Direct MALDI-TOF MS and Antimicrobial Susceptibility Testing of Positive Blood Cultures Using the FASTTM System and FAST-PBC Prep Cartridges—Performance Evaluation in a Clinical Microbiology Laboratory Serving High-Risk Patients
Abstract
:1. Introduction
2. Materials and Methods
3. Results
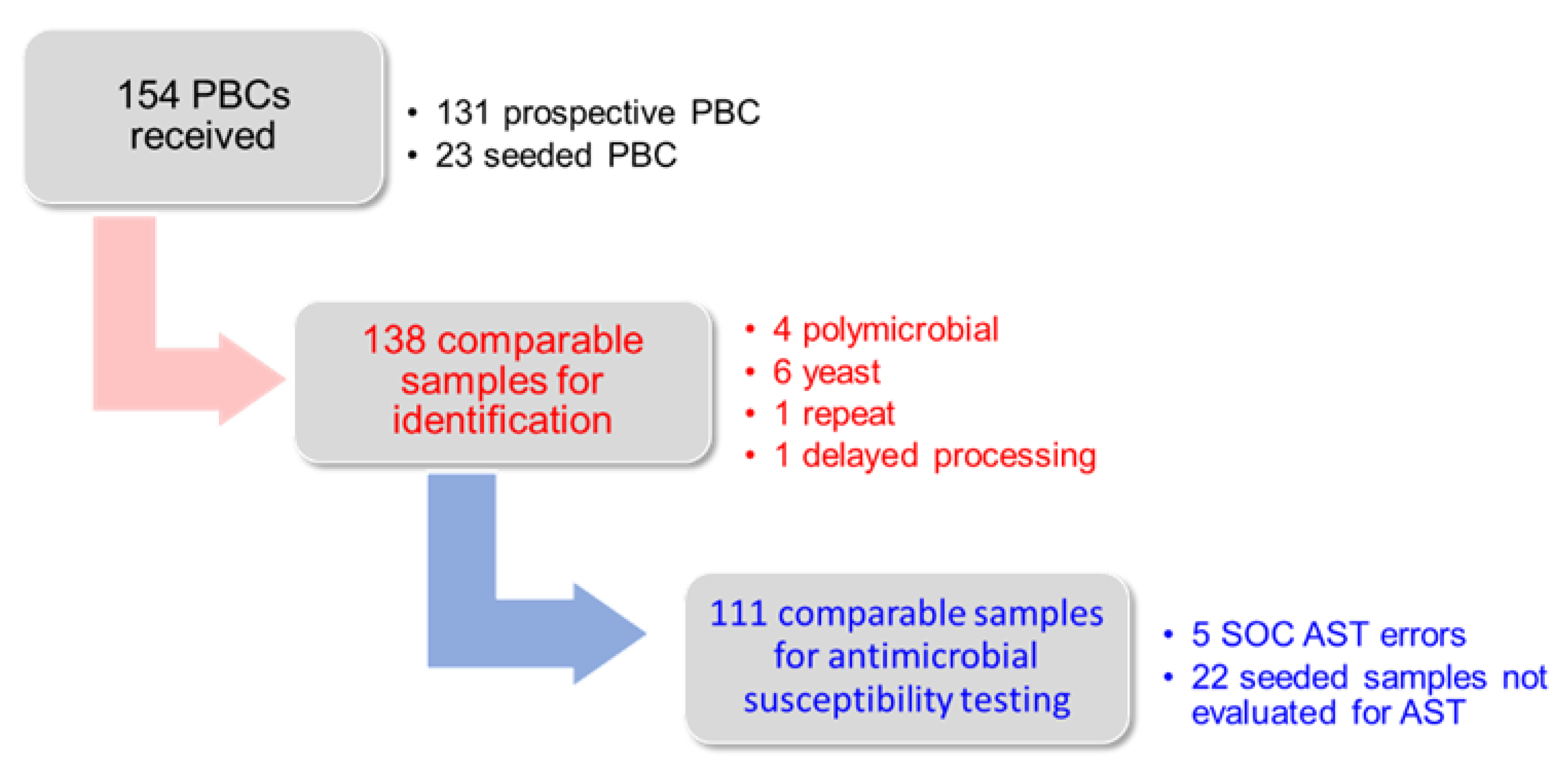
3.1. Study Samples
3.2. Identification Results
3.3. Antimicrobial Susceptibility Testing Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Nelson, R.E.; Hyun, D.; Jezek, A.; Samore, M.H. Mortality, Length of Stay, and Healthcare Costs Associated with Multidrug-Resistant Bacterial Infections among Elderly Hospitalized Patients in the United States. Clin. Infect. Dis. 2022, 74, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Diekema, D.J.; Hsueh, P.R.; Mendes, R.E.; Pfaller, M.A.; Rolston, K.V.; Sader, H.S.; Jones, R.N. The Microbiology of Bloodstream Infection: 20-Year Trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 2019, 63, e00355-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019.
