Diversity and Taxonomy of Soil Bacterial Communities in Urban and Rural Mangrove Forests of the Panama Bay
Abstract
:1. Introduction
2. Materials and Methods
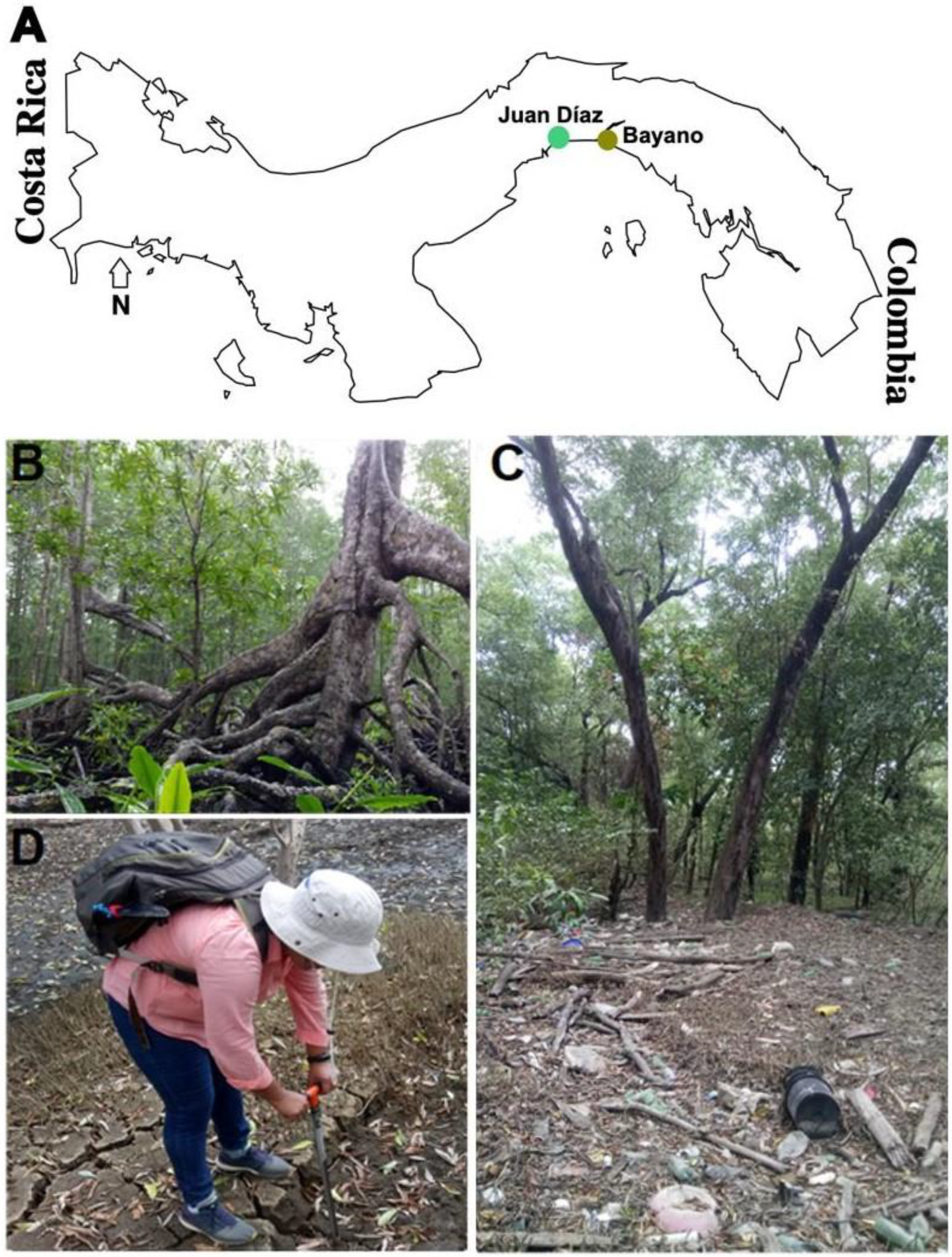
2.1. Sampling Sites
2.2. DNA Extraction and Amplification
2.3. DNA Library Preparation
2.4. Data Analysis
2.5. Bacterial Diversity and Community Composition
3. Results
3.1. Diversity and Taxonomic Composition of Soil Bacterial Communities of Bayano and Juan Díaz Mangrove Sites
3.2. Seasonality Effects on Bacterial Community Composition in Juan Díaz
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barbier, E.B.; Koch, E.W.; Silliman, B.R.; Hacker, S.D.; Wolanski, E.; Primavera, J.; Granek, E.F.; Polasky, S.; Aswani, S.; Cramer, L.A.; et al. Coastal Ecosystem-Based Management with Nonlinear Ecological Functions and Values. Science 2008, 319, 321–323. [Google Scholar] [CrossRef] [PubMed]
- Getzner, M.; Islam, M.S. Ecosystem Services of Mangrove Forests: Results of a Meta-Analysis of Economic Values. Int. J. Environ. Res. Public Health 2020, 17, 5830. [Google Scholar] [CrossRef] [PubMed]
- Giri, C.; Ochieng, E.; Tieszen, L.L.; Zhu, Z.; Singh, A.; Loveland, T.; Masek, J.; Duke, N. Status and distribution of mangrove forests of the world using earth observation satellite data. Glob. Ecol. Biogeogr. 2011, 20, 154–159. [Google Scholar] [CrossRef]
- Howard, J.; Sutton-Grier, A.; Herr, D.; Kleypas, J.; Landis, E.; Mcleod, E.; Pidgeon, E.; Simpson, S. Clarifying the role of coastal and marine systems in climate mitigation. Front. Ecol. Environ. 2017, 15, 42–50. [Google Scholar] [CrossRef]
- Guo, Y.; Gong, H.; Guo, X. Rhizosphere bacterial community of Typha angustifolia L. and water quality in a river wetland sup-plied with reclaimed water. Appl. Microbiol. Biotechnol. 2015, 99, 2883–2893. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.W.; Tsai, S.M. Distinct taxonomic and functional composition of soil microbiomes along the gradient for-est-restinga-mangrove in southeastern Brazil. Antonie van Leeuwenhoek. Int. J. Gen. Mol. Microbiol. 2018, 111, 101–114. [Google Scholar]
- Gomes, N.C.M.; Cleary, D.F.R.; Pinto, F.N.; Egas, C.; Almeida, A.; Cunha, A.; Mendonça-Hagler, L.C.S.; Smalla, K. Taking root: Enduring effect of rhizosphere bacteri-al colonization in mangroves. PLoS ONE 2010, 5, e14065. [Google Scholar] [CrossRef]
