Tea-Soybean Intercropping Improves Tea Quality and Nutrition Uptake by Inducing Changes of Rhizosphere Bacterial Communities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tea-Soybean Intercropping Experiment Design
2.2. Plant and Soil Sample Collection and Physicochemical Analysis
2.3. Competitive Ratio between Tea Plants and Soybeans in the Intercropping System
2.4. DNA Extraction and 16S rRNA Gene Sequencing
2.5. Quantitative Determination of Tea Quality Components
2.6. Statistical Analysis
3. Results and Discussion
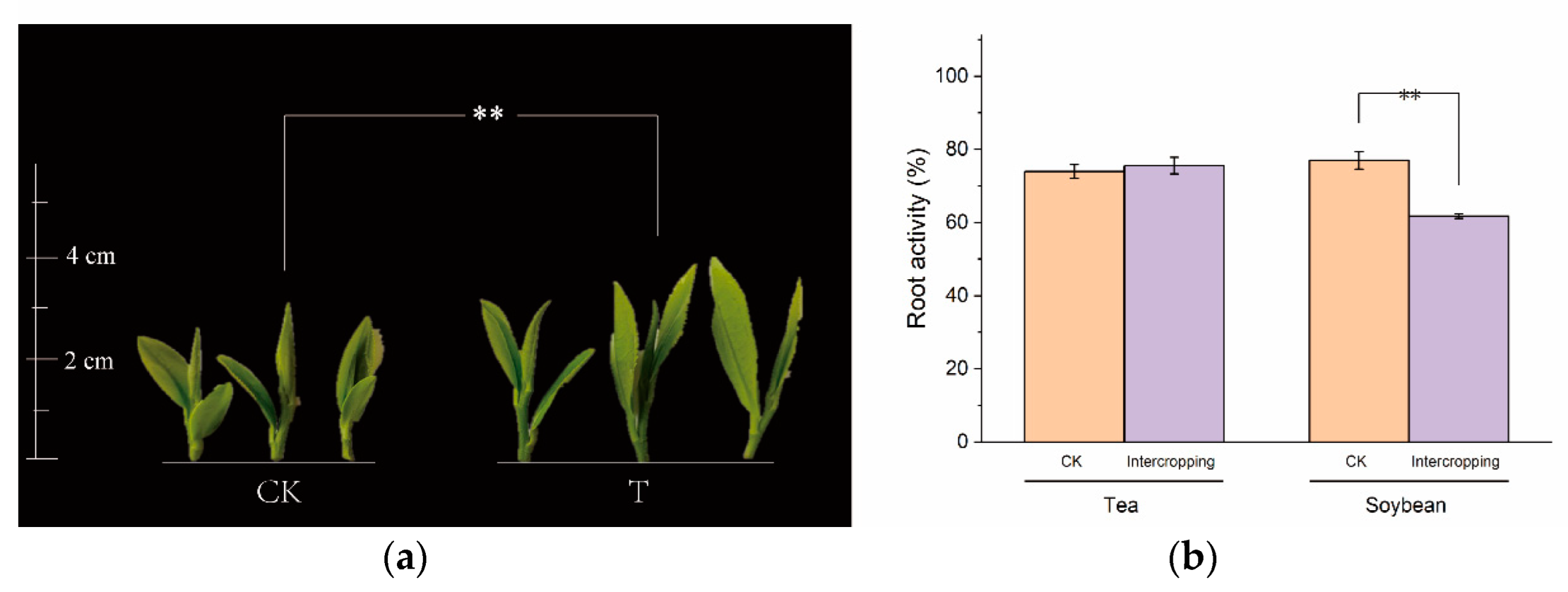
3.1. Effect of Tea Plant/Soybean Intercropping on Tea Plant Development
3.2. Effect of Tea Plant/Soybean Intercropping on N, P and K
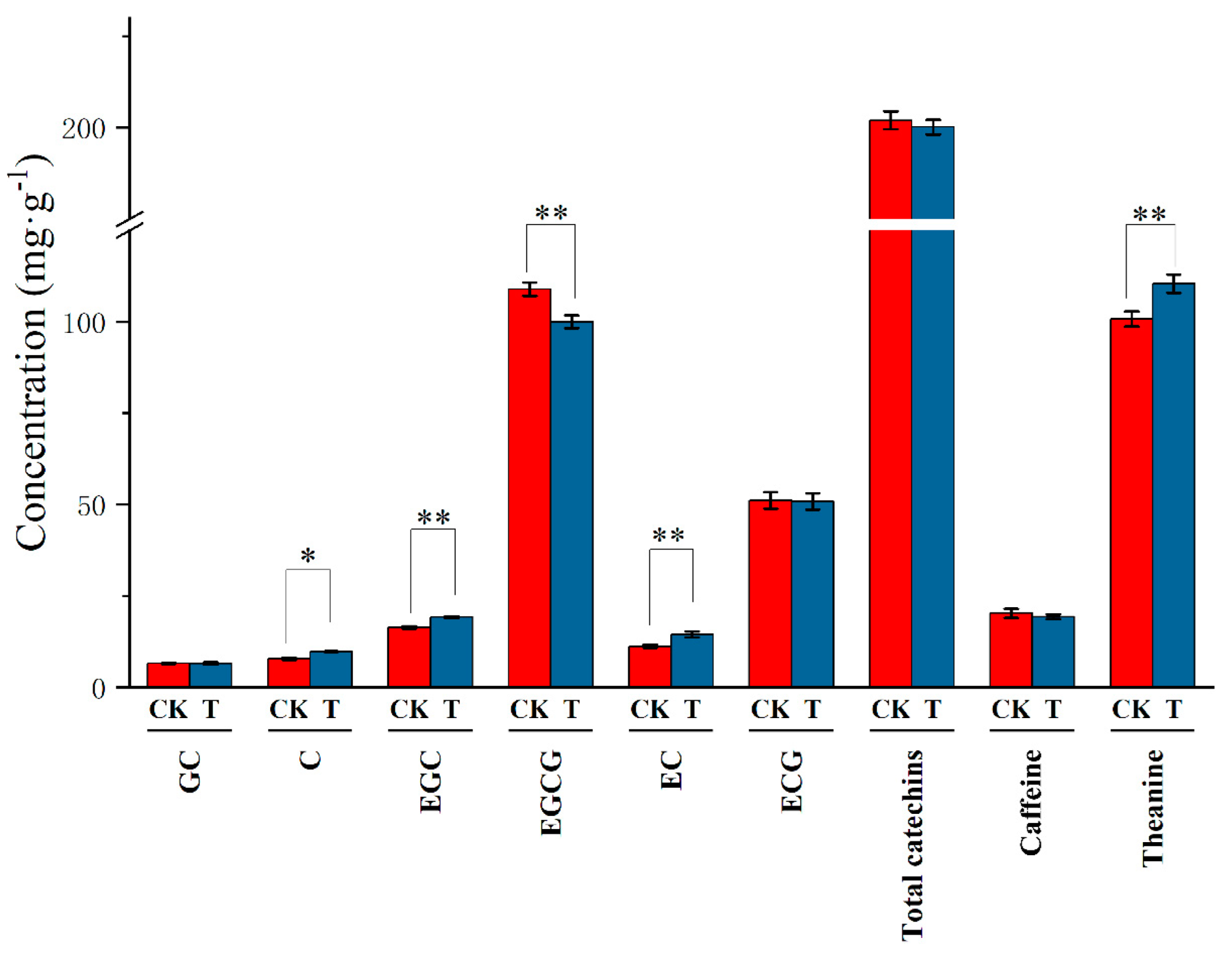
3.3. Quality-Related Components in Tea Leaves under Tea Plant-Soybean Intercropping
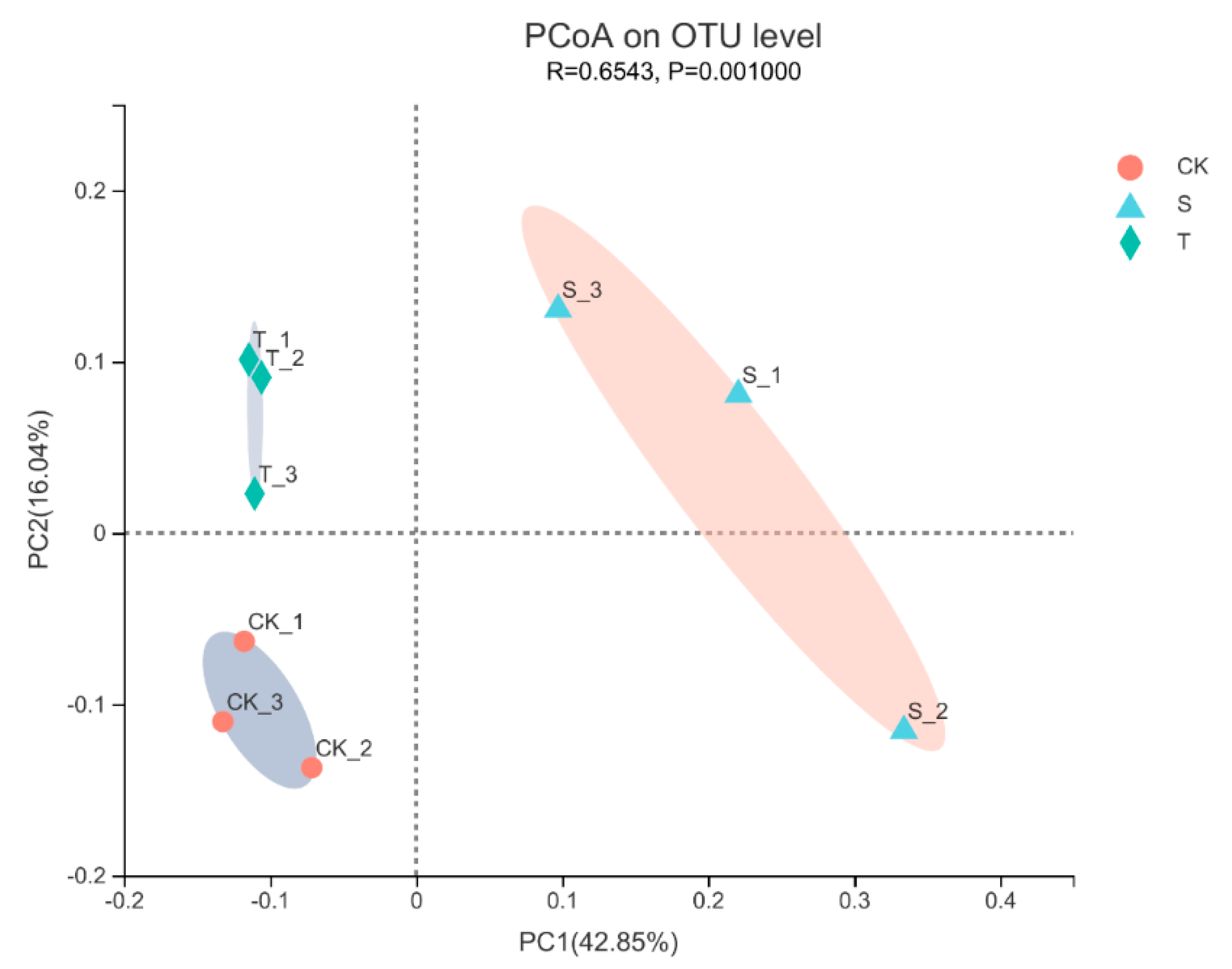
3.4. Diversity of the Bacterial Community in Rhizosphere Soil under Tea Plant-Soybean Intercropping
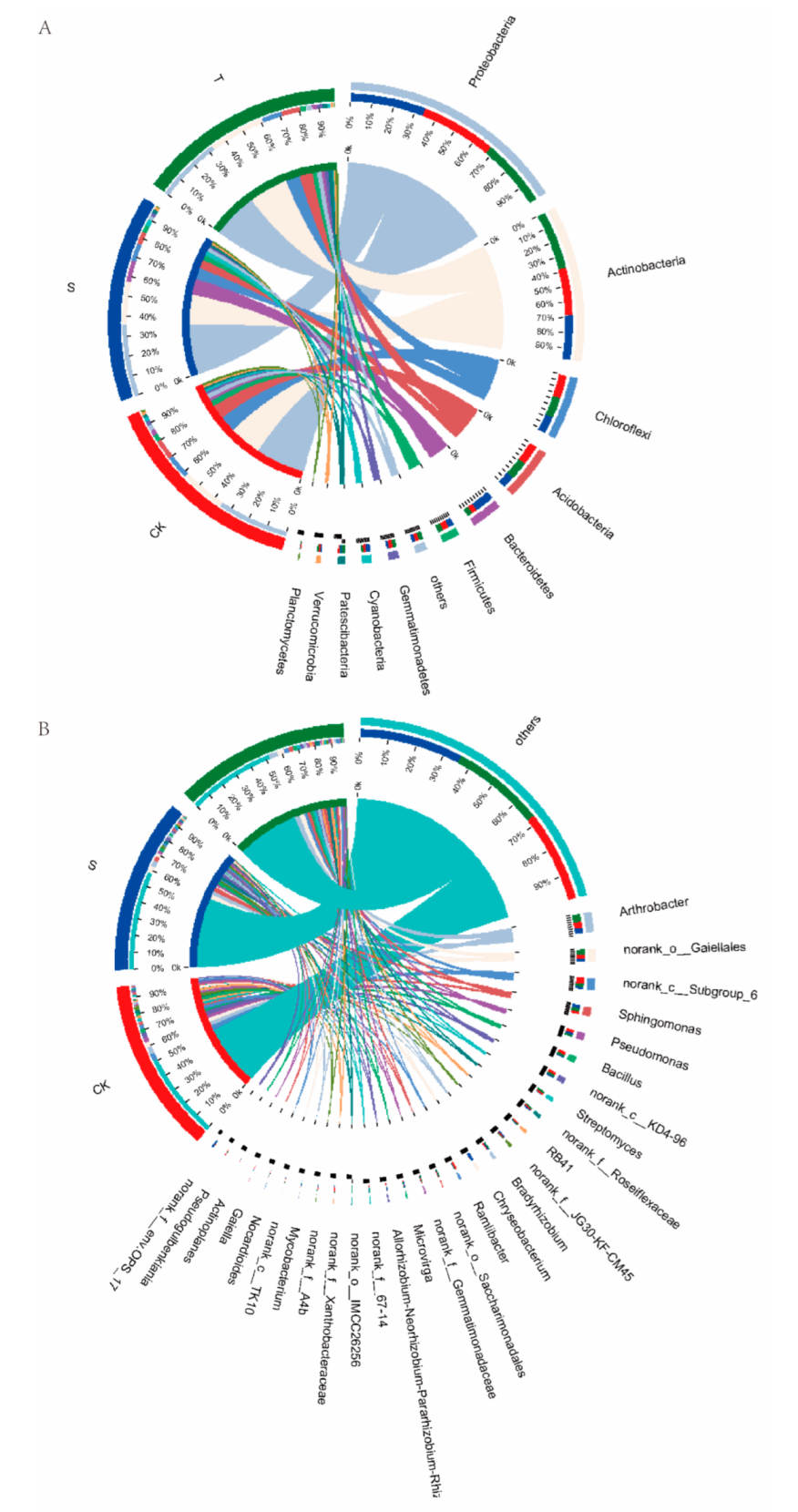
3.5. Composition of the Bacterial Community in Rhizosphere Soil under Tea Plant-Soybean Intercropping
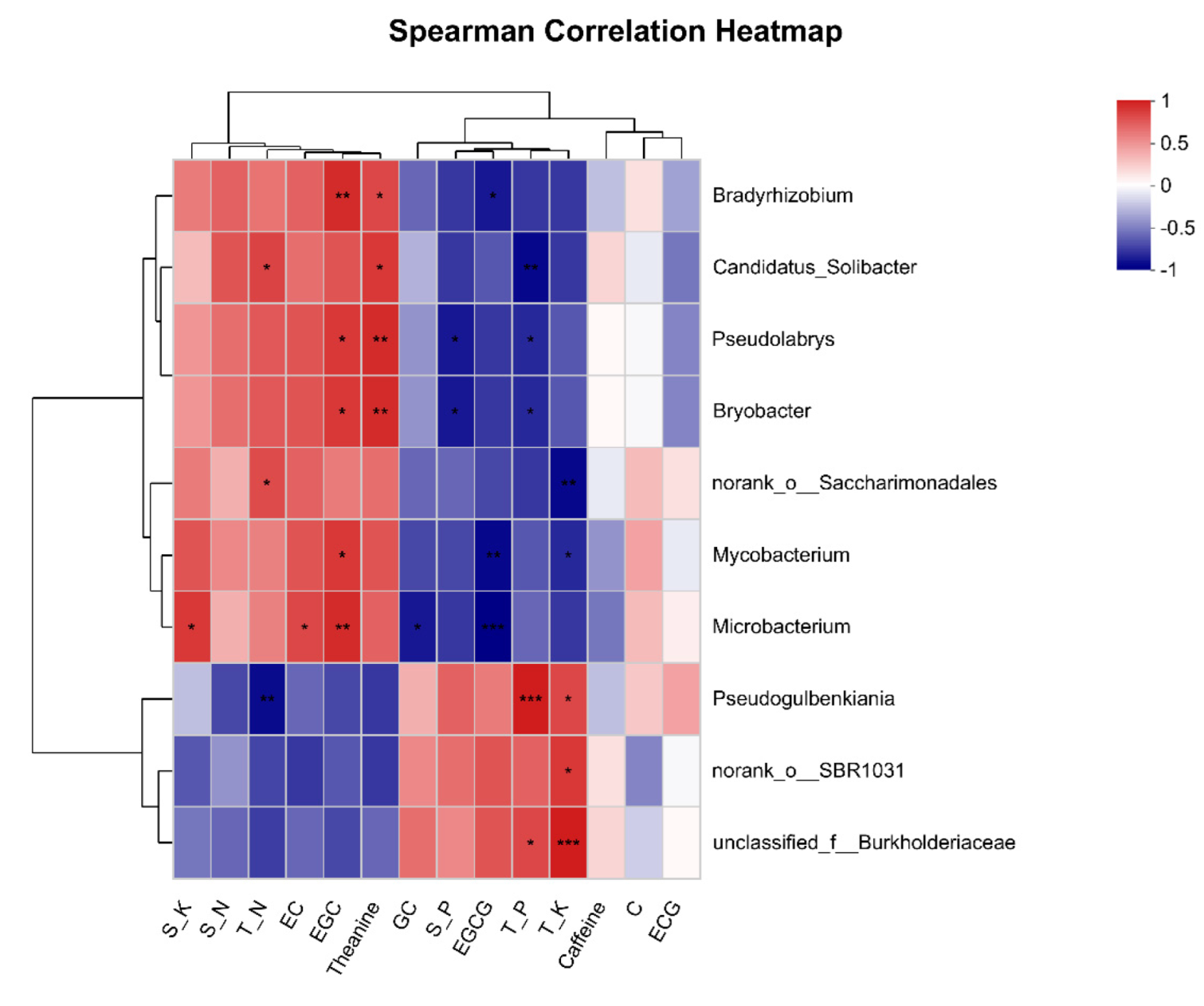
3.6. Correlation between Differential Bacteria and Soil Physicochemical Properties
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- FAO. FAOSTAT, Value of agricultural production—Gross production value of tea. In FAOSTAT: Value of Agricultural Production; FAO: Rome, Italy, 2020. [Google Scholar]
- Yan, P.; Wu, L.; Wang, D.; Fu, J.; Shen, C.; Li, X.; Zhang, L.; Zhang, L.; Fan, L.; Wenyan, H. Soil acidification in Chinese tea plantations. Sci. Total Environ. 2020, 715, 136963. [Google Scholar] [CrossRef]
- Oh, K.; Kato, T.; Zhong-Pei, L.; Fa-Yun, L. Environmental problems from tea cultivation in Japan and a control measure using calcium cyanamide. Pedosphere 2006, 16, 770–777. [Google Scholar] [CrossRef]
- Yan, P.; Shen, C.; Fan, L.; Li, X.; Zhang, L.; Zhang, L.; Han, W. Tea planting affects soil acidification and nitrogen and phosphorus distribution in soil. Agric. Ecosyst. Environ. 2018, 254, 20–25. [Google Scholar] [CrossRef]
- Willey, R. Resource use in intercropping systems. Agric. Water Manag. 1990, 17, 215–231. [Google Scholar] [CrossRef]