- Buchman, T.G.; Simpson, S.Q.; Sciarretta, K.L.; Finne, K.P.; Sowers, N.; Collier, M.; Chavan, S.; Oke, I.; Pennini, M.E.; Santhosh, A.; et al. Sepsis Among Medicare Beneficiaries: 3. The Methods, Models, and Forecasts of Sepsis, 2012–2018. Crit. Care Med. 2020, 48, 302–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timbrook, T.T.; Morton, J.B.; McConeghy, K.W.; Caffrey, A.R.; Mylonakis, E.; LaPlante, K.L. The Effect of Molecular Rapid Diagnostic Testing on Clinical Outcomes in Bloodstream Infections: A Systematic Review and Meta-Analysis. Clin. Infect. Dis. 2017, 64, 15–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogan, C.A.; Watz, N.; Budvytiene, I.; Banaei, N. Rapid antimicrobial susceptibility testing by VITEK(R)2 directly from blood cultures in patients with Gram-negative rod bacteremia. Diagn. Microbiol. Infect. Dis. 2019, 94, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Infante, A.; Ortiz de la Tabla, V.; Martin, C.; Gazquez, G.; Bunuel, F. Rapid identification and antimicrobial susceptibility testing of Gram-negative rod on positive blood cultures using MicroScan panels. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Maelegheer, K.; Nulens, E. Same-day identification and antibiotic susceptibility testing on positive blood cultures: A simple and inexpensive procedure. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Khine, A.A.; Fernandez, N.; Nair, A.; Novak-Weekley, S.; Alavie, T. Evaluation of a novel sample preparation method for rapid antimicrobial resistance testing directly from positive blood cultures. In Proceedings of the European Congress of Clinical Microbiology and Infectious Diseases, Lisbon, Portugal, 24 April 2022. [Google Scholar]
- Grinberg, S.; Schubert, S.; Hochauf-Stange, K.; Dalpke, A.H.; Narvaez Encalada, M. Saving Time in Blood Culture Diagnostics: A Prospective Evaluation of the Qvella FAST-PBC Prep Application on the Fast System. J. Clin. Microbiol. 2022, 60, e0253321. [Google Scholar] [CrossRef] [PubMed]
- CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021.
- Clark, R.B.; Lewinski, M.A.; Loeffelholz, M.J.; Tibbetts, R.J. Cumitech 31A, Verification and Validation of Procedures in the Clinical Microbiology Laboratory; Shar, S.E., Ed.; ASM Press: Washington, DC, USA, 2009. [Google Scholar]
- Congestri, F.; Pedna, M.F.; Fantini, M.; Samuelli, M.; Schiavone, P.; Torri, A.; Bertini, S.; Sambri, V. Comparison of ‘time to detection’ values between BacT/ALERT VIRTUO and BacT/ALERT 3D instruments for clinical blood culture samples. Int. J. Infect. Dis. 2017, 62, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Doi, Y.; Bonomo, R.A.; Johnson, J.K.; Simner, P.J.; Antibacterial Resistance Leadership Group. A Primer on AmpC beta-Lactamases: Necessary Knowledge for an Increasingly Multidrug-resistant World. Clin. Infect. Dis. 2019, 69, 1446–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizusawa, M.; Carroll, K.C. Novel strategies for rapid identification and susceptibility testing of MRSA. Expert Rev. Anti-Infect. Ther. 2020, 18, 759–778. [Google Scholar] [CrossRef] [PubMed]
