Plant Growth-Promoting Activities of Bacteria Isolated from an Anthropogenic Soil Located in Agrigento Province
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth-Promoting Ability Assays
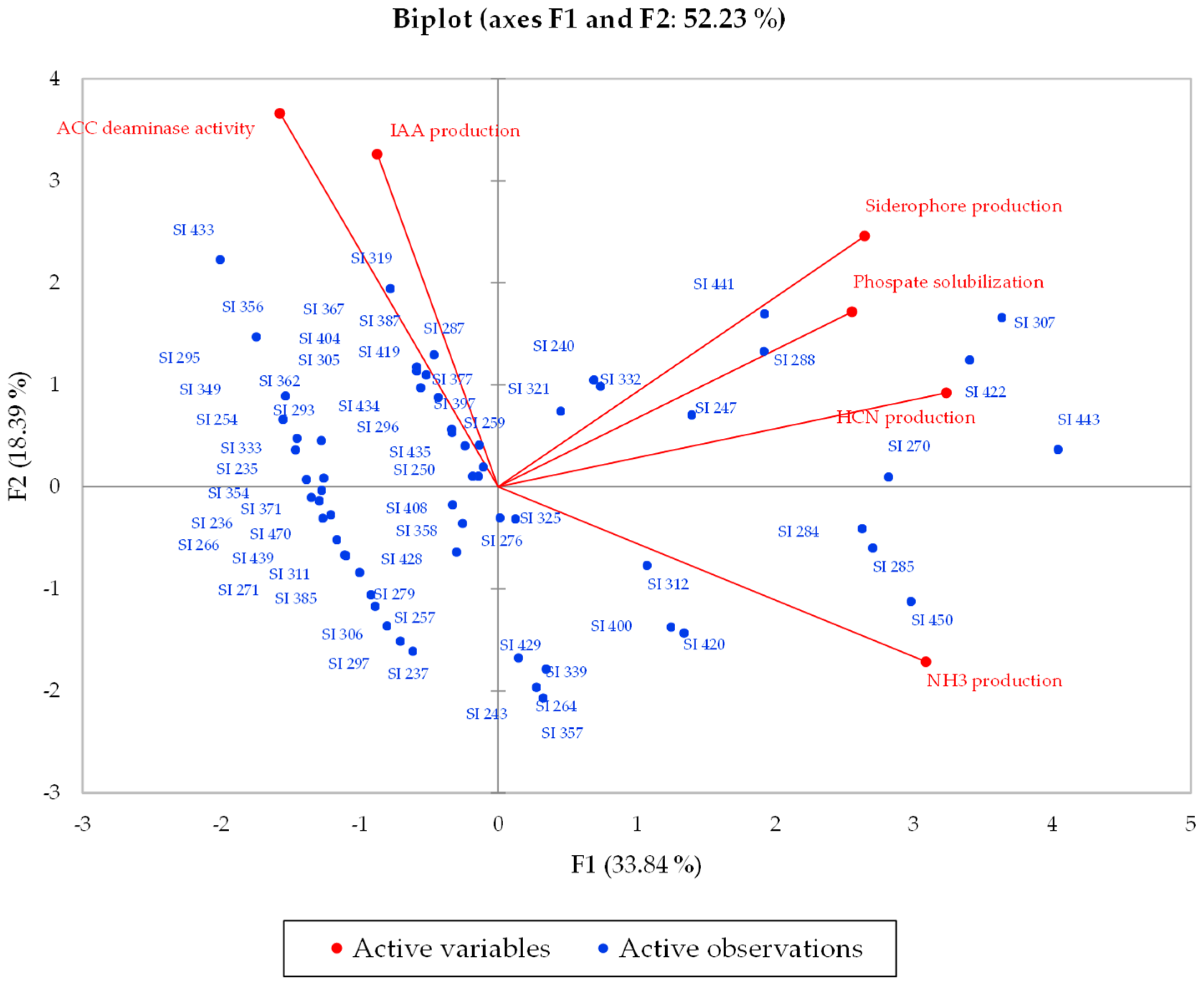
2.2. Statistical Analysis
3. Results
3.1. Plant Growth-Promoting Ability Assays
3.2. Statistical Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Howard, J. Anthropogenic soils; Springer: Berlin, Germany, 2017. [Google Scholar]
- Papa, G.L.; Palermo, V.; Dazzi, C. Is land-use change a cause of loss of pedodiversity? The case of the Mazzarrone study area, Sicily. Geomorphology 2011, 135, 332–342. [Google Scholar] [CrossRef]
- Papa, G.L.; Antisari, L.V.; Vianello, G.; Dazzi, C. Soil interpretation in the context of anthropedogenic transformations and pedotechniques application. Catena 2018, 166, 240–248. [Google Scholar] [CrossRef]
- Wang, A.S.; Angle, J.S.; Chaney, R.L.; Delorme, T.A.; McIntosh, M. Changes in soil biological activities under reduced soil pH during Thlaspi caerulescens phytoextraction. Soil Biol. Biochem. 2006, 38, 1451–1461. [Google Scholar] [CrossRef]
- Ahmad, F.; Ahmad, I.; Khan, M.S. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008, 163, 173–181. [Google Scholar] [CrossRef]
- Wu, S.C.; Cao, Z.H.; Li, Z.G.; Cheung, K.C.; Wong, M.H. Effects of biofertilizer containing N-fixer, P and K solubilizers and AM fungi on maize growth: A greenhouse trial. Geoderma 2005, 125, 155–166. [Google Scholar] [CrossRef]
- Haghighi, B.J.; Alizadeh, O.; Firoozabadi, A.H. The role of plant growth promoting rhizobacteria (PGPR) in sustainable agriculture. Adv. Environ. Biol. 2011, 5, 3079–3083. [Google Scholar]
- Antoun, H.; Kloepper, J.W. Plant growth promoting rhizobacteria. In Encyclopedia of Genetics; Brenner, S., Miller, J.H., Eds.; Academic Press: New York, NY, USA, 2001; pp. 1477–1480. [Google Scholar]
- Zakry, F.A.A.; Shamsuddin, Z.H.; Rahim, K.A.; Zakaria, Z.Z.; Rahim, A.A. Inoculation of Bacillus sphaericus UPMB-10 to young oil palm and measurement of its uptake of fixed nitrogen using the 15N isotope dilution technique. Microbes Environ. 2012, 27, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Vansuyt, G.; Robin, A.; Briat, J.F.; Curie, C.; Lemanceau, P. Iron acquisition from Fe-pyoverdine by Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2007, 20, 441–447. [Google Scholar] [CrossRef]
- Yazdani, M.; Bahmanyar, M.A.; Pirdashti, H.; Esmaili, M.A. Effect of phosphate solubilization microorganisms (PSM) and plant growth promoting rhizobacteria (PGPR) on yield and yield components of corn (Zea mays L.). WASET 2009, 49, 90–92. [Google Scholar]
- Sandhya, V.Z.A.S.; SK Z., A.; Grover, M.; Reddy, G.; Venkateswarlu, B.S.S.S. Alleviation of drought stress effects in sunflower seedlings by the exopolysaccharides producing Pseudomonas putida strain GAP-P45. Biol. Fertile. Soils 2009, 46, 17–26. [Google Scholar] [CrossRef]
- Weyens, N.; Truyens, S.; Dupae, J.; Newman, L.; Taghavi, S.; van der Lelie, D.; Carleer, R.; Vangronsveld, J. Potential of the TCE-degrading endophyte Pseudomonas putida W619-TCE to improve plant growth and reduce TCE phytotoxicity and evapotranspiration in poplar cuttings. Environ. Pollut. 2010, 158, 2915–2919. [Google Scholar] [CrossRef] [PubMed]
- Damam, M.; Kaloori, K.; Gaddam, B.; Kausar, R. Plant growth promoting substances (phytohormones) produced by rhizobacterial strains isolated from the rhizosphere of medicinal plants. Int. J. Pharm. Scie. Rev. Res. 2016, 37, 130–136. [Google Scholar]
