing capability of pure cultured cells was evaluated to smear these cultures onto R2A agar.
2.6. Whole-Genome Sequence of Isolated Strain and Genetic Characterization
Genomic DNA was extracted from the NTE-D12 strain grown aerobically in R2A medium for 24 h using a Quick-DNA HMW MagBead Kit (Zymo Research, Irvine, CA, USA). The concentration of the extracted genomic DNA was measured using a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). DNA was sequenced using the Illumina MiSeq (Illumina, San Diego, CA, USA) and the MinION (Oxford Nanopore, Oxford, UK) platforms. For Illumina sequencing, DNA was processed for shotgun library construction using the KAPA Hyper Prep kit for Illumina (KAPA Biosystems, Wilmington, MA, USA) and then sequenced using a MiSeq Reagent Kit v3 (600 cycles) to generate 2 × 300 paired-end reads. For MinION sequencing, an R9.4.1 Flow cell (FLO-MIN106) and Ligation Sequencing Kit (SQK-LSK109) were used according to the manufacturer’s instruction. Summaries of the MiSeq and MinION data were generated using FastQC and NanoStat. Hybrid de novo assembly from the MiSeq and MinION data was carried out using Unicycler ver.0.4.7 [29]. The genomes of C. algicola, C. fimi, and C. biazotea were collected from the GenBank database (the accession numbers are GCA_003851725.1, GCA_004306155.1, GCA_000212695.1, respectively). Each genome was annotated using Prokka ver. 1.14.6 [30] and FeGenie ver. 1.0 [14] for comparative genomic analysis. The subcellular location of genes with heme-binding motifs was predicted using Psortb ver. 3.0.3 [31]. We used Orthovenn2 to compare orthologous relationships for the annotated genes in the four strains [32]. Shared orthologous genes among these strains were depicted through a Venn diagram.
3. Results
3.1. Electrochemical Enrichment and Isolation of 0.22 μm Filter–Passable Bacteria
To grow and enrich 0.22 μm filter–passable bacteria electrically, we poised the electrode potential at +0.2 V and measured microbial current production (Ic), as is done when Geobacter sulfurreducens, a representative exoelectrogen for IET, is assigned to potential of EET [11]. Among the five different medium conditions (representative data of duplicates are shown in Figure 1), only one of the 1/100 R2A broths showed a gradual Ic increase to approximately 1.5 mA/m2 after 40 h (Figure 1). At the same time, the Ic decreased to about 0.7 mA/m2 after 112 h. Ic was sustained until 120 h, and an increase in turbidity was observed, indicating that electrogenic bacteria grew on the poised carbon electrode in one of the 1/100 R2A broths (1/100 R2A-2). The 16S rRNA sequence of the enriched sample showed a single peak, indicating high enrichment of clonal bacteria (Figure S1). Ic did not increase in another broth and one of the 1/100 R2A (1/100 R2A-1) broths.
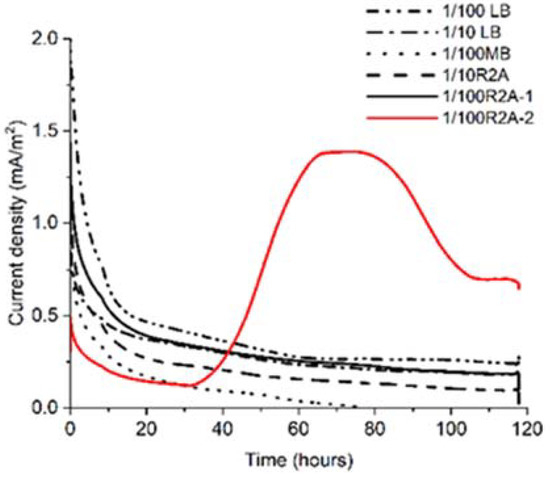
Figure 1.
Comparison of current production among five different media for incubating bacteria that can pass 0.22 μm Filter in a soil sample.
