A Cyanophage MarR-Type Transcription Factor Regulates Host RNase E Expression during Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth Conditions of Prochlorococcus MED4
2.2. Generation of Expression Vectors
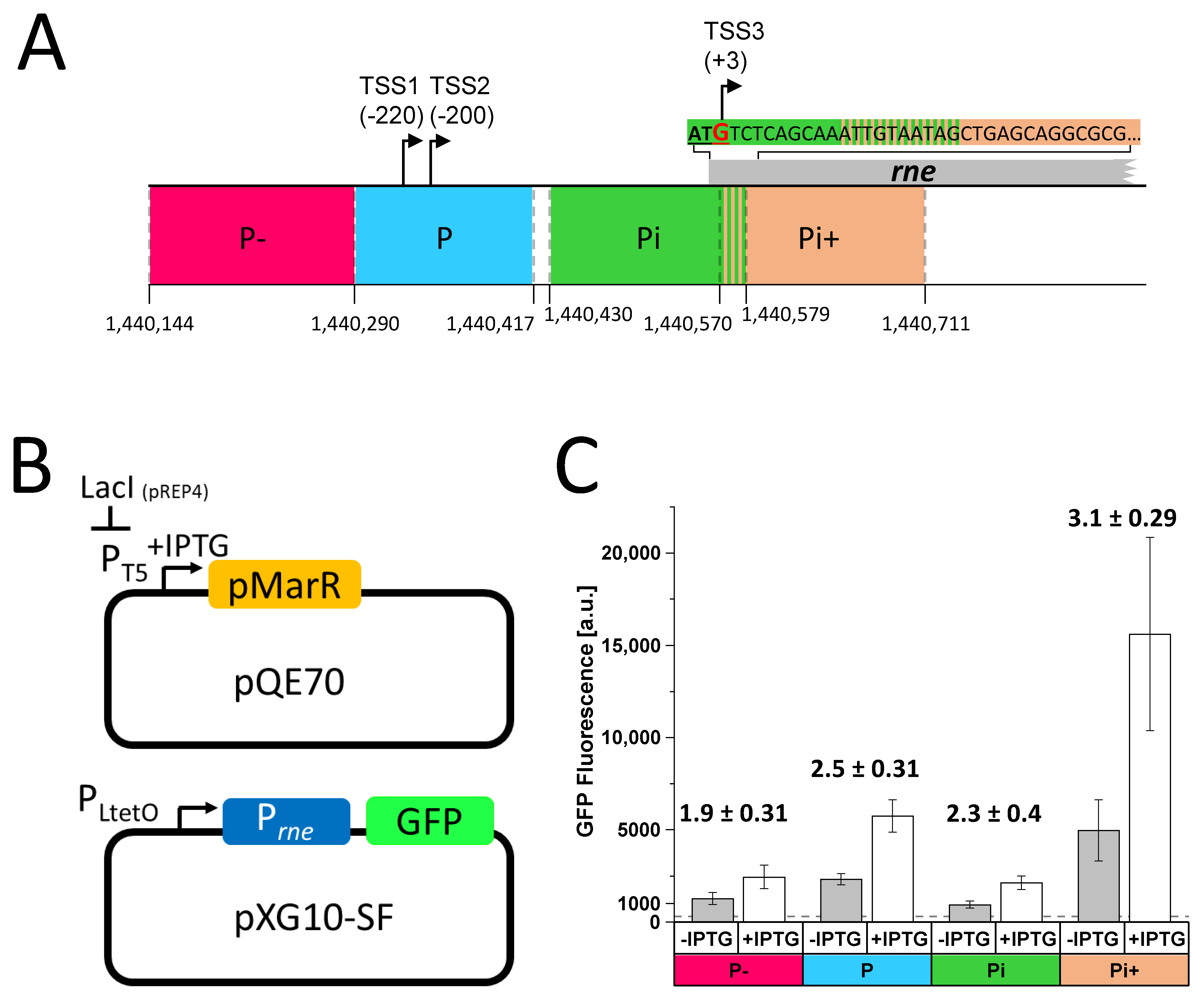
2.3. E. coli GFP In Vivo Assay
2.4. Recombinant Protein Expression and Purification
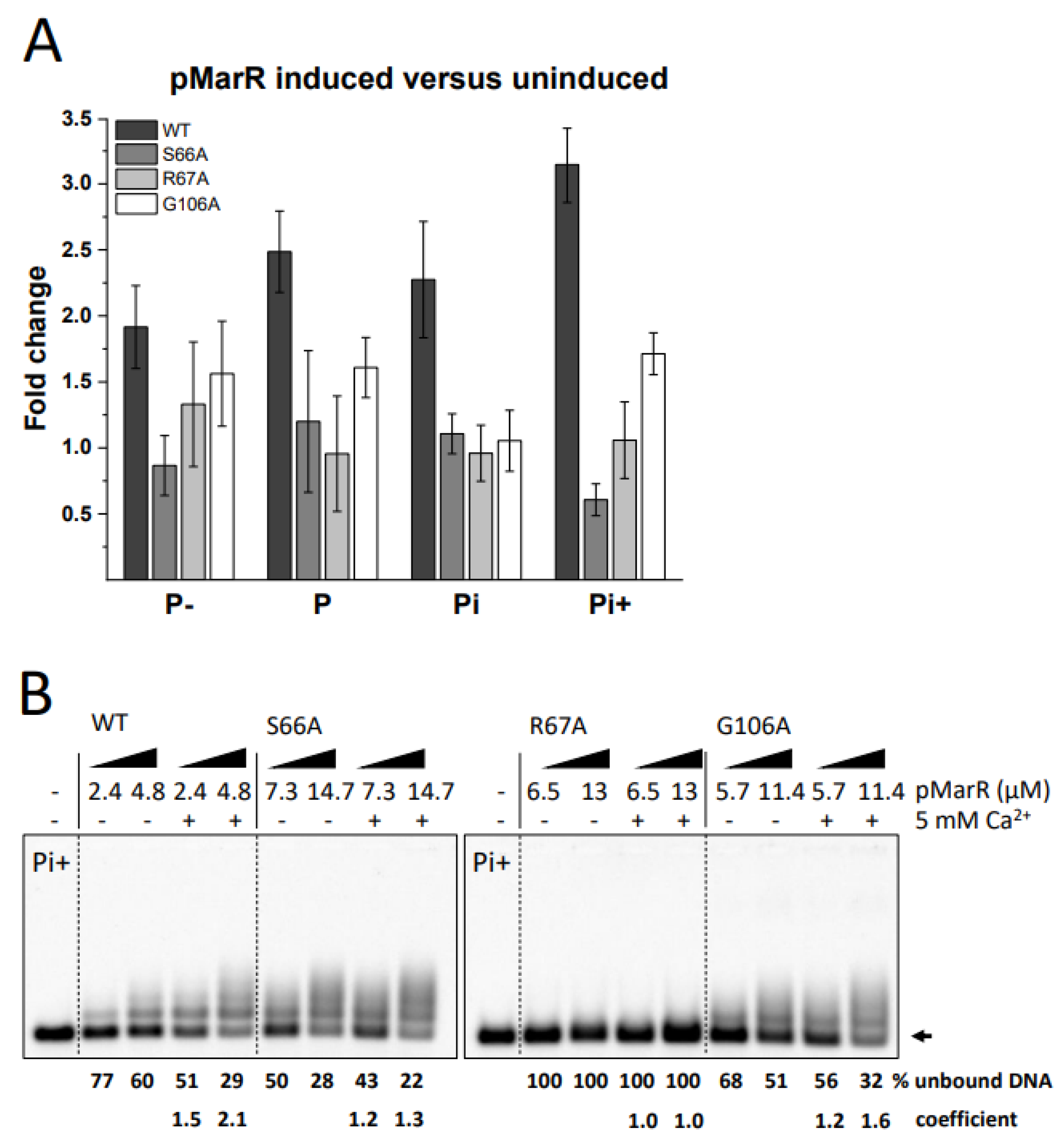
2.5. Electrophoretic Mobility Shift Assay (EMSA)
2.6. DNase I Protection Assay
2.7. Protein-Protein Affinity Chromatography and Analysis by Mass Spectrometry
2.8. Yeast Two-Hybrid Assay
3. Results
3.1. The Cyanophage Gene Product of PSSP7_011 Is Involved in the Regulation of Host RNase E Gene Expression
3.2. DNA-Binding Affinity of pMarR Is Stimulated by Calcium Ions
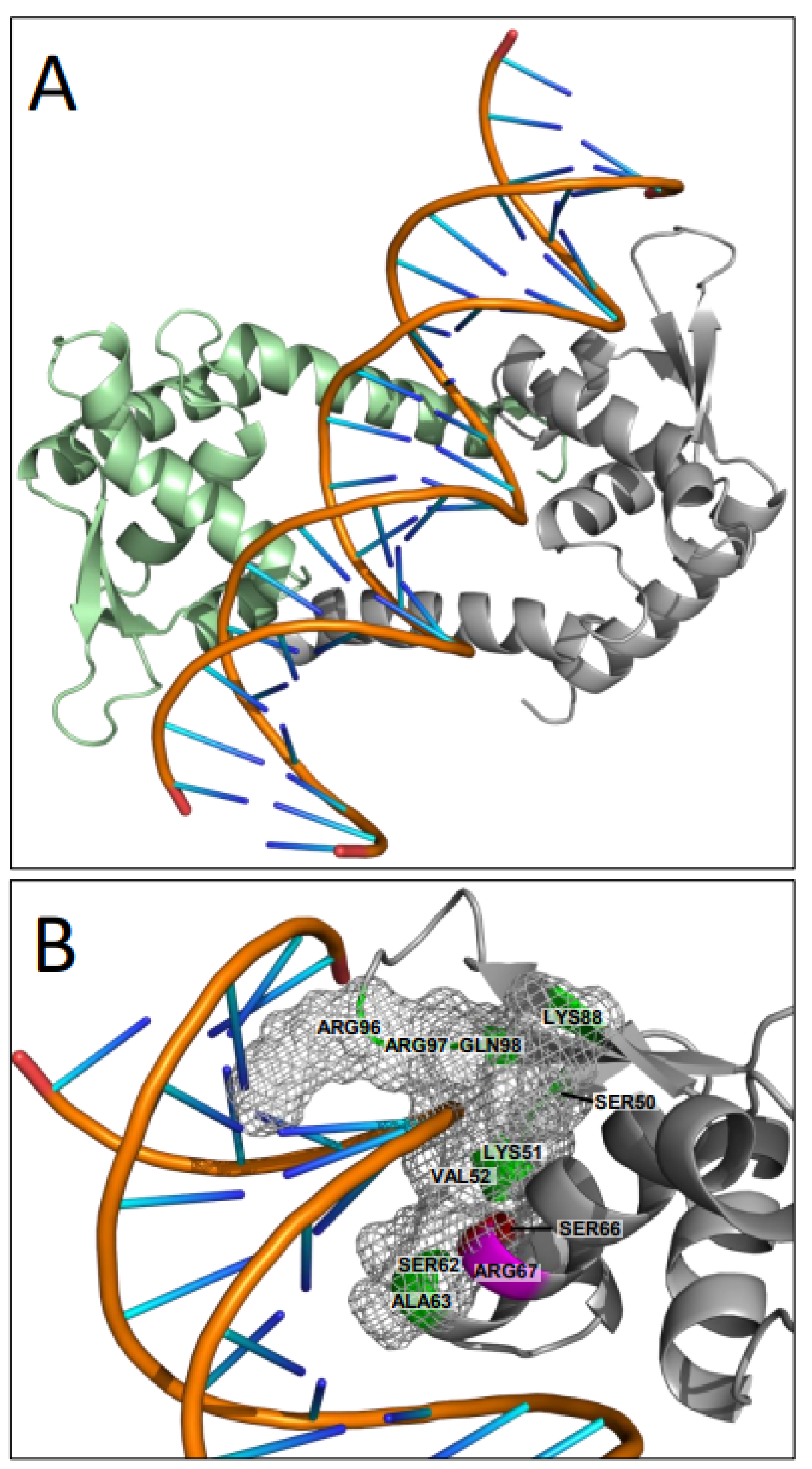
3.3. Assessment of the Functional Role of Conserved Amino Acids in pMarR
3.4. pMarR Interacts with the Alpha Subunit of the Host RNAP
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Endoribonuclease E | RNase E |
| RNA polymerase | RNAP |
| transcriptional start site | TSS |
References
- Flombaum, P.; Gallegos, J.L.; Gordillo, R.A.; Rincón, J.; Zabala, L.L.; Jiao, N.; Karl, D.M.; Li, W.K.W.; Lomas, M.W.; Veneziano, D.; et al. Present and Future Global Distributions of the Marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. USA 