Comparative Evaluation of Luminex xTAG® Gastrointestinal Pathogen Panel and Direct-From-Stool Real-Time PCR for Detection of C. difficile Toxin tcdB in Stool Samples from a Pediatric Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Nucleic Acid Extraction
2.3. Molecular Testing
2.4. Assessment of In-House tcdB RT-PCR
2.5. Preparation for Stool Specimens for Culture Enrichment
2.6. Statistical Analysis
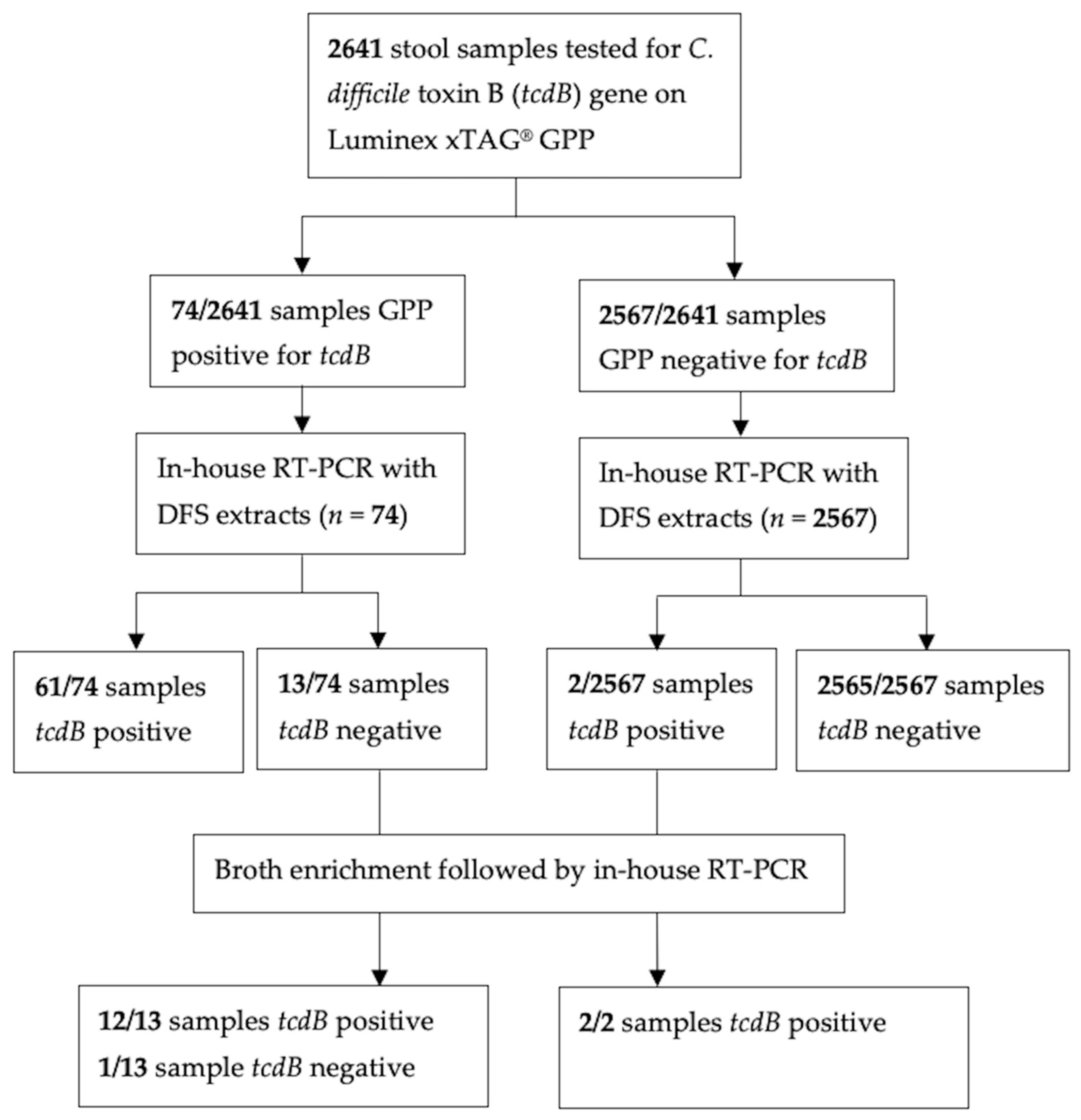
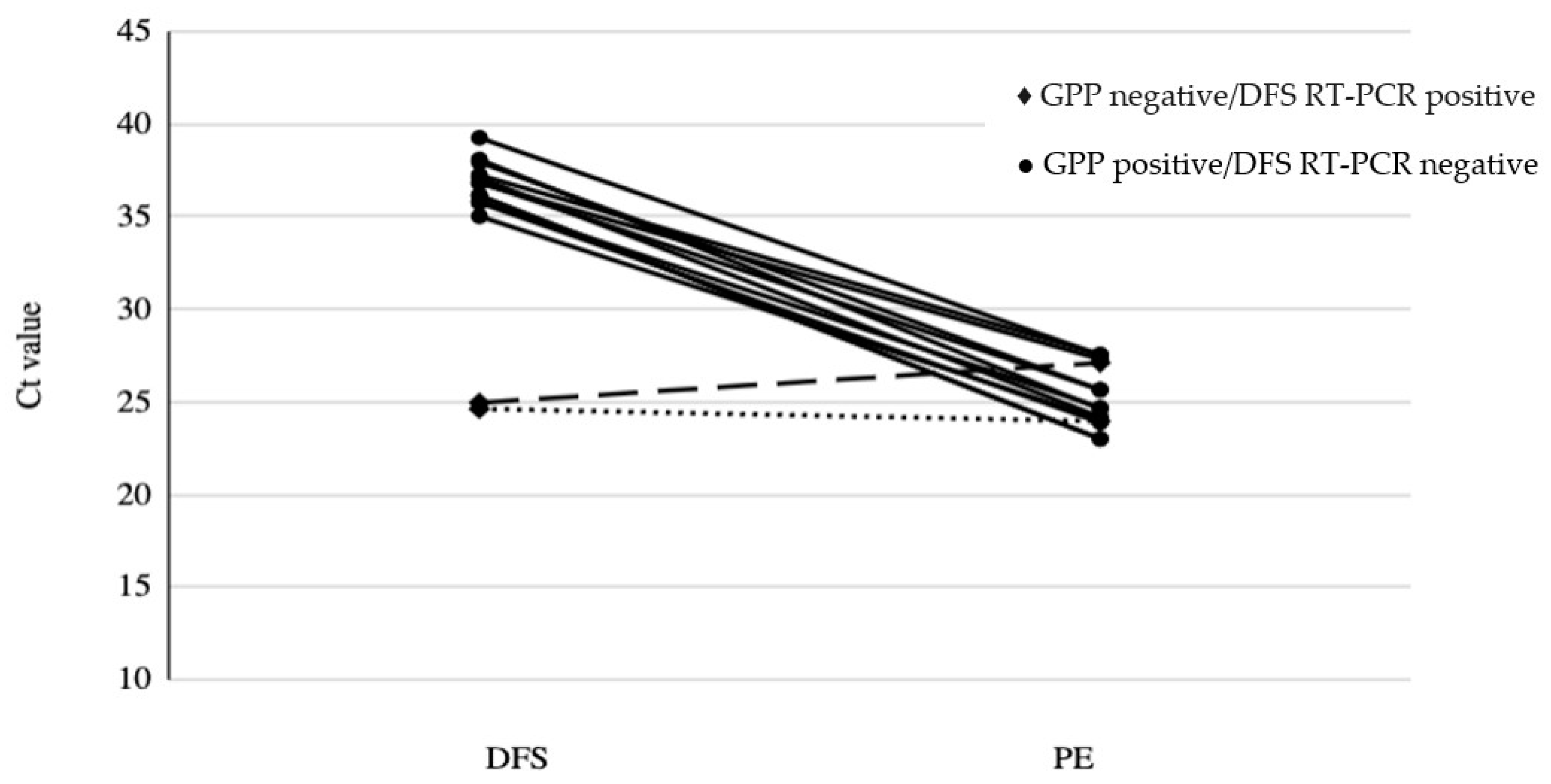
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smits, W.K.; Lyras, D.; Lacy, D.B.; Wilcox, M.H.; Kuijper, E.J. Clostridium difficile infection. Nat. Rev. Dis. Primers 2016, 2, 16020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Sorg, J.A.; Sun, X. Clostridioides difficile Biology: Sporulation, Germination, and Corresponding Therapies for C. difficile Infection. Front. Cell. Infect. Microbiol. 2018, 8, 29. [Google Scholar] [CrossRef] [Green Version]
- Burnham, C.-A.D.; Carroll, K.C. Diagnosis of Clostridium difficile infection: An ongoing conundrum for clinicians and for clinical laboratories. Clin. Microbiol. Rev. 2013, 26, 604–630. [Google Scholar] [CrossRef] [Green Version]
- Yalamanchili, H.; Dandachi, D.; Okhuysen, P.C. Use and Interpretation of Enteropathogen Multiplex Nucleic Acid Amplification Tests in Patients With Suspected Infectious Diarrhea. Gastroenterol. Hepatol. 2018, 14, 646–652. [Google Scholar]
- Bélanger, S.D.; Boissinot, M.; Clairoux, N.; Picard, F.J.; Bergeron, M.G. Rapid detection of Clostridium difficile in feces by real-time PCR. J. Clin. Microbiol. 2003, 41, 730–734. [Google Scholar] [CrossRef] [Green Version]
- Lotfian, S.; Douraghi, M.; Aliramezani, A.; Ghourchian, S.; Sarrafnejad, A.; Zeraati, H. Detection of Clostridium difficile in Fecal Specimens: A Comparative Evaluation of Nucleic Acid Amplification Test and Toxigenic Culture. Clin. Lab. 2016, 62, 1887–1892. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.W.; Tarr, P.I. Multiplex Nucleic Acid Amplification Testing to Diagnose Gut Infections: Challenges, Opportunities, and Result Interpretation. Gastroenterol. Clin. N. Am. 2018, 47, 793–812. [Google Scholar] [CrossRef] [PubMed]