After spreading the sample on the R2A agar plate, a single colony was consistently formed after five days. The 16S rRNA sequence of the isolates was identical to that of the electrically enriched sample. The 16S rRNA gene of the isolated strain named NTE-D12, is indicative of the Cellulomonas clade. The sequence similarities of the 16S rRNA gene of the three closest related species were 97.9%, 97.5%, and 97.5% to the type strains of C. algicola, C. fimi, and C. biazotea, respectively, indicating that NTE-D12 is not only a 0.22 μm filter–passable bacterium with current production capability but also a novel species (Figure 2).
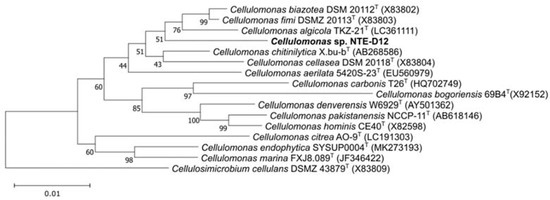
Figure 2.
Phylogenetic tree with the novel isolated strain NTE-D12. Numbers at branch points are percentages supported by bootstrap probabilities with 1000 replicates. Bar, 0.01 nucleotide substitution position.
3.2. Electrochemical Characterization of the Isolated Strain
Next, we characterized the EET capability of NTE-D12. Current production was measured in the DM with and without glucose at +0.2 V vs. Ag/AgCl and an initial OD600 of 0.5. A higher Ic was observed in the presence of glucose than in its absence. A gradual Ic increase (Figure 3A) was immediately observed upon poising the electrode potential into the cell suspension in the electrochemical system. Ic reached 9.0 mA/m2 after 12 h, whereas less than 0.4 mA/m2 was observed in the absence of glucose, indicating the association of current production capability with glucose oxidation metabolism. In the DPV measurements (Figure 3B) without cells, a specific peak was not observed. Distinct oxidation peak was observed around −0.05 V vs. standard hydrogen electrode (SHE). Cellular coverage on the electrode surface explains the alteration of the DPV profile, and the peak at −0.05 V is most likely associated with the microbial electron transport pathway.
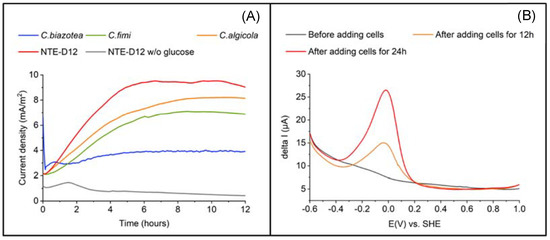
Figure 3.
(A) Comparison of current production among four strains (C. biazotea, C. fimi, C. algicola, and strain NTE-D12) using carbon electrode as a working electrode. (B) Differential pulse voltammograms of strain NTE-D12 before and after adding cells for 12 and 24 h using ITO electrode as a working electrode.
The currents measured in C. fimi, C. algicola, and NTE-D12 (n = 3 for all strains) continued to increase to a maximum of 6.9, 8.1, and 9.0 mA/ m2, respectively; it did not increase significantly after a maximum of 3.9 mA/m2 in C. biazotea compared to the others. The growth curve showed that, in the R2A medium, C. biazotea grew the fastest among the four strains (Figure S2) and was reported to have glucose oxidation capability [33], suggesting that its EET capability was lower than that of the other strains. These results showed that NTE-D12 was the most capable of utilizing glucose as an electron donor for current production among the four strains. Given that these bacterial strains have a glucose oxidation pathway from glycolysis to the TCA cycle, NTE-D12 most likely uses the same pathway to generate current from glucose oxidation. These results demonstrate that the isolated strain NTE-D12 has EET capability and grows on the electrode surface.