- Kumar, A.; Singh, M.; Singh, P.P.; Singh, S.K.; Singh, P.K.; Pandey, K.D. Isolation of plant growth promoting rhizobacteria and their impact on growth and curcumin content in Curcuma longa L. Biocatal. Agric. 2016, 8, 1–7. [Google Scholar] [CrossRef]
- García de Salamone, I.E.; Hynes, R.K.; Nelson, L.M. Cytokinin production by plant growth promoting rhizobacteria and selected mutants. Can. J. Microbiol. 2001, 47, 404–411. [Google Scholar] [CrossRef]
- Noel, T.C.; Sheng, C.; Yost, C.K.; Pharis, R.P.; Hynes, M.F. Rhizobium leguminosarum as a plant growth-promoting rhizobacterium: Direct growth promotion of canola and lettuce. Can. J. Microbiol. 1996, 42, 279–283. [Google Scholar] [CrossRef]
- Gutiérrez-Mañero, F.J.; Ramos-Solano, B.; Probanza, A.N.; Mehouachi, J.R.; Tadeo, F.; Talon, M. The plant-growth-promoting rhizobacteria Bacillus pumilus and Bacillus licheniformis produce high amounts of physiologically active gibberellins. Physiol. Plant 2001, 111, 206–211. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, A.K.; Kumar, A. Disease management of tomato through PGPB: Current trends and future perspective. 3 Biotech 2017, 7, 1–10. [Google Scholar]
- Uqab, B.; Mudasir, S.; Nazir, R. Review on bioremediation of pesticides. J. Bioremed. Biodegr. 2016, 7, 343. [Google Scholar]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Nasrulhaq Boyce, A. Role of plant growth promoting rhizobacteria in agricultural sustainability—A review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef]
- Foyer, C.H.; Rasool, B.; Davey, J.W.; Hancock, R.D. Cross-tolerance to biotic and abiotic stresses in plants: A focus on resistance to aphid infestation. J. Exp. Bot. 2016, 67, 2025–2037. [Google Scholar] [CrossRef] [PubMed]
- Consentino, B.B.; Sabatino, L.; Vultaggio, L.; Rotino, G.L.; La Placa, G.G.; D’Anna, F.; Leto, C.; Iacuzzi, N.; De Pasquale, C. Grafting Eggplant Onto Underutilized Solanum Species and Biostimulatory Action of Azospirillum brasilense Modulate Growth, Yield, NUE and Nutritional and Functional Traits. Horticulturae 2022, 8, 722. [Google Scholar] [CrossRef]
- Barbaccia, P.; Dazzi, C.; Franciosi, E.; Di Gerlando, R.; Settanni, L.; Lo Papa, G. Microbiological Analysis and Metagenomic Profiling of the Bacterial Community of an Anthropogenic Soil Modified from Typic Haploxererts. Land 2022, 11, 748. [Google Scholar] [CrossRef]
- Wohler, I. Auxin-indole derivatives in soils determined by a colorimetric method and by high performance liquid chromatography. Microbiol. Res. 1997, 152, 399–405. [Google Scholar] [CrossRef]
- Cappuccino, J.C.; Sherman, N. Negative staining. In Microbiology: A Laboratory Manual, 3rd ed.; Cappuccino, J.C., Sherman, N., Eds.; Pearson: London, UK, 1992; pp. 125–179. [Google Scholar]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Honma, M.; Shimomura, T. Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 1978, 42, 1825–1831. [Google Scholar]
- Surange, S.; Wollum Ii, A.G.; Kumar, N.; Nautiyal, C.S. Characterization of Rhizobium from root nodules of leguminous trees growing in alkaline soils. Can. J. Microbiol. 1997, 43, 891–894. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis for Special Types of Data; Springer: New York, NY, USA, 2002. [Google Scholar]
- Mazzei, P.; Francesca, N.; Moschetti, G.; Piccolo, A. NMR spectroscopy evaluation of direct relationship between soils and molecular composition of red wines from Aglianico grapes. Anal. Chim. Acta 2010, 673, 167–172. [Google Scholar] [CrossRef]
- Ahmad, I.; Pichtel, J.; Hayat, S. (Eds.) Plant-Bacteria Interactions: Strategies and Techniques to Promote Plant Growth; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Siddiqui, Z.A.; Mahmood, I. Role of bacteria in the management of plant parasitic nematodes: A review. Bioresour. Technol. 1999, 69, 167–179. [Google Scholar] [CrossRef]