- Tellez-Rio, A.; García-Marco, S.; Navas, M.; López-Solanilla, E.; Tenorio, J.L.; Vallejo, A. N2O and CH4 emissions from a fal-low-wheat rotation with low N input in conservation and conventional tillage under a Mediterranean agroecosystem. Sci. Total Environ. 2015, 508, 85–94. [Google Scholar] [CrossRef]
- Thatoi, H.; Behera, B.C.; Mishra, R.R.; Dutta, S.K. Biodiversity and biotechnological potential of microorganisms from mangrove ecosystems: A review. Ann. Microbiol. 2012, 63, 1–19. [Google Scholar] [CrossRef]
- Dourado, M.N.; Ferreira, A.; Araújo, W.L.; Azevedo, J.L.; Lacava, P.T. The Diversity of Endophytic Methylotrophic Bacteria in an Oil-Contaminated and an Oil-Free Mangrove Ecosystem and Their Tolerance to Heavy Metals. Biotechnol. Res. Int. 2012, 2012, 759865. [Google Scholar] [CrossRef] [Green Version]
- Taketani, R.G.; Franco, N.O.; Rosado, A.S.; van Elsas, J.D. Microbial community response to a simulated hydrocarbon spill in man-grove sediments. J. Microbiol. 2010, 48, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Andreote, F.D.; Jiménez, D.J.; Chaves, D.; Dias, A.C.F.; Luvizotto, D.M.; Dini-Andreote, F.; Fasanella, C.C.; Lopez, M.V.; Baena, S.; Taketani, R.G.; et al. The microbiome of Brazilian man-grove sediments as revealed by metagenomics. PLoS ONE 2012, 7, e38600. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statisti-cal, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef] [PubMed]
- Xu, J. Bioactive natural products derived from mangrove-associated microbes. RSC Adv. 2015, 5, 841–892. [Google Scholar] [CrossRef]
- Jing, H.; Cheung, S.; Zhou, Z.; Wu, C.; Nagarajan, S.; Liu, H. Spatial Variations of the Methanogenic Communities in the Sediments of Tropical Mangroves. PLoS ONE 2016, 11, e0161065. [Google Scholar] [CrossRef] [Green Version]
- Duke, N.C.; Meynecke, J.-O.; Dittmann, S.; Ellison, A.M.; Anger, K.; Berger, U.; Cannicci, S.; DieleK, K.; EwelC, C.; Field, D.; et al. A World Without Mangroves? Science 2007, 317, 41–42. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zheng, L.; Zhang, Y.; Liu, H.; Jing, H. Comparative metagenomics study reveals pollution induced changes of microbial genes in mangrove sediments. Sci. Rep. 2019, 9, 5739. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Hong, Y.G.; Cao, H.L.; Gu, J.D. Mangrove trees affect the community structure and distribution of anammox bacteria at an anthropogenic-polluted mangrove in the Pearl River Delta reflected by 16S rRNA and hydrazine oxidoreductase (HZO) en-coding gene analyses. Ecotoxicology 2011, 20, 1780–1790. [Google Scholar] [CrossRef] [Green Version]