3.3. Evaluation of Cell Size in Various Culture Conditions
The cells observed under each culture condition are shown in Figure 4A. The projected cellular area and major and minor axes were quantified from the SEM images of cells precultured under four different conditions. Cells were dehydrated under the same conditions, and we compared the size of more than 10 cells. The smallest cell areas were observed after electrochemical culture in DMG, whereas the largest areas were observed after anaerobic culture in GAM. Using the same DMG, the cellular area under aerobic culture conditions was 1.5 times larger than that under electrochemical culture conditions (Figure 4B). The major axis was the largest in the R2A aerobic culture conditions and was twice as large as that in the electrochemical culture conditions, which was the smallest. Interestingly, even under the same culture conditions, cells with approximately 10-fold different areas were observed in GAM, and the standard deviation of all measurements in GAM was larger than that in other culture conditions (Figure 4C). In contrast, cells below 0.22 μm were not observed in these culture conditions. Growth was not observed in these 0.22 μm filtrates on agar plates. These results showed that the cell size of NTE-D12 varies depending on the culture conditions and growth phase, and cultured cells of these four conditions did not pass through a 0.22 μm filter.
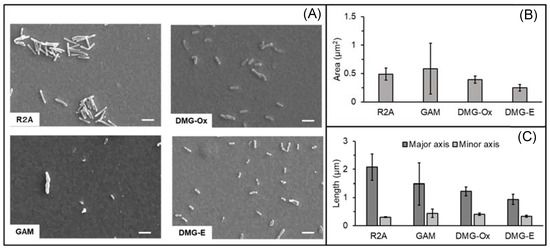
Figure 4.
(A) Cells observed using scanning electron microscopy (SEM). Scale bar = 1.0 µm. (B) Comparison of the observed area and (C) major axis and minor axis of cells after aerobic culture in R2A (R2A, n = 12), anaerobic culture in GAM (GAM, n = 15), aerobic culture in DMG (DMG; DMG-Ox, n = 10), and electrochemical culture in DMG (DMG-E, n = 18). Error bars represent standard deviation.
3.4. Genomic Features of the Isolated Strain and Comparison with Related Species
We determined the whole-genome sequence of strain NTE-D12. The MiSeq sequencing run generated 21,560,786 reads, with a length of 301 bases. The data size corresponds to a coverage depth of over 1000. Despite using data with sufficient coverage, Unicycler assembly using MiSeq data generated 14 contigs. Therefore, long-read sequencing was performed using MinION. The MinION sequencing run generated 280,000 reads, totaling 3,641,335,730 bases after base-calling. Before hybrid assembly, it was downsampled to 83,829,559 bases consisting of 4000 reads, because the data size was too large to perform Unicycler assembly. Hybrid Unicycler assembly was performed using all available MiSeq data and the downsampled MinION read data, resulting in two circular genomes.
The genomic features of NTE-D12 and the three related species are summarized in Table 1, and the features table created using Prokka is shown in Table S1. The genome of strain NTE-D12 consists of 3,684,286 bp circular chromosomes and a 5386 bp plasmid encoding 3333 protein-coding sequences (CDS), 54 transfer RNAs, 1 transfer–messenger RNA, and 6 ribosomal RNAs. The G–C content was 73.06%. The total genome sequence length and CDS number of NTE-D12 were smaller than those of the three related species. Type IV prepilin-like protein leader peptide-processing enzymes were encoded in the three related species, but none of the genes constituting T4P were encoded. Flavoprotein is important for the EET model in Listeria monocytogenes [34], a Gram-positive bacteria similar to Cellulomonas. Each genome encodes an electron transfer flavoprotein, succinate dehydrogenase flavoprotein, and flavohemoprotein. Only C. algicola encoded the sulfite reductase (NADPH) flavoprotein, whereas only NTE-D12 encoded FixA, B, and X. None of these proteins were predicted to be localized to the cell wall (Table S2). We could not annotate any quinone biosynthesis genes in NTE-D12, suggesting that other quinone-related species are involved in the electron transport chain in strain NTE-D12 that cannot be annotated using Prokka. Analysis of iron-related genes using FeGenie showed that none of four strains encoded iron-reduction genes. We also detected genes with a heme-binding motif and predicted their subcellular localization (Table S3). Among these four strains, NTE-D12 had genes with a heme-binding motif predicted to localize most frequently in the cytoplasmic membrane and cell wall.

Table 1.
Genomic features of Cellulomonas sp. strain NTE-D12 and three related species.