- Gray, E.J.; Smith, D.L. Intracellular and extracellular PGPR: Commonalities and distinctions in the plant–bacterium signaling processes. Soil Biol. Biochem. 2005, 37, 395–412. [Google Scholar] [CrossRef]
- Molina, L.; Constantinescu, F.; Michel, L.; Reimmann, C.; Duffy, B.; Défago, G. Degradation of pathogen quorum-sensing molecules by soil bacteria: A preventive and curative biological control mechanism. FEMS Microbiol. 2003, 45, 71–81. [Google Scholar] [CrossRef]
- Compant, S.; Duffy, B.; Nowak, J.; Clément, C.; Barka, E.A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [PubMed]
- Spadaro, D.; Gullino, M.L. Improving the efficacy of biocontrol agents against soilborne pathogens. Crop Prot. 2005, 24, 601–613. [Google Scholar] [CrossRef]
- Rodríguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Bargabus, R.L.; Zidack, N.K.; Sherwood, J.E.; Jacobsen, B.J. Screening for the identification of potential biological control agents that induce systemic acquired resistance in sugar beet. Biol. Control 2004, 30, 342–350. [Google Scholar] [CrossRef]
- Berkeley, R.; Heyndrickx, M.; Logan, N.; De Vos, P. (Eds.) Applications and Systematics of Bacillus and Relatives; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Belimov, A.A.; Kunakova, A.M.; Kozhemiakov, A.P.; Stepanok, V.V.; Yudkin, L.Y. Effect of associative bacteria on barley grown in heavy metal contaminated soil. In Proceedings of the International Symposium on Agro-Environmental Issues and Future Strategies: Towards the 21st Century, Faisalakad, Pakistan, May 1998. [Google Scholar]
- Antoun, H.; Prévost, D. Ecology of plant growth promoting rhizobacteria. In PGPR: Biocontrol and Biofertilization; Siddiqui, Z.A., Ed.; Springer: Berlin, Germany, 2005; pp. 1–38. [Google Scholar]
- Rosas, S.; Rovera, M.; Andrés, J.A.; Pastor, N.A.; Guiñazú, L.B.; Carlier, E.; Correa, N.S. Characterization of Pseudomonas aurantiaca as biocontrol and PGPR agent. Endophytic properties. In Proceedings of the Prospects and Applications for Plant Associated Microbes, 1st International Conference on Plant–Microbe Interactions: Endophytes and Biocontrol Agents, Finland, Lapland, 18–22 April 2005. [Google Scholar]
- Gamalero, E.; Glick, B.R. Mechanisms used by plant growth-promoting bacteria. In Bacteria in Agrobiology: Plant Nutrient Management; Springer: Berlin/Heidelberg, Germany, 2011; pp. 17–46. [Google Scholar]
- Seidel, C.; Walz, A.; Park, S.; Cohen, J.D.; Ludwig-Muller, J. Indole-3-acetic acid protein conjugates: Novel players in auxin homeostasis. Plant Biol. 2006, 8, 340–345. [Google Scholar] [CrossRef]
- Apine, O.A.; Jadhav, J.P. Optimization of medium for indole-3-acetic acid production using Pantoea agglomerans strain PVM. J. Appl. Microbiol. 2011, 110, 1235–1244. [Google Scholar] [CrossRef]
- Egamberdieva, D. Alleviation of salt stress by plant growth regulators and IAA producing bacteria in wheat. Acta Physiol. Plant 2009, 31, 861–864. [Google Scholar] [CrossRef]
- Tara, N.; Saharan, B.S. Plant growth promoting traits shown by bacteria Brevibacterium frigrotolerans SMA23 isolated from Aloe vera rhizosphere. Agric. Sci. Dig.-A Res. J. 2017, 37, 226–231. [Google Scholar]
- Wahyudi, A.T.; Priyanto, J.A.; Afrista, R.; Kurniati, D.; Astuti, R.I.; Akhdiya, A. Plant growth promoting activity of actinomycetes isolated from soybean rhizosphere. Online J. Biol. Sci. 2019, 19, 1–8. [Google Scholar] [CrossRef]
- Susilowati, D.N.; Sudiana, I.M.; Mubarik, N.R.; Suwanto, A. Species and functional diversity of rhizobacteria of rice plant in the coastal soils of Indonesia. Indones. J. Agric. Sci. 2015, 16, 39–50. [Google Scholar] [CrossRef]