2013, 110, 9824–9829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biller, S.J.; Berube, P.M.; Lindell, D.; Chisholm, S.W. Prochlorococcus: The Structure and Function of Collective Diversity. Nat. Rev. Microbiol. 2015, 13, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Partensky, F.; Hess, W.R.; Vaulot, D. Prochlorococcus, a Marine Photosynthetic Prokaryote of Global Significance. Microbiol. Mol. Biol. Rev. 1999, 63, 106–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilhelm, S.W.; Suttle, C.A. Viruses and Nutrient Cycles in the Sea. BioScience 1999, 49, 781–788. [Google Scholar] [CrossRef] [Green Version]
- Carlson, M.C.G.; Ribalet, F.; Maidanik, I.; Durham, B.P.; Hulata, Y.; Ferrón, S.; Weissenbach, J.; Shamir, N.; Goldin, S.; Baran, N.; et al. Viruses Affect Picocyanobacterial Abundance and Biogeography in the North Pacific Ocean. Nat. Microbiol. 2022, 7, 570–580. [Google Scholar] [CrossRef]
- Roucourt, B.; Lavigne, R. The Role of Interactions between Phage and Bacterial Proteins within the Infected Cell: A Diverse and Puzzling Interactome. Environ. Microbiol. 2009, 11, 2789–2805. [Google Scholar] [CrossRef]
- Marchand, I.; Nicholson, A.W.; Dreyfus, M. Bacteriophage T7 Protein Kinase Phosphorylates RNase E and Stabilizes MRNAs Synthesized by T7 RNA Polymerase. Mol. Microbiol. 2001, 42, 767–776. [Google Scholar] [CrossRef] [Green Version]