Orthologous gene family analysis using Orthovenn2 resulted in 3740 clusters (C. biazotea, 3318; C. fimi, 3348; C. algicola, 3402; strain NTE-D12, 2313 orthologous clusters) (Figure 5). A total of 1936 orthologs were shared by all four strains, with 75 strain-specific gene clusters. Among the four strains, strain NTE-D12 had the highest number of specific gene clusters (50). Interestingly, an electron carrier activity functional gene cluster was specifically annotated in the NTE-D12. This indicated that strain NTE-D12 has a unique electron transfer system compared to that in other related species.
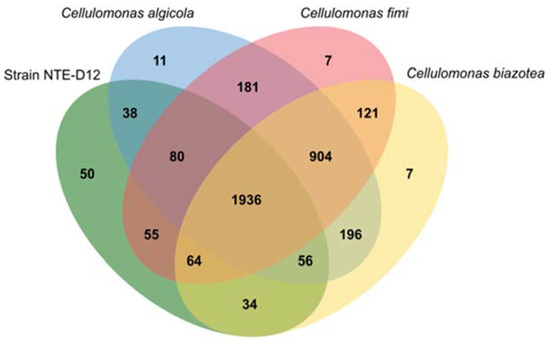
Figure 5.
Venn diagram showing orthologues gene clusters among four strains (Cellulomonas biazotea, Cellulomonas fimi, Cellulomonas algicola, and Cellulomonas sp. strain NTE-D12).
4. Discussion
In the present study, we investigated the EET capability of UMB grown on an electrode after passing through a 0.22 μm filter, based on the hypothesis that some UMB have a symbiotic relationship associated with IET. Ic increased only in 1/100 R2A-2 but not in 1/100 R2A-1 though they are the same medium. Since 0.22 μm filterable bacteria are rare in the environment [35], strain NTE-D12 could have also been present as quite a small amount in the preculture with GAM broth and was contingently only introduced into the one 1/100 R2A-2 culture. The isolation of Cellulomonas sp. strain NTE-D12, a candidate novel species, from only one among five different electro-culture systems, verified our scheme for enriching highly novel UMB species. The current production of NTE-D12 was similar compared that of Shewanella oneidensis strain MR-1 using the same carbon electrode, which is a typical EET-capable bacteria [36]. NTE-D12 generated the highest current among the four related species. C. fimi and C. algicola also generated current, whereas C. biazotea did not. This result indicated NTE-D12 has higher EET capability than the other related species.
The cell size of strain NTE-D12 varied in the four evaluated culture conditions, and after culturing, the cells did not pass through a 0.22 μm filter. Some of the UMB are considered to have an ultra-microcell morphology to resist stress from the surrounding environment [37,38,39]. Although the effect of stress on cell morphology remains controversial, metabolites from other soil-derived bacteria may create a stressful environment for NTE-D12 in GAM culture.
From genomic feature analysis results, iron-reducing genes involved in EET were not detected in the genomes of the analyzed four strains. Flavoprotein, which is considered to be important for EET strategy in Gram-positive bacteria [34], was not predicted to be localized in the cell wall. These results suggest that these four strains capable of current generation have an unknown EET strategy. The EET mechanism of these Cellulomonas needs to be investigated in detail in the future. The genome size of strain NTE-D12 is close to that of the Sphingopyxis alaskensis strain RB2256 (3.35 Mb), which is relatively larger than the UMB reported so far [40]. Despite having the smallest genome and number of CDSs compared to the three other related species, strain NTE-D12 has the most unique CDSs, including genes predicted to have heme-binding motifs localized at the cell wall and electron carrier activity. Outer-membrane heme-binding proteins have been speculated to participate in EET by typical iron-reducing bacteria and have been previously suggested to be important [19,41]. The fact that there was only one gene cluster with electron transfer activity indicated that this strain has a unique electron transfer system compared to other related species. This unique electron transfer system is suggested to be associated with higher current production capability. Combined with the results of smaller genome size, this strain may have evolved depending on IET with other species in the environment.