- Rood, I.G.H.; Li, Q. Review: Molecular detection of extended spectrum-beta-lactamase- and carbapenemase-producing Enterobacteriaceae in a clinical setting. Diagn. Microbiol. Infect. Dis. 2017, 89, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Antimicrobial resistance: The example of Staphylococcus aureus. J. Clin. Investig. 2003, 111, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Winstanley, T.; Courvalin, P. Expert systems in clinical microbiology. Clin. Microbiol. Rev. 2011, 24, 515–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leclercq, R.; Canton, R.; Brown, D.F.; Giske, C.G.; Heisig, P.; MacGowan, A.P.; Mouton, J.W.; Nordmann, P.; Rodloff, A.C.; Rossolini, G.M.; et al. EUCAST expert rules in antimicrobial susceptibility testing. Clin. Microbiol. Infect. 2013, 19, 141–160. [Google Scholar] [CrossRef] [PubMed]

| Gram-Negative | Prospective | Spiked | Total | No-ID | Discordant |
|---|---|---|---|---|---|
| Escherichia coli | 18 | 1 | 19 | 2 | 0 |
| Klebsiella pneumoniae | 11 | 1 | 12 | 0 | 0 |
| Klebsiella aerogenes | 3 | 0 | 3 | 0 | 0 |
| Klebsiella oxytoca | 2 | 0 | 2 | 0 | 0 |
| Proteus mirabilis | 1 | 1 | 2 | 0 | 0 |
| Enterobacter cloacae | 3 | 1 | 4 | 0 | 0 |
| Serratia marcescens | 1 | 1 | 2 | 0 | 0 |
| Raoultella ornithinolytica | 0 | 1 | 1 | 0 | 0 |
| Morganella morganii | 0 | 1 | 1 | 0 | 0 |
| Citrobacter freundii | 0 | 1 | 1 | 0 | 0 |
| Pseudomonas aeruginosa | 6 | 5 | 11 | 1 | 0 |
| Stenotrophomonas maltophilia | 2 | 0 | 2 | 1 | 1 |
| Campylobacter fetus | 1 | 0 | 1 | 1 | 0 |
| Cardiobacterium hominis | 1 | 0 | 1 | 0 | 0 |
| Bacteroides fragilis | 1 | 0 | 1 | 0 | 0 |
| Bacteroides thetaiotaomicron | 1 | 0 | 1 | 0 | 0 |
| Subtotals | 51 | 13 | 64 | 5 | 1 |
| Gram Positive | Prospective | Spiked | Total | No-ID | Discordant |
| Staphylococcus epidermidis | 21 | 0 | 21 | 5 | 1 |
| Staphylococcus capitis | 3 | 0 | 3 | 1 | 0 |
| Staphylococcus caprae | 1 | 0 | 1 | 0 | 0 |
| Staphylococcus haemolyticus | 2 | 0 | 2 | 0 | 0 |
| Staphylococcus hominis | 3 | 0 | 3 | 0 | 0 |
| Staphylococcus aureus | 13 | 4 | 17 | 0 | 0 |
| Streptococcus anginosus | 1 | 0 | 1 | 1 | 0 |
| Streptococcus mitis/oralis | 3 | 0 | 3 | 2 | 0 |
| Streptococcus parasanguinis | 2 | 0 | 2 | 0 | 1 |
| Streptococcus vestibularis | 1 | 0 | 1 | 0 | 0 |
| Streptococcus pyogenes | 1 | 0 | 1 | 0 | 0 |
| Enterococcus faecalis | 7 | 2 | 9 | 0 | 0 |
| Enterococcus faecium | 5 | 2 | 7 | 0 | 0 |
| Enterococcus avium | 0 | 1 | 1 | 0 | 0 |
| Clostridium ramnosum | 1 | 0 | 1 | 0 | 0 |
| Clostridium tertium | 1 | 0 | 1 | 0 | 0 |
| Subtotals | 65 | 9 | 74 | 9 | 2 |
| Totals | 116 | 22 | 138 | 14 | 3 |
| LC + MALDI MS | ||||
|---|---|---|---|---|
| No ID | Discordant | Species ID | Total | |
| Gram-negative | 9.8% (5) | 2.0% (1) | 88.2% (45) | 100% (51) |
| Gram-positive | 13.8% (9) | 3.1% (2) | 83.1% (54) | 100% (65) |
| Total | 12.1% (14) | 2.6% (3) | 85.3% (99) | 100% (116) |
| Verigene | ||||
| No ID | Genus ID Only | Species ID | Total | |
| Gram-negative | 13.7% (7) | 11.8% (6) | 74.5% (38) | 100% (51) |
| Gram-positive | 6.2% (4) | 21.5% (14) | 72.3% (47) | 100% (65) |