- Robertson, E.S.; Aggison, L.A.; Nicholson, A.W. Phosphorylation of Elongation Factor G and Ribosomal Protein S6 in Bacteriophage T7-Infected Escherichia Coli. Mol. Microbiol. 1994, 11, 1045–1057. [Google Scholar] [CrossRef]
- Tabib-Salazar, A.; Liu, B.; Shadrin, A.; Burchell, L.; Wang, Z.; Wang, Z.; Goren, M.G.; Yosef, I.; Qimron, U.; Severinov, K.; et al. Full Shut-off of Escherichia Coli RNA-Polymerase by T7 Phage Requires a Small Phage-Encoded DNA-Binding Protein. Nucleic Acids Res. 2017, 45, 7697–7707. [Google Scholar] [CrossRef]
- Condon, C. The Phylogenetic Distribution of Bacterial Ribonucleases. Nucleic Acids Res. 2002, 30, 5339–5346. [Google Scholar] [CrossRef]
- Stazic, D.; Lindell, D.; Steglich, C. Antisense RNA Protects MRNA from RNase E Degradation by RNA-RNA Duplex Formation during Phage Infection. Nucleic Acids Res. 2011, 39, 4890–4899. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, M.B.; Coleman, M.L.; Weigele, P.; Rohwer, F.; Chisholm, S.W. Three Prochlorococcus Cyanophage Genomes: Signature Features and Ecological Interpretations. PLoS Biol. 2005, 3, e144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; Zhang, S.; Jiao, N.; Chen, F. Comparative Genomic and Phylogenomic Analyses Reveal a Conserved Core Genome