- Claas, E.C.; Burnham, C.-A.D.; Mazzulli, T.; Templeton, K.; Topin, F. Performance of the xTAG® gastrointestinal pathogen panel, a multiplex molecular assay for simultaneous detection of bacterial, viral, and parasitic causes of infectious gastroenteritis. J. Microbiol. Biotechnol. 2013, 23, 1041–1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navidad, J.F.; Griswold, D.J.; Gradus, M.S.; Bhattacharyya, S. Evaluation of Luminex xTAG gastrointestinal pathogen analyte-specific reagents for high-throughput, simultaneous detection of bacteria, viruses, and parasites of clinical and public health importance. J. Clin. Microbiol. 2013, 51, 3018–3024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stockmann, C.; Rogatcheva, M.; Harrel, B.; Vaughn, M.; Crisp, R.; Poritz, M.; Thatcher, S.; Korgenski, E.K.; Barney, T.; Daly, J.; et al. How well does physician selection of microbiologic tests identify Clostridium difficile and other pathogens in paediatric diarrhoea? Insights using multiplex PCR-based detection. Clin. Microbiol. Infect. 2015, 21, 179.e9–179.e15. [Google Scholar] [CrossRef] [Green Version]
- Khare, R.; Espy, M.J.; Cebelinski, E.; Boxrud, D.; Sloan, L.M.; Cunningham, S.A.; Pritt, B.S.; Patel, R.; Binnicker, M.J. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J. Clin. Microbiol. 2014, 52, 3667–3673. [Google Scholar] [CrossRef] [PubMed]
- Albert, M.J.; Rotimi, V.O.; Iqbal, J.; Chehadeh, W. Evaluation of the xTAG Gastrointestinal Pathogen Panel Assay for the Detection of Enteric Pathogens in Kuwait. Med. Princ. Pract. 2016, 25, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Wessels, E.; Rusman, L.G.; Van Bussel, M.J.A.W.M.; Claas, E.C.J. Added value of multiplex Luminex Gastrointestinal Pathogen Panel (xTAG® GPP) testing in the diagnosis of infectious gastroenteritis. Clin. Microbiol. Infect. 2014, 20, O182–O187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckmann, C.; Heininger, U.; Marti, H.; Hirsch, H.H. Gastrointestinal pathogens detected by multiplex nucleic acid amplification testing in stools of pediatric patients and patients returning from the tropics. Infection 2014, 42, 961–970. [Google Scholar] [CrossRef] [Green Version]