- Zahid, M.; Abbasi, M.K.; Hameed, S.; Rahim, N. Isolation and identification of indigenous plant growth promoting rhizobacteria from Himalayan region of Kashmir and their effect on improving growth and nutrient contents of maize (Zea mays L.). Front. Microbiol. 2015, 6, 207. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.P.; Yadav, J.; Tiwari, K.N. Enhancement of nodulation and yield of chickpea by co-inoculation of indigenous mesorhizobium spp. and Plant Growth–Promoting Rhizobacteria in Eastern Uttar Pradesh. Commun. Soil Sci. Plant Anal. 2012, 43, 605–621. [Google Scholar] [CrossRef]
- Verma, J.P.; Yadav, J.; Tiwari, K.N.; Kumar, A. Effect of indigenous Mesorhizobium spp. and plant growth promoting rhizobacteria on yields and nutrients uptake of chickpea (Cicer arietinum L.) under sustainable agriculture. Ecol. Eng. 2013, 51, 282–286. [Google Scholar] [CrossRef]
- Duca, D.; Lorv, J.; Patten, C.L.; Rose, D.; Glick, B.R. Indole-3-acetic acid in plant–microbe interactions. Anton. Leeuw. 2014, 106, 85–125. [Google Scholar] [CrossRef]
- Jha, B.; Gontia, I.; Hartmann, A. The roots of the halophyte Salicornia brachiata are a source of new halotolerant diazotrophic bacteria with plant growth-promoting potential. Plant Soil 2012, 356, 265–277. [Google Scholar] [CrossRef]
- Howell, C.R.; Beier, R.C.; Stipanovic, R.D. Production of ammonia by Enterobacter cloacae and its possible role in the biological control of Pythium preemergence damping-off by the bacterium. Phytopathology 1988, 78, 1075–1078. [Google Scholar] [CrossRef]
- Joseph, B.; Patra, R.R.; Lawerence, R. Characterization of plant growth promoting rhizobacteria with chickpea (Cicer arietinum L.). Int. J. Plant Prod. 2007, 1, 141–151. [Google Scholar]
- Flaishman, M.A.; Eyal, Z.A.; Zilberstein, A.; Voisard, C.; Hass, D. Suppression of Septoria tritici blotch and leaf rust of wheat by recombinant cyanide producing strains of Pseudomonas putida. Mol. Plant Microbe Int. 1996, 9, 642–645. [Google Scholar] [CrossRef]
- Marques, A.P.; Pires, C.; Moreira, H.; Rangel, A.O.; Castro, P.M. Assessment of the plant growth promotion abilities of six bacterial isolates using Zea mays as indicator plant. Soil Biol. Biochem. 2010, 42, 1229–1235. [Google Scholar] [CrossRef]
- Kanchiswamy, C.N.; Malnoy, M.; Maffei, M.E. Chemical diversity of microbial volatiles and their potential for plant growth and productivity. Front. Plant Sci. 2015, 6, 151. [Google Scholar] [CrossRef] [PubMed]
- Prasad, M.; Srinivasan, R.; Chaudhary, M.; Choudhary, M.; Jat, L.K. Plant growth promoting rhizobacteria (PGPR) for sustainable agriculture: Perspectives and challenges. In PGPR Amelioration in Sustainable Agriculture; Singh, A.K., Kumar, A., Singh, P.K., Eds.; Elsevier: Amsterdam, The Netherland, 2019; pp. 129–157. [Google Scholar]
- Ammari, T.; Mengel, K. Total soluble Fe in soil solutions of chemically different soils. Geoderma 2006, 136, 876–885. [Google Scholar] [CrossRef]
- Whipps, J.M. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 2001, 52, 487–511. [Google Scholar] [CrossRef]
- Li, M.; Ahammed, G.J.; Li, C.; Bao, X.; Yu, J.; Huang, C.; Yin, H.; Zhou, J. Brassinosteroid ameliorates zinc oxide nanoparticles-induced oxidative stress by improving antioxidant potential and redox homeostasis in tomato seedling. Front. Plant Sci. 2016, 7, 615. [Google Scholar] [CrossRef]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef]
- Rasool, A.; Mir, M.I.; Zulfajri, M.; Hanafiah, M.M.; Unnisa, S.A.; Mahboob, M. Plant growth promoting and antifungal asset of indigenous rhizobacteria secluded from saffron (Crocus sativus L.) rhizosphere. Microb. Pathog. 2021, 150, 104734. [Google Scholar] [CrossRef] [PubMed]