- Wang, Z.-G.; Jin, X.; Bao, X.-G.; Li, X.-F.; Zhao, J.-H.; Sun, J.-H.; Christie, P.; Li, L. Intercropping enhances productivity and maintains the most soil fertility properties relative to sole cropping. PLoS ONE 2014, 9, e113984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zhou, C.; Xu, Y.; Huang, X.; Zhang, L.; Mu, W. Effects of intercropping tea with aromatic plants on population dynamics of arthropods in Chinese tea plantations. J. Pest Sci. 2017, 90, 227–237. [Google Scholar] [CrossRef]
- Sedaghathoor, S.; Janatpoor, G. Study on effect of soybean and tea intercropping on yield and yield components of soybean and tea. J. Agric. Biol. Sci. 2012, 7, 664–671. [Google Scholar]
- Duan, Y.; Shang, X.; Liu, G.; Zou, Z.; Zhu, X.; Ma, Y.; Li, F.; Fang, W. The effects of tea plants-soybean intercropping on the secondary metabolites of tea plants by metabolomics analysis. BMC Plant Biol. 2021, 21, 482. [Google Scholar] [CrossRef]
- Brooker, R.W.; Bennett, A.E.; Cong, W.F.; Daniell, T.J.; George, T.S.; Hallett, P.D.; Hawes, C.; Iannetta, P.P.; Jones, H.G.; Karley, A.J. Improving intercropping: A synthesis of research in agronomy, plant physiology and ecology. New Phytol. 2015, 206, 107–117. [Google Scholar] [CrossRef]
- Tang, X.; Bernard, L.; Brauman, A.; Daufresne, T.; Deleporte, P.; Desclaux, D.; Souche, G.; Placella, S.A.; Hinsinger, P. Increase in microbial biomass and phosphorus availability in the rhizosphere of intercropped cereal and legumes under field conditions. Soil Biol. Biochem. 2014, 75, 86–93. [Google Scholar] [CrossRef]
- Lian, T.; Mu, Y.; Jin, J.; Ma, Q.; Cheng, Y.; Cai, Z.; Nian, H. Impact of intercropping on the coupling between soil microbial community structure, activity, and nutrient-use efficiencies. PeerJ 2019, 7, e6412. [Google Scholar] [CrossRef]
- Tang, X.; Zhong, R.; Jiang, J.; He, L.; Huang, Z.; Shi, G.; Wu, H.; Liu, J.; Xiong, F.; Han, Z. Cassava/peanut intercropping improves soil quality via rhizospheric microbes increased available nitrogen contents. BMC Biotechnol. 2020, 20, 13. [Google Scholar] [CrossRef] [Green Version]
- Jain, V.; Gupta, K. The flavonoid naringenin enhances intercellular colonization of rice roots by Azorhizobium caulinodans. Biol. Fertil. Soils 2003, 38, 119–123. [Google Scholar] [CrossRef]
- Schelud’ko, A.V.; Makrushin, K.V.; Tugarova, A.V.; Krestinenko, V.A.; Panasenko, V.I.; Antonyuk, L.P.; Katsy, E.I. Changes in motility of the rhizobacterium Azospirillum brasilense in the presence of plant lectins. Microbiol. Res. 2009, 164, 149–156. [Google Scholar] [CrossRef]