- Bulow, E.; Ferdinand, T.J. The Effect of Consumptive Waste on Mangrove Functionality: A Comparative Analysis. Cent. De Incid. Ambient. Panamá 2013, 33. Available online: https://www.mcgill.ca/pfss/files/pfss/the_effect_of_consumptive_waste_on_mangrove_functionality-_a_comparative_analysis.pdf (accessed on 8 September 2022).
- Kaufmann, K.W. Nuestros Humedales, Nuestro Futuro: Plan de Conservación para los Humedales de la Bahía de Panamá; Sociedad Audubon de Panamá: Ciudad de Panamá, Panama, 2012; pp. 1–73. [Google Scholar]
- Suman, D. Panama Bay Wetlands: Case Study of a Threatened Ecosystem. Water Resour. Wetl. 2014, 366–371. Available online: https://www.limnology.ro/water2014/proceedings/49_Suman.pdf (accessed on 12 April 2022).
- Autoridad Nacional del Ambiente. Informe de Monitoreo de la Calidad del Agua en las Cuencas Hidrográficas de Panamá Compendio de Resultados 2009–2012. Autoridad Nacional del Ambiente. 2015, 7, 37–72. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diver-sity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4516–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paton, S. Meteorological and Hydrological Summary for Barro Colorado Island; Smithsonian Tropical Research Institute: Ciudad de Panamá, Panama, 2017; p. 42. [Google Scholar]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of mark-er-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- R Development Core Team. R a Language and Environment for Statistical Computing: Reference Index; R Foundation for Statistical Computing, 2010; Available online: http://softlibre.unizar.es/manuales/aplicaciones/r/fullrefman.pdf (accessed on 8 September 2022).
- Nowinski, B.; Smith, C.B.; Thomas, C.M.; Esson, K.; Marin, R.; Preston, C.M.; Birch, J.M.; Scholin, C.A.; Huntemann, M.; Clum, A.; et al. Microbial metagenomes and metatranscriptomes during a coastal phytoplankton bloom. Sci. Data 2019, 6, 129. [Google Scholar] [CrossRef] [Green Version]
- Suenami, S.; Nobu, M.K.; Miyazaki, R. Community analysis of gut microbiota in hornets, the largest eusocial wasps, Vespa mandarinia and V. simillima. Sci. Rep. 2019, 9, 9830. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Mcglinn, D.; Stevens, M.H.H.; Solymos, P.; Borcard, D.; Chirico, D.; et al. Vegan: Community Ecology Package. Software. Available online: http://CRAN.R-project.org/package=vegan (accessed on 25 March 2022).