- Wani, P.A.; Khan, M.S. Bacillus species enhance growth parameters of chickpea (Cicer arietinum L.) in chromium stressed soils. Food Chem. Toxicol. 2010, 48, 3262–3267. [Google Scholar] [CrossRef] [PubMed]
- Koo, S.Y.; Cho, K.S. Isolation and characterization of a plant growth-promoting rhizobacterium, Serratia sp. SY5. J. Microbiol. Biotechnol. 2009, 19, 1431–1438. [Google Scholar] [PubMed]
- Sriprang, R.; Hayashi, M.; Ono, H.; Takagi, M.; Hirata, K.; Murooka, Y. Enhanced accumulation of Cd21 by a Mesorhizobium sp. transformed with a gene from Arabidopsis thaliana coding for phytochelatin synthase. Appl. Environ. Microbiol. 2003, 69, 1791–1796. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, M.; Freitas, H. Influence of metal resistant-plant growth-promoting bacteria on the growth of Ricinus communis in soil contaminated with heavy metals. Chemosphere 2008, 71, 834–842. [Google Scholar] [CrossRef]
- Ma, Y.; Rajkumar, M.; Freitas, H. Improvement of plant growth and nickel uptake by nickel resistant-plant-growth promoting bacteria. J. Hazard. Mater. 2009, 166, 1154–1161. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.V.; Glick, B.R. Increased plant fitness by rhizobacteria. In Molecular Ecotoxicology of Plants; Sabderman, H., Jr., Ed.; Springer: Berlin/Heidelberg, Germany, 2004; Volume 170, pp. 177–205. [Google Scholar]
- Glick, B.R.; Cheng, Z.; Czarny, J.; Duan, J. Promotion of plant growth by ACC deaminase-producing soil bacteria. In New Perspectives and Approaches in Plant Growth-Promoting Rhizobacteria Research; Bakker, P.A., Raaijmakers, J.M., Bloemberg, G., Höfte, M., Lemanceau, P., Cooke, B.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 329–339. [Google Scholar]
- Bayliss, C.; Bent, E.; Culham, D.E.; MacLellan, S.; Clarke, A.J.; Brown, G.L.; Wood, J.M. Bacterial genetic loci implicated in the Pseudomonas putida GR12-2R3-canola mutualism: Identification of an exudate-inducible sugar transporter. Can. J. Microbiol. 1997, 43, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Penrose, D.M.; Glick, B.R. Levels of ACC and related compounds in exudate and extracts of canola seeds treated with ACC deaminase-containing plant growth-promoting bacteria. Can. J. Microbiol. 2001, 47, 368–372. [Google Scholar] [CrossRef]
- Tiryaki, D.; Aydın, İ.; Atıcı, Ö. Psychrotolerant bacteria isolated from the leaf apoplast of cold-adapted wild plants improve the cold resistance of bean (Phaseolus vulgaris L.) under low temperature. Cryobiology 2019, 86, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Palacio-Rodríguez, R.; Coria-Arellano, J.L.; López-Bucio, J.; Sánchez-Salas, J.; Muro-Pérez, G.; Castañeda-Gaytán, G.; Sáenz-Mata, J. Halophilic rhizobacteria from Distichlis spicata promote growth and improve salt tolerance in heterologous plant hosts. Symbiosis 2017, 73, 179–189. [Google Scholar] [CrossRef]
- Misra, S.; Dixit, V.K.; Khan, M.H.; Mishra, S.K.; Dviwedi, G.; Yadav, S.; Lehri, A.; Chauhan, P.S. Exploitation of agro-climatic environment for selection of 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase producing salt tolerant indigenous plant growth promoting rhizobacteria. Microbiol. Res. 2017, 205, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Anand, K.; Kumari, B.; Mallick, M. Phosphate solubilizing microbes: An effective and alternative approach as biofertilizers. Int. J. Pharm. Pharm. 2016, 8, 37–40. [Google Scholar]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. Springer Plus 2013, 2, 587. [Google Scholar] [CrossRef]