- Marschner, P.; Yang, C.-H.; Lieberei, R.; Crowley, D.E. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 2001, 33, 1437–1445. [Google Scholar] [CrossRef]
- Bai, A.J.; Rai, V.R. Bacterial quorum sensing and food industry. Compr. Rev. Food Sci. Food Saf. 2011, 10, 183–193. [Google Scholar] [CrossRef]
- Duchene, O.; Vian, J.-F.; Celette, F. Intercropping with legume for agroecological cropping systems: Complementarity and facilitation processes and the importance of soil microorganisms. A review. Agric. Ecosyst. Environ. 2017, 240, 148–161. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Kuzyakov, Y. Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: Critical review. Biol. Fertil. Soils 2008, 45, 115–131. [Google Scholar] [CrossRef]
- Kuzyakov, Y. Priming effects: Interactions between living and dead organic matter. Soil Biol. Biochem. 2010, 42, 1363–1371. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, X.; Nie, C.; Wu, T.; Dai, W.; Liu, H.; Yang, R. Analysis of unculturable bacterial communities in tea orchard soils based on nested PCR-DGGE. World J. Microbiol. Biotechnol. 2012, 28, 1967–1979. [Google Scholar] [CrossRef]
- Tan, L.; Gu, S.; Li, S.; Ren, Z.; Deng, Y.; Liu, Z.; Gong, Z.; Xiao, W.; Hu, Q. Responses of microbial communities and interaction networks to different management practices in tea plantation soils. Sustainability 2019, 11, 4428. [Google Scholar] [CrossRef]
- Li, Y.; Li, Z.; Li, Z.; Jiang, Y.; Weng, B.; Lin, W. Variations of rhizosphere bacterial communities in tea (Camellia sinensis L.) continuous cropping soil by high-throughput pyrosequencing approach. J. Appl. Microbiol. 2016, 121, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Arafat, Y.; Wei, X.; Jiang, Y.; Chen, T.; Saqib, H.S.A.; Lin, S.; Lin, W. Spatial distribution patterns of root-associated bacterial communities mediated by root exudates in different aged ratooning tea monoculture systems. Int. J. Mol. Sci. 2017, 18, 1727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nerva, L.; Sandrini, M.; Moffa, L.; Velasco, R.; Balestrini, R.; Chitarra, W. Breeding toward improved ecological plant–microbiome interactions. Trends Plant Sci. 2022, 26, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.-T.; Lin, S.-H. Priming effects of cover cropping on bacterial community in a tea plantation. Sustainability 2021, 13, 4345. [Google Scholar] [CrossRef]