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor workflow for microbiome data analysis: From raw reads to community analyses [version 1; referees: 3 approved]. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef]
- Lim, J.-M.; Jeon, C.O.; Lee, S.S.; Park, D.-J.; Xu, L.-H.; Jiang, C.-L.; Kim, C.-J. Reclassification of Salegentibacter catena Ying et al. 2007 as Salinimicrobium catena gen. nov., comb. nov. and description of Salinimicrobium xinjiangense sp. nov., a halophilic bacterium isolated from Xinjiang province in China. Int. J. Syst. Evol. Microbiol. 2008, 58, 438–442. [Google Scholar] [CrossRef]
- Allard, S.M.; Costa, M.T.; Bulseco, A.N.; Helfer, V.; Wilkins, L.G.E.; Hassenrück, C.; Zengler, K.; Zimmer, M.; Erazo, N.; Rodrigues, J.L.M.; et al. Introducing the Mangrove Microbiome Initia-tive: Identifying Microbial Research Priorities and Approaches to Better Understand, Protect, and Rehabilitate Mangrove Ecosystems. mSystems 2020, 5, e00658-20. [Google Scholar] [CrossRef] [PubMed]
- Cabral, L.; Júnior, G.V.L.; de Sousa, S.T.P.; Dias, A.C.F.; Cadete, L.L.; Andreote, F.D.; Hess, M.; de Oliveira, V.M. Anthropogenic impact on mangrove sediments triggers differential responses in the heavy metals and antibiotic resistomes of microbial communities. Environ. Pollut. 2016, 216, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Feller, I.C.; Friess, D.A.; Krauss, K.W.; Lewis, R.R. The state of the world’s mangroves in the 21st century under climate change. Hydrobiologia 2017, 803, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Peixoto, R.; Chaer, G.; Carmo, F.L.; de Araujo, F.V.; Paes, J.E.; Volpon, A.; Santiago, G.A.; Rosado, A. Bacterial communities reflect the spatial variation in pollutant levels in Brazilian mangrove sediment. Antonie Leeuwenhoek 2010, 99, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Saintilan, N.; Khan, N.S.; Ashe, E.; Kelleway, J.J.; Rogers, K.; Woodroffe, C.D.; Horton, B.P. Thresholds of mangrove survival under rapid sea level rise. Science 2020, 368, 1118–1121. [Google Scholar] [CrossRef]
- Waite, D.W.; Vanwonterghem, I.; Rinke, C.; Parks, D.H.; Zhang, Y.; Takai, K.; Sievert, S.M.; Simon, J.; Campbell, B.J.; Hanson, T.E.; et al. Comparative genomic analysis of the class Epsi-lonproteobacteria and proposed reclassification to epsilonbacteraeota (phyl. nov.). Front Microbiol. 2017, 8, 682. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.-J.; Chen, Y.-L.; Sun, Y.-H.; Pan, J.; Cai, M.-W.; Li, M. Diversity, metabolism and cultivation of archaea in mangrove ecosystems. Mar. Life Sci. Technol. 2020, 3, 252–262. [Google Scholar] [CrossRef]
- A Hug, L.; Castelle, C.J.; Wrighton, K.C.; Thomas, B.C.; Sharon, I.; Frischkorn, K.R.; Williams, K.H.; Tringe, S.G.; Banfield, J.F. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 2013, 1, 22. [Google Scholar] [CrossRef] [Green Version]