- Chen, Y.P.; Rekha, P.D.; Arun, A.B.; Shen, F.T.; Lai, W.A.; Young, C.C. Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl. Soil Ecol. 2006, 34, 33–41. [Google Scholar] [CrossRef]
- Illmer, P.; Schinner, F. Solubilization of inorganic calcium phosphates-solubilization mechanisms. Soil Biol. Biochem. 1995, 27, 265–270. [Google Scholar] [CrossRef]
- Sperber, J.I. The incidence of apatite-solubilizing organisms in the rhizosphere and soil. Aust. J. Aric. Res. 1958, 9, 778–781. [Google Scholar] [CrossRef]
- Duff, R.B.; Webley, D.M. 2-Ketogluconic acid as a natural chelator produced by soil bacteria. Chem. Ind. 1959, 1959, 1376–1377. [Google Scholar]
- Di Simine, C.D.; Sayer, J.A.; Gadd, G.M. Solubilization of zinc phosphate by a strain of Pseudomonas fluorescens isolated from a forest soil. Biol. Fert. Soils 1998, 28, 87–94. [Google Scholar] [CrossRef]
- Gulati, A.; Rahi, P.; Vyas, P. Characterization of phosphate-solubilizing fluorescent Pseudomonas from the rhizosphere of seabuckthorn growing in the cold deserts of Himalayas. Curr. Microbiol. 2008, 56, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Park, K.H.; Lee, C.Y.; Son, H.J. Mechanism of insoluble phosphate solubilization by Pseudomonas fluorescens RAF15 isolated from ginseng rhizosphere and its plant growth-promoting activities. Lett. Appl. Microbiol. 2009, 49, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Malboobi, M.A.; Behbahani, M.; Madani, H.; Owlia, P.; Deljou, A.; Yakhchali, B.; Moradi, M.; Hassanabadi, H. Performance evaluation of potent phosphate solubilizing bacteria in potato rhizosphere. World J. Microbiol. Biotechnol. 2009, 25, 1479–1484. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, L.; Hao, R.; Bai, X.; Wang, Y.; Yu, X. Drought-tolerant plant growth-promoting rhizobacteria isolated from jujube (Ziziphus jujuba) and their potential to enhance drought tolerance. Plant Soil 2020, 452, 423–440. [Google Scholar] [CrossRef]
- Molla, M.A.Z.; Chowdhury, A.A.; Islam, A.; Hoque, S. Microbial mineralization of organic phosphate in soil. Plant Soil 1984, 78, 393–399. [Google Scholar] [CrossRef]
- Mba, C.C. Rock phosphate solubilizing and cellulolytic actinomycetes isolates of earthworm casts. Environ. Manag. 1994, 18, 257–261. [Google Scholar] [CrossRef]
- Hamdali, H.; Hafidi, M.; Virolle, M.J.; Ouhdouch, Y. Rock phosphate-solubilizing Actinomycetes: Screening for plant growth-promoting activities. World J. Microbiol. Biotechnol. 2008, 24, 2565–2575. [Google Scholar] [CrossRef]
- Chang, C.H.; Yang, S.S. Thermo-tolerant phosphate-solubilizing microbes for multi-functional biofertilizer preparation. Bioresour. Technol. 2009, 100, 1648–1658. [Google Scholar] [CrossRef] [PubMed]
- de Freitas, J.R.; Banerjee, M.R.; Germida, J.J. Phosphate-solubilizing rhizobacteria enhance the growth and yield but not phosphorus uptake of canola (Brassica napus L.). Biol. Fertil. Soils 1997, 24, 358–364. [Google Scholar] [CrossRef]
- Toro, M.; Azcon, R.; Barea, J. Improvement of arbuscular mycorrhiza development by inoculation of soil with phosphate-solubilizing rhizobacteria to improve rock phosphate bioavailability ((sup32) P) and nutrient cycling. Appl. Environ. Microbiol. 1997, 63, 4408–4412. [Google Scholar] [CrossRef] [PubMed]
- Rojas, A.; Holguin, G.; Glick, B.R.; Bashan, Y. Synergism between Phyllobacterium sp. (N2-fixer) and Bacillus licheniformis (P-solubilizer), both from a semiarid mangrove rhizosphere. FEMS Microbiol. Ecol. 2001, 35, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Sahin, F.; Cakmakci, R.; Kantar, F. Sugar beet and barley yields in relation to inoculation with N2-fixing and phosphate solubilizing bacteria. Plant Soil 2004, 265, 123–129. [Google Scholar] [CrossRef]