- Huang, Z.; Cui, C.; Cao, Y.; Dai, J.; Cheng, X.; Hua, S.; Wang, W.; Duan, Y.; Petropoulos, E.; Wang, H. Tea plant–legume intercropping simultaneously improves soil fertility and tea quality by changing Bacillus species composition. Hortic. Res. 2022, 9, uhac046. [Google Scholar] [CrossRef]
- McSpadden Gardener, B.B. Ecology of Bacillus and Paenibacillus spp. in agricultural systems. Phytopathology 2004, 94, 1252–1258. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Qin, Y.; Li, M. Intercropping of tea (Camellia sinensis L.) and Chinese chestnut: Variation in the structure of rhizosphere bacterial communities. J. Plant. Nutr. Soil Sci. 2021, 21, 2178–2190. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, Q.; Dai, X.; He, M. Effects of potassium chloride and nitric oxide on growth and physiological characteristics of winter wheat under salt stress. Biol. Plant. 2020, 64, 258–265. [Google Scholar] [CrossRef] [Green Version]
- Willey, R.; Rao, M. A competitive ratio for quantifying competition between intercrops. Exp. Agric. 1980, 16, 117–125. [Google Scholar] [CrossRef]
- Fan, F.; Zhang, F.; Song, Y.; Sun, J.; Bao, X.; Guo, T.; Li, L. Nitrogen fixation of faba bean (Vicia faba L.) interacting with a non-legume in two contrasting intercropping systems. Plant Soil 2006, 283, 275–286. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Sun, L.; Qiu, C.; Ding, Y.; Gu, H.; Wang, L.; Wang, Z.; Ding, Z. Organic mulching positively regulates the soil microbial communities and ecosystem functions in tea plantation. BMC Microbiol. 2020, 20, 103. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, Y.; Ma, D.; Wang, L.; Zhang, X.; Ding, Y.; Fan, K.; Xu, Z.; Yuan, C.; Jia, H. Differential responses of the rhizosphere microbiome structure and soil metabolites in tea (Camellia sinensis) upon application of cow manure. BMC Microbiol. 2020, 20, 55. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, Y.; Ding, Z.; Liu, F. The dynamic changes of catechins and related genes in tea (Camellia sinensis) flowers. Acta Physiol. Plant. 2019, 41, 30. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, C.; Wang, Y.; Shen, J.; Ding, Z. Conserved microRNA act boldly during sprout development and quality formation in Pingyang Tezaocha (Camellia sinensis). Front. Genet. 2019, 10, 237. [Google Scholar] [CrossRef]
- Duan, Y.; Shen, J.; Zhang, X.; Wen, B.; Ma, Y.; Wang, Y.; Fang, W.; Zhu, X. Effects of soybean–tea intercropping on soil-available nutrients and tea quality. Acta Physiol. Plant 2019, 41, 140. [Google Scholar] [CrossRef]