- Onori, M.; Coltella, L.; Mancinelli, L.; Argentieri, M.; Menichella, D.; Villani, A.; Grandin, A.; Valentini, D.; Raponi, M.; Russo, C. Evaluation of a multiplex PCR assay for simultaneous detection of bacterial and viral enteropathogens in stool samples of paediatric patients. Diagn. Microbiol. Infect. Dis. 2014, 79, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.L.; Chatigre, J.K.; Gohou, J.P.; Coulibaly, J.T.; Leuppi, R.; Polman, K.; Chappuis, F.; Mertens, P.; Herrmann, M.; N’goran, E.K.; et al. Combined stool-based multiplex PCR and microscopy for enhanced pathogen detection in patients with persistent diarrhoea and asymptomatic controls from Côte d’Ivoire. Clin. Microbiol. Infect. 2015, 21, 591.e1–591.e10. [Google Scholar] [CrossRef] [Green Version]
- Freedman, S.B.; Lee, B.E.; Louie, M.; Pang, X.-L.; Ali, S.; Chuck, A.; Chui, L.; Currie, G.R.; Dickinson, J.; Drews, S.J.; et al. Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE): Epidemiology, emerging organisms, and economics. BMC Pediatr. 2015, 15, 89. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, C.D.; Shah-Gandhi, B.; Parsons, B.D.; Morin, S.B.; Du, T.; Golding, G.R.; Chui, L. Direct Clostridioides difficile ribotyping from stool using capillary electrophoresis. Diagn. Microbiol. Infect. Dis. 2021, 99, 115259. [Google Scholar] [CrossRef]
- Kubota, H.; Sakai, T.; Gawad, A.; Makino, H.; Akiyama, T.; Ishikawa, E.; Oishi, K. Development of TaqMan-based quantitative PCR for sensitive and selective detection of toxigenic Clostridium difficile in human stools. PLoS ONE 2014, 9, e111684. [Google Scholar] [CrossRef]
- Center for Devices and Radiological Health. Statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests—Guidance for Industry and FDA Staff; U.S. Food and Drug Administration (FDA): Silver Spring, MD, USA, 2020. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/statistical-guidance-reporting-results-studies-evaluating-diagnostic-tests-guidance-industry-and-fda (accessed on 1 September 2021).