5. Conclusions
In the present study, a novel Cellulomonas sp. strain NTE-D12 was isolated from microbial consortia filtered through a 0.22-μm-pore filter membrane. The isolated strain has higher EET capability, smaller genome size, and unique electron transfer genes compared to related species. This cultivation scheme, a combination of 0.22 μm filtrate and electrochemical culture, can be used to isolate IET-dependent UMB. In the future, this scheme may be applied to isolate or enrich cultures for CPR, which are substantial unculturable phylogenetic groups that account for most of the UMB. These bacteria have not yet been isolated because of their symbiotic relationships with other bacterial species. Because EET-capable microorganisms are extremely diverse [17,18,34,42], it is conceivable that CPR have a symbiotic relationship based on IET. The cultivation scheme established in the present study will enable the isolation of these uncultivated microorganisms and is expected to provide innovative knowledge on microbial symbiosis.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/microorganisms10102051/s1: Figure S1: 16S rRNA gene sequencing results; Figure S2: Growth curve of strain NTE-D12 and three related species; Table S1: Genomic features using Prokka; Table S2: Flavoprotein-related genes; Table S3: Genes with heme-binding motif using FeGenie.
Author Contributions
Conceptualization, S.I. and A.O.; data curation, S.I. and S.W.; formal analysis, S.I.; funding acquisition, A.O.; investigation, S.I., S.W. and T.M.; methodology, S.I. and A.O.; project administration, A.O.; resources, A.O.; software, S.I. and S.W.; supervision, A.O.; validation, S.I., S.W. and T.M.; visualization, S.I.; writing—original draft, S.I.; writing—review and editing, S.W. and A.O. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by a Grant-in-Aid for Research from the Japan Society for Promotion of Science KAKENHI (Grant Nos. 20H05590 and 22H02265) and PRIME from the Japan Agency for Medical Research and Development (19gm6010002h0004). This work was also supported by JST, PRESTO Grant Number JPMJPR19H1, Japan, and by the JAEA Nuclear Energy S&T and Human Resource Development Project through concentrating wisdom Grant Number JPJA20P20333393.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Whole-genome and plasmid sequences were deposited as AP026442 and AP026443, respectively, in the DNA Data Bank of Japan (DDBJ). In addition, the datasets used for the present study, with accession codes DRR392336 and DRR392337, were obtained from NCBI.
Acknowledgments
We thank Shin-ichi Hirano, Yuma Dotsuta, Kyohei Otani, Toru Kitagaki, and Fumiyoshi Ueno for the sample collection.
Conflicts of Interest
The authors declare no conflict of interest.
References
- MacDonell, M.T.; Hood, M.A. Isolation and Characterization of Ultramicrobacteria from a Gulf Coast Estuary. Appl. Environ. Microbiol. 1982, 43, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, T.; Miyoshi, T.; Kimura, H. Phylotype diversity of deep-sea hydrothermal vent prokaryotes trapped by 0.2- and 0.1-μm-pore-size filters. Extremophiles 2007, 11, 637–646. [Google Scholar]
- Nakai, R.; Abe, T.; Takeyama, H.; Naganuma, T. Metagenomic Analysis of 0.2-μm-Passable Microorganisms in Deep-Sea Hydrothermal Fluid. Mar. Biotechnol. 2011, 13, 900–908. [Google Scholar]