- Ma, Y.-h.; Fu, S.-l.; Zhang, X.-p.; Zhao, K.; Chen, H.Y. Intercropping improves soil nutrient availability, soil enzyme activity and tea quantity and quality. Appl. Soil Ecol. 2017, 119, 171–178. [Google Scholar] [CrossRef]
- Xiao, Y.; Li, L.; Zhang, F. Effect of root contact on interspecific competition and N transfer between wheat and fababean using direct and indirect 15N techniques. Plant Soil 2004, 262, 45–54. [Google Scholar] [CrossRef]
- Wang, D.; Marschner, P.; Solaiman, Z.; Rengel, Z. Growth, P uptake and rhizosphere properties of intercropped wheat and chickpea in soil amended with iron phosphate or phytate. Soil Biol. Biochem. 2007, 39, 249–256. [Google Scholar] [CrossRef]
- Farooq, T.H.; Kumar, U.; Mo, J.; Shakoor, A.; Wang, J.; Rashid, M.H.U.; Tufail, M.A.; Chen, X.; Yan, W. Intercropping of peanut–tea enhances soil enzymatic activity and soil nutrient status at different soil profiles in subtropical southern China. Plants 2021, 10, 881. [Google Scholar] [CrossRef]
- Nakagawa, M. Constituents in tea leaf and their contribution to the taste of green tea liquor. Jpn. Agric. Res. Q. 1970, 5, 43–47. [Google Scholar]
- Ye, Y.; Yan, J.; Cui, J.; Mao, S.; Li, M.; Liao, X.; Tong, H. Dynamic changes in amino acids, catechins, caffeine and gallic acid in green tea during withering. J. Food Compos. Anal. 2018, 66, 98–108. [Google Scholar] [CrossRef]
- Kaneko, S.; Kumazawa, K.; Masuda, H.; Henze, A.; Hofmann, T. Molecular and sensory studies on the umami taste of Japanese green tea. J. Agric. Food Chem. 2006, 54, 2688–2694. [Google Scholar] [CrossRef]
- Fu, H.; Li, H.; Yin, P.; Mei, H.; Li, J.; Zhou, P.; Wang, Y.; Ma, Q.; Jeyaraj, A.; Thangaraj, K. Integrated application of rapeseed cake and green manure enhances soil nutrients and microbial communities in tea garden soil. Sustainability 2021, 13, 2967. [Google Scholar] [CrossRef]
- Deeg, C.M.; Zimmer, M.M.; George, E.E.; Husnik, F.; Keeling, P.J.; Suttle, C.A. Chromulinavorax destructans, a pathogen of microzooplankton that provides a window into the enigmatic candidate phylum Dependentiae. PLoS Pathog. 2019, 15, e1007801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, R.; Cui, X.; Luo, X.; Mao, P.; Zhuang, P.; Li, Y.; Li, Y.; Li, Z. Effects of plant growth regulator and chelating agent on the phytoextraction of heavy metals by Pfaffia glomerata and on the soil microbial community. Environ. Pollut. 2021, 283, 117159. [Google Scholar] [CrossRef] [PubMed]