- Mujakić, I.; Piwosz, K.; Koblížek, M. Phylum Gemmatimonadota and Its Role in the Environment. Microorganisms 2022, 10, 151. [Google Scholar] [CrossRef]
- Kerou, M.; Offre, P.; Valledor, L.; Abby, S.S.; Melcher, M.; Nagler, M.; Weckwerth, W.; Schleper, C. Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc. Natl. Acad. Sci. USA 2016, 113, E7937–E7946. [Google Scholar] [CrossRef] [Green Version]
- Huergo, L.F.; Rissi, D.V.; Elias, A.S.; Gonçalves, M.V.; Gernet, M.V.; Barreto, F.; Dahmer, G.W.; Reis, R.A.; Pedrosa, F.O.; Souza, E.M.; et al. Influence of ancient anthropogenic activities on the mangrove soil microbiome. Sci. Total Environ. 2018, 645, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Yamaguchi, K.; Hanada, S. Sulfurovum denitrificans sp. nov., an obligately chemolithoautotrophic sulfur-oxidizing epsilonproteobacterium isolated from a hydrothermal field. Int. J. Syst. Evol. Microbiol. 2018, 68, 2183–2187. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, V.L.; Rocha, L.L.; Colares, G.B.; Angelim, A.L.; Normando, L.R.; Cantão, M.E.; Agnez-Lima, L.F.; Andreote, F.D.; Melo, V.M. Microbiomes and potential metabolic pathways of pristine and anthropized Brazilian mangroves. Reg. Stud. Mar. Sci. 2015, 2, 56–64. [Google Scholar] [CrossRef]
- Naushad, H.S.; Gupta, R.S. Phylogenomics and Molecular Signatures for Species from the Plant Pathogen-Containing Order Xanthomonadales. PLoS ONE 2013, 8, e55216. [Google Scholar] [CrossRef]
- An, X.; Wang, Z.; Teng, X.; Zhou, R.; Wang, X.; Xu, M.; Lian, B. Corrigendum: “Rhizosphere bacterial diversity and environmental function prediction of wild salt-tolerant plants in coastal silt soil”. Ecol. Indic. 2022, 134, 108503. [Google Scholar] [CrossRef]




| Relative Abundance ± SD | Relative Abundance ± SD | |
|---|---|---|
| Bayano, N = 9 | Juan Díaz, N = 50 | |
| Phylum (Kingdom) | ||
| Acidobacteriota (Bacteria) | 4.84 ± 1.19 | 5.00 ± 1.70 |
| Actinobacteriota (Bacteria) | 4.11 ± 0.66 | 8.640 ± 3.71 |
| Bacteroidota (Bacteria) | 7.89 ± 1.59 | 7.67 ± 4.45 |
| Campilobacterota (Bacteria) | 0.85 ± 0.88 | 2.37 ± 2.61 |
| Chloroflexi (Bacteria) | 8.78 ± 1.68 | 11.45 ± 3.60 |
| Crenarchaeota (Archaea) | 1.99 ± 0.62 | 1.76 ± 1.02 |
| Desulfobacterota (Bacteria) | 13.69 ± 3.97 | 8.34 ± 4.23 |
| Gemmatimonadota (Bacteria) | 3.61 ± 0.62 | 5.69 ± 1.80 |
| Myxococcota (Bacteria) | 2.48 ± 0.46 | 3.97 ± 1.44 |
| Planctomycetota (Bacteria) | 2.74 ± 0.43 | 4.02 ± 1.66 |
| Proteobacteria (Bacteria) | 33.83 ± 4.93 | 29.11 ± 4.82 |
| Class (Phylum) | ||
| Acidimicrobiia (Actinobacteriota) | 1.26 ± 0.26 | 3.07 ± 1.43 |
| Actinobacteria (Actinobacteriota) | 2.29 ± 0.68 | 4.53 ± 2.74 |
| Alphaproteobacteria (Proteobacteria) | 11.65 ± 2.16 | 14.46 ± 3.10 |
| Anaerolineae (Chloroflexi) | 6.56 ± 1.21 | 8.86 ± 3.09 |