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. 2012, 22, 376–382. [Google Scholar] [CrossRef]
- Tougas, S.R.; Lodha, N.; Vandermeer, B.; Lorenzetti, D.L.; Tarr, P.I.; Tarr, G.A.M.; Chui, L.; Vanderkooi, O.G.; Freedman, S.B. Prevalence of Detection of Clostridiodes difficile Among Asymptomatic Children. JAMA Pediatr. 2021, 175, e212328. [Google Scholar] [CrossRef] [PubMed]
- Freeman, K.; Tsertsvadze, A.; Taylor-Phillips, S.; McCarthy, N.; Mistry, H.; Manuel, R.; Mason, J. Agreement between gastrointestinal panel testing and standard microbiology methods for detecting pathogens in suspected infectious gastroenteritis: Test evaluation and meta-analysis in the absence of a reference standard. PLoS ONE 2017, 12, e0173196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borriello, S.P.; Honour, P. Simplified procedure for the routine isolation of Clostridium difficile from faeces. J. Clin. Pathol. 1981, 34, 1124–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riley, T.V.; Brazier, J.S.; Hassan, H.; Williams, K.; Phillips, K.D. Comparison of alcohol shock enrichment and selective enrichment for the isolation of Clostridium difficile. Epidemiol. Infect. 1987, 99, 355–359. [Google Scholar] [CrossRef] [Green Version]
- Kellner, T.; Parsons, B.; Chui, L.; Berenger, B.M.; Xie, J.; Burnham, C.-A.D.; Tarr, P.I.; Lee, B.E.; Nettel-Aguirre, A.; Szelewicki, J.; et al. Comparative Evaluation of Enteric Bacterial Culture and a Molecular Multiplex Syndromic Panel in Children with Acute Gastroenteritis. J. Clin. Microbiol. 2019, 57, e00205-19. [Google Scholar] [CrossRef]
| Target, Primer/Probe | Sequence 5′ → 3′ |
|---|---|
| tcdB-F | TACAAACAGGTGTATTTAGTACAGAAGATGGA |
| tcdB-R | CACCTATTTGATTTAGMCCTTTAAAAGC |
| tcdB-P | /56-FAM/TTTKCCAGT/ZEN/AAAATCAATTGCTTC/3IABkFQ/ |
| GPP | DFS RT-PCR | PE RT-PCR | No. Samples (n = 15) |
|---|---|---|---|
| Positive | Negative | Positive | 12 |
| Negative | Positive | Positive | 2 |
| Positive | Negative | Negative | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tyrrell, H.; Morin, S.B.N.; Lloyd, C.D.; Parsons, B.; Stokowski, T.; Xie, J.; Zhuo, R.; Lee, B.E.; Pang, X.-L.; Freedman, S.B.; et al. Comparative Evaluation of Luminex xTAG® Gastrointestinal Pathogen Panel and Direct-From-Stool Real-Time PCR for Detection of C. difficile Toxin tcdB in Stool Samples from a Pediatric Population. Microorganisms 2022, 10, 2214. https://doi.org/10.3390/microorganisms10112214
Tyrrell H, Morin SBN, Lloyd CD, Parsons B, Stokowski T, Xie J, Zhuo R, Lee BE, Pang X-L, Freedman SB, et al. Comparative Evaluation of Luminex xTAG® Gastrointestinal Pathogen Panel and Direct-From-Stool Real-Time PCR for Detection of C. difficile Toxin tcdB in Stool Samples from a Pediatric Population. Microorganisms. 2022; 10(11):2214. https://doi.org/10.3390/microorganisms10112214
Chicago/Turabian StyleTyrrell, Hannah, Sarah B. N. Morin, Colin D. Lloyd, Brendon Parsons, Taryn Stokowski, Jianling Xie, Ran Zhuo, Bonita E. Lee, Xiao-Li Pang, Stephen B. Freedman, and et al. 2022. "Comparative Evaluation of Luminex xTAG® Gastrointestinal Pathogen Panel and Direct-From-Stool Real-Time PCR for Detection of C. difficile Toxin tcdB in Stool Samples from a Pediatric Population" Microorganisms 10, no. 11: 2214. https://doi.org/10.3390/microorganisms10112214