- Gottel, N.R.; Castro, H.F.; Kerley, M.; Yang, Z.; Pelletier, D.A.; Podar, M.; Karpinets, T.; Uberbacher, E.; Tuskan, G.A.; Vilgalys, R. Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl. Environ. Microbiol. 2011, 77, 5934–5944. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Su, W.; Chen, H.; Barberán, A.; Zhao, H.; Yu, M.; Yu, L.; Brookes, P.C.; Schadt, C.W.; Chang, S.X. Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of Actinobacteria and Proteobacteria in agro-ecosystems across the globe. Glob. Chang. Biol. 2018, 24, 3452–3461. [Google Scholar] [CrossRef]
- Shamseldin, A.; Abdelkhalek, A.; Sadowsky, M.J. Recent changes to the classification of symbiotic, nitrogen-fixing, legume-associating bacteria: A review. Symbiosis 2017, 71, 91–109. [Google Scholar] [CrossRef]
- Xue, Z.; Zhang, T.; Sun, Y.; Yin, T.; Cao, J.; Fang, F.; Feng, Q.; Luo, J. Integrated moving bed biofilm reactor with partial denitrification-anammox for promoted nitrogen removal: Layered biofilm structure formation and symbiotic functional microbes. Sci. Total Environ. 2022, 839, 156339. [Google Scholar] [CrossRef]
- Pan, L.; Chen, J.; Ren, S.; Shen, H.; Rong, B.; Liu, W.; Yang, Z. Complete genome sequence of Mycobacterium Mya-zh01, an endophytic bacterium, promotes plant growth and seed germination isolated from flower stalk of Doritaenopsis. Arch. Microbiol. 2020, 202, 1965–1976. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.J.d.S.; Dias-Filho, M.B.; Gurgel, E.S.C. Successful plant growth-promoting microbes: Inoculation methods and abiotic factors. Front. Sustain. Food Syst. 2021, 5, 606454. [Google Scholar] [CrossRef]
- Mantelin, S.; Touraine, B. Plant growth-promoting bacteria and nitrate availability: Impacts on root development and nitrate uptake. J. Exp. Bot. 2004, 55, 27–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Jin, Z.; Wang, X.; George, T.S.; Feng, G.; Zhang, L. Simulated root exudates stimulate the abundance of Saccharimonadales to improve the alkaline phosphatase activity in maize rhizosphere. Appl. Soil Ecol. 2022, 170, 104274. [Google Scholar] [CrossRef]
- Liu, K.-l.; Han, T.-f.; Huang, J.; Asad, S.; Li, D.-M.; Yu, X.-C.; Huang, Q.-H.; Ye, H.-C.; Hu, H.-W.; Hu, Z.-H. Links between potassium of soil aggregates and pH levels in acidic soils under long-term fertilization regimes. Soil Tillage Res. 2020, 197, 104480. [Google Scholar] [CrossRef]