- Castellano-Hinojosa, A.; Pérez-Tapia, V.; Bedmar, E.J.; Santillana, N. Purple corn-associated rhizobacteria with potential for plant growth promotion. J. Appl. Microbiol. 2018, 124, 1254–1264. [Google Scholar] [CrossRef]
- Fan, Z.Y.; Miao, C.P.; Qiao, X.G.; Zheng, Y.K. Diversity, distribution, and antagonistic activities of rhizobacteria of Panax notoginseng. J. Ginseng Res. 2016, 40, 97–104. [Google Scholar] [CrossRef]
- Bender, R.R.; Haegele, J.W.; Ruffo, M.L.; Below, F.E. Nutrient uptake, partitioning, and remobilization in modern, transgenic insect-protected maize hybrids. Agron. J. 2013, 105, 161–170. [Google Scholar] [CrossRef]
- Kifle, M.H.; Laing, M.D. Isolation and screening of bacteria for their diazotrophic potential and their influence on growth promotion of maize seedlings in greenhouses. Front. Plant Sci. 2016, 6, 1225. [Google Scholar] [CrossRef]

| Strains | Species | IAA Production (mg/L) | NH3 Production | HCN Production | ACC Deaminase Activity (nmol α-Ketobutyrate/g h) | Siderophore Production | Phospate Solubilization |
|---|---|---|---|---|---|---|---|
| SI 257 | Br. frigoritolerans | 2.50 ± 0.3 ghijk | − | − | 0 w | − | − |
| SI 264 | Br. frigoritolerans | 2.50 ± 0.4 ghijk | + | − | 0 w | − | − |
| SI 325 | Br. frigoritolerans | 0 k | − | − | 37.17 ± 2 hijk | + | − |
| SI 312 | Br. frigoritolerans | 1.76 ± 0.2 ijk | + | − | 22.72 ± 5 mnopqr | + | − |
| SI 385 | Br. frigoritolerans | 3.46 ± 0.2 defghij | − | − | 9.05 ± 3 stuvw | − | − |
| SI 333 | Br. frigoritolerans | 3.75 ± 0.61 cdefghij | − | − | 40.60 ± 6 fghi | − | − |
| SI 349 | Br. frigoritolerans | 6.72 ± 0.65 ab | − | − | 28.91 ± 6 jklm | − | − |
| SI 400 | Br. frigoritolerans | 1.76 ± 0.1 ijk | + | − | 0 w | + | − |
| SI 433 | Br. frigoritolerans | 7.37 ± 0.5 a | − | − | 80.58 ± 4 a | − | − |
| SI 387 | Br. frigoritolerans | 6.72 ± 0.6 ab | − | − | 16.18 ± 2 opqrst | + | − |
| SI 293 | R. erythropolis | 2.14 ± 0.2 hijk | − | − | 72.56 ± 4 ab | − | − |
| SI 250 | R. equi | 3.15 ± 0.15 efghij | − | − | 17.52 ± 7 nopqrs | + | − |
| SI 271 | N. globerula | 4.81 ± 0.2 abcdefgh | − | − | 0 w | − | − |
| SI 279 | Str. mauvecolor | 3.15 ± 0.32 efghij | − | − | 4.20 ± 1 uvw | − | − |
| SI 332 | Str. Silaceus | 5.33 ± 0.34 abcdefg | − | − | 0 w | + | 3.01 |
| SI 362 | M. hydrocarboxydans | 5.57 ± 0.52 abcdef | − | − | 34.85 ± 5 ijkl | − | − |
| SI 371 | M. oxydans | 4.83 ± 0.38 abcdefgh | − | − | 23.98 ± 5 lmnopq | − | − |
| SI 295 | A. nitrophenolicus | 5.08 ± 0 abcdeg | − | − | 55.96 ± 6 cde | − | − |
| SI 429 | P. aurescens | 3.46 ± 0.14 defghij | + | − | 0 w | − | − |
| SI 236 | I. cucumis | 5.81 ± 0.31 abcde | − | − | 9.05 ± 3 stuvw | − | − |
| SI 254 | Pb. simplex | 6.50 ± 0.37 abc | − | − | 20.15 ± 3 mnopqrs | − | − |
| SI 397 | Pb. simplex | 4.03 ± 0.07 bcdefghij | − | − | 7.51 ± 2 tuvw | + | − |
| SI 259 | Pb. simplex | 1.33 ± 0.3 jk | − | − | 49.49 ± 5 efg | + | − |
| SI 306 | B. tequilensis | 3.15 ± 0.22 efghij | − | − | 0 w | − | − |
| SI 296 | B. tequilensis | 3.46 ± 0.35 defghij | − | − | 25.23 ± 4 lmnop | + | − |
| SI 319 | B. tequilensis | 6.27 ± 0.3 abcd | − | − | 51.66 ± 5 def | + | − |
| SI 354 | B. tequilensis | 6.72 ± 0.1 ab | − | − | 0 w | − | − |
| SI 305 | B. megaterium | 5.57 ± 0.3 abcdef | − | − | 27.69 ± 4 jklmn | + | − |
| SI 404 | B. megaterium | 6.94 ± 0.88 ab | − | − | 7.51 ± 0.7 tuvw | + | − |
| SI 408 | B. megaterium | 5.08 ± 0.11 abcdefg | + | − | 38.32 ± 6 ghij | − | − |
| SI 470 | B. megaterium | 4.83 ± 0.04 abcdefgh | − | − | 14.81 ± 2 pqrstu | − | − |