Shared by Estuarine and Oceanic Cyanopodoviruses. PLoS ONE 2015, 10, e0142962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambrecht, S.J.; Steglich, C.; Hess, W.R. A Minimum Set of Regulators to Thrive in the Ocean. FEMS Microbiol. Rev. 2020, 44, 232–252. [Google Scholar] [CrossRef] [PubMed]
- Lindell, D.; Jaffe, J.D.; Coleman, M.L.; Futschik, M.E.; Axmann, I.M.; Rector, T.; Kettler, G.; Sullivan, M.B.; Steen, R.; Hess, W.R.; et al. Genome-Wide Expression Dynamics of a Marine Virus and Host Reveal Features of Co-Evolution. Nature 2007, 449, 83–86. [Google Scholar] [CrossRef]
- Deochand, D.K.; Grove, A. MarR Family Transcription Factors: Dynamic Variations on a Common Scaffold. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 595–613. [Google Scholar] [CrossRef]
- Stazic, D.; Pekarski, I.; Kopf, M.; Lindell, D.; Steglich, C. A Novel Strategy for Exploitation of Host RNase E Activity by a Marine Cyanophage. Genetics 2016, 203, 1149–1159. [Google Scholar] [CrossRef] [Green Version]
- Jain, C.; Belasco, J.G. RNase E Autoregulates Its Synthesis by Controlling the Degradation Rate of Its Own MRNA in Escherichia Coli: Unusual Sensitivity of the Rne Transcript to RNase E Activity. Genes Dev. 1995, 9, 84–96. [Google Scholar] [CrossRef] [Green Version]