- He, X.; Chi, Q.; Zhao, C.; Cheng, Y.; Huang, X.; Zhao, J.; Cai, Z.; Zhang, J.; Müller, C. Plants with an ammonium preference affect soil N transformations to optimize their N acquisition. Soil Biol. Biochem. 2021, 155, 108158. [Google Scholar] [CrossRef]
- Li, X.; Qiao, J.; Li, S.; Haggblom, M.M.; Li, F.; Hu, M. Bacterial communities and functional genes stimulated during anaerobic arsenite oxidation and nitrate reduction in a paddy soil. Environ. Sci. Technol. 2019, 54, 2172–2181. [Google Scholar] [CrossRef]
- Compant, S.; Nowak, J.; Coenye, T.; Clément, C.; Ait Barka, E. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 2008, 32, 607–626. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.-C.; Ko, C.-H.; Su, Y.-S.; Lai, W.-A.; Shen, F.-T. Metabolic potential and community structure of bacteria in an organic tea plantation. Appl. Soil Ecol. 2021, 157, 103762. [Google Scholar] [CrossRef]
- Seldina, Y. The Effect of Epigallocatechin Gallate (EGCG) on Mycobacterium Tuberculosis and Mycobacterium Smegmatis. Ph.D. Thesis, University of Pittsburgh, Pittsburgh, PA, USA, 2011. [Google Scholar]
- Kim, E.J.; Fathoni, A.; Jeong, G.-T.; Do Jeong, H.; Nam, T.-J.; Kong, I.-S.; Kim, J.K. Microbacterium oxydans, a novel alginate-and laminarin-degrading bacterium for the reutilization of brown-seaweed waste. J. Environ. Manag. 2013, 130, 153–159. [Google Scholar] [CrossRef]
- Cordovez, V.; Schop, S.; Hordijk, K.; Dupré de Boulois, H.; Coppens, F.; Hanssen, I.; Raaijmakers, J.M.; Carrión, V.J. Priming of plant growth promotion by volatiles of root-associated Microbacterium spp. Appl. Environ. Microb. 2018, 84, e01818–e01865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuong, Q.V.; Bowyer, M.C.; Roach, P.D. L-Theanine: Properties, synthesis and isolation from tea. J. Sci. Food Agric. 2011, 91, 1931–1939. [Google Scholar] [CrossRef]
- Meena, R.S.; Vijayakumar, V.; Yadav, G.S.; Mitran, T. Response and interaction of Bradyrhizobium japonicum and arbuscular mycorrhizal fungi in the soybean rhizosphere. Plant Growth Regul. 2018, 84, 207–223. [Google Scholar] [CrossRef]