| SI 266 | B. megaterium | 6.04 ± 0.1 abcde | − | − | 0 w | − | − |
| SI 339 | B. halotolerans | 0 k | + | − | 34.85 ± 3 ijkl | − | − |
| SI 419 | B. halotolerans | 4.83 ± 0 abcdefgh | − | − | 27.69 ± 4 jklmn | + | − |
| SI 297 | B. mohavensis | 1.33 ± 0.13 jk | − | − | 7.51 ± 2 tuvw | − | − |
| SI 311 | B. cabrialensis | 4.83 ± 0.4 abcdefgh | − | − | 0 w | − | − |
| SI 428 | B. cabrialesii | 6.94 ± 0.04 ab | + | − | 0 w | − | − |
| SI 356 | T. saccharophilus | 6.27 ± 0.21 abcd | − | − | 64.36 ± 6 bc | − | − |
| SI 243 | E. adherens | 2.50 ± 0.3 ghijk | + | − | 0 w | − | − |
| SI 240 | Sn. meliloti | 5.57 ± 0.3 abcdef | − | − | 0 w | + | 2.96 |
| SI 235 | Sn. meliloti | 6.50 ± 0.3 abc | − | − | 9.05 ± 1 stuvw | − | − |
| SI 420 | S. quinivorans | 0 k | + | − | 17.52 ± 3 nopqrs | + | − |
| SI 237 | C. respiraculi | 0 k | − | − | 18.85 ± 3 mnopqrs | − | − |
| SI 435 | V. paradoxus | 5.33 ± 0.4 abcdefg | − | − | 0 w | − | 3.1 |
| SI 439 | V. paradoxus | 5.33 ± 0.42 abcdefg | − | − | 0 w | − | − |
| SI 321 | St. indicatrix | 6.50 ± 0.3 abc | + | − | 26.47 ± 4 klmno | + | − |
| SI 358 | L. soli | 4.57 ± 0.37 abcdefghij | + | − | 37.17 ± 3 hijk | − | − |
| SI 357 | L. soli | 2.14 ± 0.02 hijk | + | − | 0 w | − | − |
| SI 377 | St. rhizophila | 4.57 ± 0.23 abcdefghi | − | − | 18.85 ± 2 mnopqrs | + | − |
| SI 367 | Ps. Plecoglossicida | 6.50 ± 0.28 abc | − | − | 20.15 ± 3 mnopqrs | + | − |
| SI 247 | Ps. brassicacearum | 3.75 ± 0.6 cdefghij | − | + | 16.18 ± 2 opqrst | + | − |
| SI 307 | Ps. frederiksbergensis | 3.15 ± 0.2 efghij | + | + | 48.40 ± 5 efgh | + | 3.80 |
| SI 441 | Ps. reinekei | 6.50 ± 1 abc | + | + | 45.09 ± 6.4 efghi | + | − |
| SI 443 | Ps. atacamensis | 2.14 ± 0.2 ghijk | + | + | 10.54 ± 1 stuvw | + | 3.85 |
| SI 422 | Ps. granadensis | 5.08 ± 1 abcdefg | + | + | 16.18 ± 3 opqrst | + | 3.2 |
| SI 450 | Ps. granadensis | 0 k | + | + | 12.00 ± 1 rstuv | + | − |
| SI 434 | Ps. moorei | 4.83 ± 0.65 abcdefgh | − | − | 14.81 ± 3 pqrstu | + | − |
| SI 270 | Ps. lini | 2.83 ± 0.1 fghijk | + | − | 0 w | + | 4.99 |
| SI 285 | Ps. lini | 2.83 ± 0.22 fghijk | + | + | 0 w | + | − |
| SI 276 | Ps. lini | 2.14 ± 0.12 hijk | − | − | 13.42 ± 2.4 qrstu | + | − |
| SI 284 | Ps. lini | 3.46 ± 0.2 defghij | + | + | 0 w | + | − |
| SI 288 | Ps. lini | 6.27 ± 0.34 abcd | + | − | 16.2 ± 3 opqrst | + | 4 |
| SI 287 | Ps. lini | 3.15 ± 0.1 efghij | − | − | 62.28 ± 4.8 bcd | + | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbaccia, P.; Gaglio, R.; Dazzi, C.; Miceli, C.; Bella, P.; Lo Papa, G.; Settanni, L. Plant Growth-Promoting Activities of Bacteria Isolated from an Anthropogenic Soil Located in Agrigento Province. Microorganisms 2022, 10, 2167. https://doi.org/10.3390/microorganisms10112167
Barbaccia P, Gaglio R, Dazzi C, Miceli C, Bella P, Lo Papa G, Settanni L. Plant Growth-Promoting Activities of Bacteria Isolated from an Anthropogenic Soil Located in Agrigento Province. Microorganisms. 2022; 10(11):2167. https://doi.org/10.3390/microorganisms10112167
Chicago/Turabian StyleBarbaccia, Pietro, Raimondo Gaglio, Carmelo Dazzi, Claudia Miceli, Patrizia Bella, Giuseppe Lo Papa, and Luca Settanni. 2022. "Plant Growth-Promoting Activities of Bacteria Isolated from an Anthropogenic Soil Located in Agrigento Province" Microorganisms 10, no. 11: 2167. https://doi.org/10.3390/microorganisms10112167
APA StyleBarbaccia, P., Gaglio, R., Dazzi, C., Miceli, C., Bella, P., Lo Papa, G., & Settanni, L. (2022). Plant Growth-Promoting Activities of Bacteria Isolated from an Anthropogenic Soil Located in Agrigento Province. Microorganisms, 10(11), 2167. https://doi.org/10.3390/microorganisms10112167