| Bacteroidia (Bacteroidota) | 4.99 ± 1.68 | 5.20 ± 3.87 |
| BD2-11_terrestrial_group (Gemmatimonadota) | 1.68 ± 0.31 | 2.61 ± 1.08 |
| Campylobacteria (Campilobacterota) | 0.85 ± 0.88 | 2.37 ± 2.61 |
| Desulfobacteria (Desulfobacterota) | 6.19 ± 2.34 | 2.46 ± 1.64 |
| Desulfobulbia (Desulfobacterota) | 4.24 ± 1.60 | 3.72 ± 2.15 |
| Gammaproteobacteria (Proteobacteria) | 21.46 ± 3.42 | 14.58 ± 4.35 |
| Nitrososphaeria (Crenarchaeota) | 1.22 ± 0.70 | 1.36 ± 1.02 |
| Planctomycetes (Planctomycetota) | 0.95 ± 0.34 | 2.01 ± 1.25 |
| Polyangia (Myxococcota) | 1.87 ± 0.37 | 3.05 ± 1.41 |
| Rhodothermia (Bacteroidota) | 0.79 ± 0.24 | 1.62 ± 1.25 |
| Vicinamibacteria (Acidobacteriota) | 0.89 ± 0.38 | 1.61 ± 0.94 |
| Genus (Class) | ||
| BD2-11_terrestrial_group (BD2-11_terrestrial_group) | 1.68 ± 0.31 | 2.61 ± 1.08 |
| Desulfatiglans (Desulfobacteria) | 1.55 ± 0.54 | 0.30 ± 0.26 |
| EPR3968-O8a-Bc78 (Gammaproteobacteria) | 1.87 ± 0.71 | 2.38 ± 1.08 |
| Ignavibacterium (Ignavibacteria) | 1.11 ± 0.60 | 0.43 ± 0.44 |
| MBMPE27 (Gammaproteobacteria) | 1.05 ± 0.99 | 0.37 ± 0.40 |
| MBNT15 (MBNT15) | 2.14 ± 0.68 | 1.88 ± 1.12 |
| Methyloceanibacter (Alphaproteobacteria) | 0.98 ± 0.26 | 1.23 ± 0.89 |
| NB1-j (NB1-j) | 1.05 ± 0.45 | 1.51 ± 0.76 |
| Pseudolabrys (Alphaproteobacteria) | 1.15 ± 0.42 | 0.92 ± 0.50 |
| Salinimicrobium | 0.30 ± 0.18 | 0.88 ± 1.86 |
| SBR1031 (Anaerolineae) | 1.30 ± 0.31 | 1.71 ± 0.77 |
| SEEP-SRB1 (Desulfobacteria) | 1.01 ± 0.32 | 0.10 ± 0.21 |
| Sva0081_sediment_group (Desulfobacteria) | 1.76 ± 0.51 | 0.66 ± 0.68 |
| S0134_terrestrial_group (S0134_terrestrial_group) | 0.71 ± 0.28 | 1.54 ± 0.83 |
| Sulfurovum (Campylobacteria) | 0.56 ± 0.36 | 2.14 ± 2.40 |
| Woeseia (Gammaproteobacteria) | 1.66 ± 0.94 | 1.04 ± 0.57 |
| Relative Abundance ± SD | Relative Abundance ± SD | Relative Abundance ± SD | |
|---|---|---|---|
| Dec 17, N = 13 | Feb 18, N = 14 | May 18, N = 14 | |
| Phylum (Kingdom) | |||
| Acidobacteriota (Bacteria) | 4.64 ± 1.28 | 5.87 ± 2.27 | 4.91 ± 1.67 |
| Actinobacteriota (Bacteria) | 9.49 ± 3.97 | 7.52 ± 4.45 | 7.38 ± 2.36 |
| Bacteroidota (Bacteria) | 6.08 ± 2.38 | 4.95 ± 3.71 | 12.40 ± 4.55 |
| Campilobacterota (Bacteria) | 3.88 ± 2.93 | 2.20 ± 2.51 | 1.87 ± 2.69 |
| Chloroflexi (Bacteria) | 9.96 ± 2.73 | 11.58 ± 4.58 | 10.88 ± 3.26 |
| Crenarchaeota (Archaea) | 2.11 ± 0.91 | 1.99 ± 1.19 | 0.81 ± 0.32 |
| Desulfobacterota (Bacteria) | 11.14 ± 3.11 | 8.52 ± 5.19 | 5.05 ± 3.09 |
| Gemmatimonadota (Bacteria) | 4.70 ± 2.10 | 5.40 ± 1.56 | 6.30 ± 1.65 |
| Myxococcota (Bacteria) | 3.43 ± 0.80 | 3.88 ± 0.95 | 5.39 ± 1.47 |
| Planctomycetota (Bacteria) | 3.70 ± 0.80 | 5.42 ± 2.33 | 3.51 ± 0.87 |
| Proteobacteria (Bacteria) | 27.85 ± 4.64 | 30.36 ± 4.68 | 30.70 ± 4.95 |
| Class (Phylum) | |||
| Acidimicrobiia (Actinobacteriota) | 3.56 ± 1.50 | 3.51 ± 1.96 | 2.55 ± 0.88 |