- Brown, C.T.; Hug, L.A.; Thomas, B.C.; Sharon, I.; Castelle, C.J.; Singh, A.; Wilkins, M.J.; Wrighton, K.C.; Williams, K.H.; Banfield, J.F. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 2015, 523, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Baker, B.; Anantharaman, K.; Brown, C.T.; Probst, A.; Castelle, C.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Duda, V.I.; Suzina, N.E.; Polivtseva, V.N.; Boronin, A.M. Ultramicrobacteria: Formation of the concept and contribution of ultramicrobacteria to biology. Microbiology 2012, 81, 379–390. [Google Scholar] [CrossRef]
- Suzuki, S.; Horinouchi, S.; Beppu, T. Growth of a Tryptophanase-producing Thermophile, Symbiobacterium thermophilum gen. nov., sp. nov., Is Dependent on Co-culture with a Bacillus Sp. Microbiology 1988, 134, 2353–2362. [Google Scholar]
- Morris, J.J.; Lenski, R.E.; Zinser, E.R. The black queen hypothesis: Evolution of dependencies through adaptive gene loss. mBio 2012, 3. [Google Scholar] [CrossRef]
- Watanabe, K.; Manefield, M.; Lee, M.; Kouzuma, A. Electron shuttles in biotechnology. Curr. Opin. Biotechnol. 2009, 20, 633–641. [Google Scholar] [CrossRef]
- Summers, Z.M.; Fogarty, H.E.; Leang, C.; Franks, A.E.; Malvankar, N.S.; Lovley, D.R. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 2010, 330, 1413–1415. [Google Scholar] [CrossRef]
- Kato, S.; Hashimoto, K.; Watanabe, K. Microbial interspecies electron transfer via electric currents through conductive minerals. Proc. Natl. Acad. Sci. USA 2012, 109, 10042–10046. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R. Syntrophy Goes Electric: Direct Interspecies Electron Transfer. Annu. Rev. Microbiol. 2017, 71, 643–664. [Google Scholar] [CrossRef]
- Luef, B.; Frischkorn, K.R.; Wrighton, K.C.; Holman, H.-Y.N.; Birarda, G.; Thomas, B.C.; Singh, A.; Williams, K.H.; Siegerist, C.E.; Tringe, S.G.; et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 2015, 6, 6372. [Google Scholar] [CrossRef] [PubMed]
- Garber, A.I.; Nealson, K.H.; Okamoto, A.; McAllister, S.M.; Chan, C.S.; Barco, R.A.; Merino, N. FeGenie: A Comprehensive Tool for the Identification of Iron Genes and Iron Gene Neighborhoods in Genome and Metagenome Assemblies. Front. Microbiol. 2020, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Conley, B.E.; Intile, P.J.; Bond, D.R.; Gralnick, J.A. Divergent Nrf family proteins and MtrCAB homologs facilitate extracellular electron transfer in Aeromonas hydrophila. Appl. Environ. Microbiol. 2018, 84, e02134-18. [Google Scholar] [CrossRef] [PubMed]
- Conley, B.E.; Weinstock, M.T.; Bond, D.R.; Gralnick, J.A. A hybrid extracellular electron transfer pathway enhances the survival of vibrio natriegens. Appl. Environ. Microbiol. 2020, 86, e01253-20. [Google Scholar] [CrossRef]
- Rowe, A.R.; Yoshimura, M.; LaRowe, D.E.; Bird, L.J.; Amend, J.P.; Hashimoto, K.; Nealson, K.H.; Okamoto, A. In situ electrochemical enrichment and isolation of a magnetite-reducing bacterium from a high pH serpentinizing spring. Environ. Microbiol. 2017, 19, 2272–2285. [Google Scholar] [CrossRef]
- Naradasu, D.; Miran, W.; Sakamoto, M.; Okamoto, A. Isolation and characterization of human gut bacteria capable of extracellular electron transport by electrochemical techniques. Front. Microbiol. 2019, 10, 3267. [Google Scholar] [CrossRef]
- Okamoto, A.; Nakamura, R.; Hashimoto, K. In-vivo identification of direct electron transfer from Shewanella oneidensis MR-1 to electrodes via outer-membrane OmcA-MtrCAB protein complexes. Electrochim. Acta 2011, 56, 5526–5531. [Google Scholar] [CrossRef]
- Miran, W.; Long, X.; Huang, W.; Okamoto, A. Current Production Capability of Drug-Resistant Pathogen Enables Its Rapid Label-Free Detection Applicable to Wastewater-Based Epidemiology. Microorganisms 2022, 10, 472. [Google Scholar] [CrossRef]