- Rana, N.K.; Mohanpuria, P.; Yadav, S.K. Cloning and characterization of a cytosolic glutamine synthetase from Camellia sinensis (L.) O. Kuntze that is upregulated by ABA, SA, and H2O2. Mol. Biotechnol. 2008, 39, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Kulichevskaya, I.S.; Huber, K.J.; Overmann, J. Defining the taxonomic status of described subdivision 3 Acidobacteria: Proposal of Bryobacteraceae fam. nov. Int. J. Syst. Evol. 2017, 67, 498–501. [Google Scholar] [CrossRef]
- Miao, Y.; Johnson, N.W.; Gedalanga, P.B.; Adamson, D.; Newell, C.; Mahendra, S. Response and recovery of microbial communities subjected to oxidative and biological treatments of 1,4-dioxane and co-contaminants. Water Res. 2019, 149, 74–85. [Google Scholar] [CrossRef]
- Lee, L.S.; Choi, J.H.; Son, N.; Kim, S.H.; Park, J.D.; Jang, D.J. Metabolomic analysis of the effect of shade treatment on the nutritional and sensory qualities of green tea. J. Agric. Food Chem. 2013, 61, 332–338. [Google Scholar] [CrossRef]


| Treatments | N (mg/kg) | P (mg/kg) | K (mg/kg) | |
|---|---|---|---|---|
| Soil | Tea plant monoculture soil | 1010.00 ± 133.17 a | 416.18 ± 18.15 a | 202.64 ± 1.00 a |
| Soy bean monoculture soil | 903.33 ± 43.33 a | 410.50 ± 48.79 ab | 199.96 ± 3.19 a | |
| Intercropping soil | 1190.00 ± 100.17 a | 277.79 ± 15.81 b | 207.63 ± 1.92 a | |
| Plants | Tea plant monoculture | 1656.69 ± 102.38 d | 947.50 ± 31.40 c | 7967.78 ± 260.30 b |
| Soy bean monoculture | 2633.46 ± 3.43 b | 1707.18 ± 44.54 b | 17749.63 ± 456.59 a | |
| Intercropping tea plant | 2160.15 ± 101.96 c | 678.81 ± 32.66 d | 6823.71 ± 341.90 b | |
| Intercropping soy bean | 3035.21 ± 3.69 a | 1996.30 ± 54.51 a | 19554.57 ± 943.15 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Dong, X.; Wang, Y.; Maker, G.; Agarwal, M.; Ding, Z. Tea-Soybean Intercropping Improves Tea Quality and Nutrition Uptake by Inducing Changes of Rhizosphere Bacterial Communities. Microorganisms 2022, 10, 2149. https://doi.org/10.3390/microorganisms10112149
Sun L, Dong X, Wang Y, Maker G, Agarwal M, Ding Z. Tea-Soybean Intercropping Improves Tea Quality and Nutrition Uptake by Inducing Changes of Rhizosphere Bacterial Communities. Microorganisms. 2022; 10(11):2149. https://doi.org/10.3390/microorganisms10112149
Chicago/Turabian StyleSun, Litao, Xue Dong, Yu Wang, Garth Maker, Manjree Agarwal, and Zhaotang Ding. 2022. "Tea-Soybean Intercropping Improves Tea Quality and Nutrition Uptake by Inducing Changes of Rhizosphere Bacterial Communities" Microorganisms 10, no. 11: 2149. https://doi.org/10.3390/microorganisms10112149