| Actinobacteria (Actinobacteriota) | 4.64 ± 2.73 | 2.81 ± 2.26 | 4.33 ± 2.10 |
| Alphaproteobacteria (Proteobacteria) | 14.16 ± 2.73 | 14.85 ± 3.82 | 14.69 ± 3.59 |
| Anaerolineae (Chloroflexi) | 7.50 ± 2.31 | 8.55 ± 3.83 | 8.75 ± 2.77 |
| Bacteroidia (Bacteroidota) | 4.23 ± 2.23 | 3.17 ± 3.15 | 8.86 ± 4.63 |
| BD2-11_terrestrial_group (Gemmatimonadota) | 2.13 ± 1.01 | 2.66 ± 0.94 | 2.97 ± 1.33 |
| Campylobacteria (Campilobacterota) | 3.88 ± 2.93 | 2.20 ± 2.51 | 1.87 ± 2.69 |
| Desulfobacteria (Desulfobacterota) | 3.51 ± 1.62 | 2.46 ± 1.99 | 1.47 ± 1.11 |
| Desulfobulbia (Desulfobacterota) | 4.89 ± 1.40 | 4.09 ± 2.92 | 1.93 ± 1.31 |
| Gammaproteobacteria (Proteobacteria) | 13.68 ± 3.26 | 15.36 ± 5.54 | 15.97 ± 4.01 |
| Nitrososphaeria (Crenarchaeota) | 1.57 ± 0.97 | 1.58 ± 1.34 | 0.54 ± 0.22 |
| Planctomycetes (Planctomycetota) | 1.94 ± 0.70 | 2.88 ± 2.04 | 1.57 ± 0.37 |
| Polyangia (Myxococcota) | 2.53 ± 0.69 | 2.97 ± 1.07 | 4.39 ± 1.47 |
| Rhodothermia (Bacteroidota) | 1.05 ± 0.29 | 1.37 ± 0.81 | 2.48 ± 1.98 |
| Vicinamibacteria (Acidobacteriota) | 1.61 ± 0.51 | 2.16 ± 1.38 | 1.23 ± 0.85 |
| Genus (Class) | |||
| BD2-11_terrestrial_group (BD2-11_terrestrial_group) | 2.12 ± 1.00 | 2.66 ± 0.93 | 2.97 ± 1.33 |
| BIrii41 (Polyangia) | 0.58 ± 0.42 | 0.64 ± 0.44 | 1.32 ± 0.57 |
| EPR3968-O8a-Bc78 (Gammaproteobacteria) | 2.11 ± 0.61 | 2.57 ± 1.05 | 2.69 ± 1.09 |
| MBNT15 (MBNT15) | 2.09 ± 1.06 | 1.75 ± 0.90 | 1.38 ± 1.19 |
| Methyloceanibacter (Alphaproteobacteria) | 1.41 ± 0.86 | 1.37 ± 0.92 | 0.65 ± 0.15 |
| NB1-j (NB1-j) | 1.37 ± 0.55 | 2.06 ± 0.65 | 1.48 ± 0.79 |
| Salinimicrobium | 0.18 ± 0.22 | 0.06 ± 0.09 | 2.57 ± 2.92 |
| SBR1031 (Anaerolineae) | 1.54 ± 0.69 | 1.55 ± 0.99 | 1.77 ± 0.73 |
| S0134_terrestrial_group (S0134_terrestrial_group) | 1.21 ± 0.69 | 1.37 ± 0.87 | 1.55 ± 0.79 |
| Sulfurovum (Campylobacteria) | 3.42 ± 2.55 | 2.05 ± 2.37 | 1.67 ± 2.65 |
| Subgroup_10 (Thermoanaerobaculia) | 0.77 ± 0.32 | 1.30 ± 0.51 | 0.93 ± 0.50 |
| Woeseia (Gammaproteobacteria) | 1.15 ± 0.48 | 1.39 ± 0.64 | 0.83 ± 0.40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quintero, I.J.; Castillo, A.M.; Mejía, L.C. Diversity and Taxonomy of Soil Bacterial Communities in Urban and Rural Mangrove Forests of the Panama Bay. Microorganisms 2022, 10, 2191. https://doi.org/10.3390/microorganisms10112191
Quintero IJ, Castillo AM, Mejía LC. Diversity and Taxonomy of Soil Bacterial Communities in Urban and Rural Mangrove Forests of the Panama Bay. Microorganisms. 2022; 10(11):2191. https://doi.org/10.3390/microorganisms10112191
Chicago/Turabian StyleQuintero, Indira J., Anakena M. Castillo, and Luis C. Mejía. 2022. "Diversity and Taxonomy of Soil Bacterial Communities in Urban and Rural Mangrove Forests of the Panama Bay" Microorganisms 10, no. 11: 2191. https://doi.org/10.3390/microorganisms10112191