| Total | 9.5% (11) | 17.2% (20) | 73.3% (85) | 100% (116) |
| Gram-Positive AST Summary | ||
|---|---|---|
| CA | 639/652 | 98.0% |
| EA | 642/652 | 98.4% |
| VME | 4/210 | 1.9% |
| ME | 1/431 | 0.2% |
| mE | 8/652 | 1.2% |
| Gram-Negative AST Summary | ||
| CA | 601/613 | 98.0% |
| EA | 603/613 | 98.4% |
| VME | 1/96 | 1.0% |
| ME | 0/506 | 0.0% |
| mE | 11/613 | 1.8% |
| Antimicrobial | Essential Agreement | Categorical Agreement | Minor Errors | Major Errors | Very Major Errors |
|---|---|---|---|---|---|
| Amikacin | 39/39 (100%) | 39/39 (100%) | 0/39 | 0/39 | 0/0 |
| Ampicillin | 30/31 (96.8%) | 29/31 (93.5%) | 2/31 (6.5%) | 0/10 | 0/21 |
| Ampicillin/Sulbactam | 30/31 (96.8%) | 27/31 (87.1%) | 4/31 (12.9%) | 0/19 | 0/10 |
| Cefazolin | 39/39 (100%) | 36/39 (92.3%) | 3/39 (7.8%) | 0/22 | 0/17 |
| Cefepime | 39/39 (100%) | 39/39 (100%) | 0/39 | 0/36 | 0/1 |
| Cefoxitin | 38/39 (97.4%) | 38/39 (97.4%) | 1/39 (2.6%) | 0/29 | 0/9 |
| Ceftazidime | 39/39 (100%) | 39/39 (100%) | 0/39 | 0/33 | 0/5 |
| Ceftriaxone | 38/39 (97.4%) | 38/39 (97.4%) | 0/39 | 0/29 | 1/10 (10.0%) |
| Ciprofloxacin | 39/39 (100%) | 39/39 (100%) | 1/39 | 0/26 | 0/12 |
| Ertapenem | 39/39 (100%) | 39/39 (100%) | 0/39 | 0/38 | 0/1 |
| Gentamicin | 39/39 (100%) | 39/39 (100%) | 0/39 | 0/37 | 0/2 |
| Meropenem | 39/39 (100%) | 39/39 (100%) | 0/39 | 0/39 | 0/0 |
| Piperacillin/Tazobactam | 37/37 (100%) | 37/37 (100%) | 0/37 | 0/35 | 0/1 |
| Tobramycin | 39/39 (100%) | 39/39 (100%) | 0/39 | 0/35 | 0/1 |
| Trimethoprim/Sulfamethoxazole | 39/39 (100%) | 39/39 (100%) | 0/39 | 0/25 | 0/14 |
| Total | 558/567 (98.4%) | 555/567 (97.9%) | 11/567 (1.9%) | 0/452 | 1/104 (1.0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ugaban, K.; Pak, P.; She, R.C. Direct MALDI-TOF MS and Antimicrobial Susceptibility Testing of Positive Blood Cultures Using the FASTTM System and FAST-PBC Prep Cartridges—Performance Evaluation in a Clinical Microbiology Laboratory Serving High-Risk Patients. Microorganisms 2022, 10, 2076. https://doi.org/10.3390/microorganisms10102076
Ugaban K, Pak P, She RC. Direct MALDI-TOF MS and Antimicrobial Susceptibility Testing of Positive Blood Cultures Using the FASTTM System and FAST-PBC Prep Cartridges—Performance Evaluation in a Clinical Microbiology Laboratory Serving High-Risk Patients. Microorganisms. 2022; 10(10):2076. https://doi.org/10.3390/microorganisms10102076
Chicago/Turabian StyleUgaban, Khay, Pil Pak, and Rosemary C. She. 2022. "Direct MALDI-TOF MS and Antimicrobial Susceptibility Testing of Positive Blood Cultures Using the FASTTM System and FAST-PBC Prep Cartridges—Performance Evaluation in a Clinical Microbiology Laboratory Serving High-Risk Patients" Microorganisms 10, no. 10: 2076. https://doi.org/10.3390/microorganisms10102076
APA StyleUgaban, K., Pak, P., & She, R. C. (2022). Direct MALDI-TOF MS and Antimicrobial Susceptibility Testing of Positive Blood Cultures Using the FASTTM System and FAST-PBC Prep Cartridges—Performance Evaluation in a Clinical Microbiology Laboratory Serving High-Risk Patients. Microorganisms, 10(10), 2076. https://doi.org/10.3390/microorganisms10102076