- Hiraishi, A.; Shin, Y.K.; Ueda, Y.; Sugiyama, J. Automated sequencing of PCR-amplified 16S rDNA on ‘Hydrolink’ gels. J. Microbiol. Methods 1994, 19, 145–154. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tokunou, Y.; Hashimoto, K.; Okamoto, A. Acceleration of Extracellular Electron Transfer by Alternative Redox-Active Molecules to Riboflavin for Outer-Membrane Cytochrome c of Shewanella oneidensis MR-1. J. Phys. Chem. C 2016, 120, 16168–16173. [Google Scholar] [CrossRef]
- Okamoto, A.; Hashimoto, K.; Nealson, K.H.; Nakamura, R. Rate enhancement of bacterial extracellular electron transport involves bound flavin semiquinones. Proc. Natl. Acad. Sci. USA 2013, 110, 7856. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, H.; Hayashi, T.; Hamada, M.; Kohda, T.; Serisawa, Y.; Matsuyama-Serisawa, K.; Nakagawa, Y.; Otoguro, M.; Yanagida, F.; Tamura, T.; et al. Cellulomonas algicola sp. nov., an actinobacterium isolated from a freshwater alga. Int. J. Syst. Evol. Microbiol. 2019, 69, 2723–2728. [Google Scholar] [CrossRef] [PubMed]
- Light, S.H.; Su, L.; Rivera-Lugo, R.; Cornejo, J.A.; Louie, A.; Iavarone, A.T.; Ajo-Franklin, C.M.; Portnoy, D.A. A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature 2018, 562, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Fukuba, T.; Sato, M.; Elsaied, E.H.; Naganuma, T. Overlooked Microbial Agents in Aquaculture: Nanobacteria. In Microbial Approaches to Aquatic Nutrition within Environmentally Sound Aquaculture Production Systems; World Aquaculture Society: Becton Rouge, LA, USA, 2002; pp. 99–107. [Google Scholar]
- Ho, C.; Emran, M.Y.; Ihara, S.; Huang, W.; Wakai, S.; Li, W.P.; Okamoto, A. Osmium-Grafted Magnetic Nanobeads Increase Microbial Current Generation Via Culture-Free and Quick Enrichment of Electrogenic Bacteria. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4177599 (accessed on 1 August 2022).
- Nakai, R.; Shibuya, E.; Justel, A.; Rico, E.; Quesada, A.; Kobayashi, F.; Iwasaka, Y.; Shi, G.-Y.; Amano, Y.; Iwatsuki, T.; et al. Phylogeographic analysis of filterable bacteria with special reference to Rhizobiales strains that occur in cryospheric habitats. Antarct. Sci. 2013, 25, 219–228. [Google Scholar] [CrossRef]
- Kjelleberg, S.; Albertson, N.; Flärdh, K.; Holmquist, L.; Jouper-Jaan, Å.; Marouga, R.; Östling, J.; Svenblad, B.; Weichart, D. How do non-differentiating bacteria adapt to starvation? Antonie Van Leeuwenhoek 1993, 63, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Monier, J.M.; Lindow, S.E. Pseudomonas syringae Responds to the Environment on Leaves by Cell Size Reduction. Phytopathology 2003, 93, 1209–1216. [Google Scholar] [CrossRef]
- Eguchi, M.; Nishikawa, T.; Macdonald, K.; Cavicchioli, R.; Gottschal, J.C.; Kjelleberg, S. Responses to Stress and Nutrient Availability by the Marine Ultramicrobacterium Sphingomonas sp. Strain RB2256. Appl. Environ. Microbiol. 1996, 62, 1287–1294. [Google Scholar] [CrossRef]
- Shi, L.; Richardson, D.J.; Wang, Z.; Kerisit, S.N.; Rosso, K.M.; Zachara, J.M.; Fredrickson, J.K. The roles of outer membrane cytochromes of Shewanella and Geobacter in extracellular electron transfer. Environ. Microbiol. Rep. 2009, 1, 220–227. [Google Scholar] [CrossRef]
- Deng, X.; Dohmae, N.; Nealson, K.H.; Hashimoto, K.; Okamoto, A. Multi-heme cytochromes provide a pathway for survival in energy-limited environments. Sci. Adv. 2018, 4, eaao5682. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).