- Diwa, A.; Bricker, A.L.; Jain, C.; Belasco, J.G. An Evolutionarily Conserved RNA Stem-Loop Functions as a Sensor That Directs Feedback Regulation of RNase E Gene Expression. Genes Dev. 2000, 14, 1249–1260. [Google Scholar] [CrossRef]
- Schuck, A.; Diwa, A.; Belasco, J.G. RNase E Autoregulates Its Synthesis in Escherichia Coli by Binding Directly to a Stem-Loop in the Rne 5′ Untranslated Region. Mol. Microbiol. 2009, 72, 470–478. [Google Scholar] [CrossRef]
- Moore, L.R.; Coe, A.; Zinser, E.R.; Saito, M.A.; Sullivan, M.B.; Lindell, D.; Frois-Moniz, K.; Waterbury, J.; Chisholm, S.W. Culturing the Marine Cyanobacterium Prochlorococcus: Prochlorococcus Culturing. Limnol. Oceanogr. Methods 2007, 5, 353–362. [Google Scholar] [CrossRef] [Green Version]
- Lambrecht, S.J.; Wahlig, J.M.L.; Steglich, C. The GntR Family Transcriptional Regulator PMM1637 Regulates the Highly Conserved Cyanobacterial SRNA Yfr2 in Marine Picocyanobacteria. DNA Res. 2018, 25, 489–497. [Google Scholar] [CrossRef] [Green Version]
- Beyer, H.M.; Gonschorek, P.; Samodelov, S.L.; Meier, M.; Weber, W.; Zurbriggen, M.D. AQUA Cloning: A Versatile and Simple Enzyme-Free Cloning Approach. PLoS ONE 2015, 10, e0137652. [Google Scholar] [CrossRef] [Green Version]
- Wessel, D.; Flügge, U.I. A Method for the Quantitative Recovery of Protein in Dilute Solution in the Presence of Detergents and Lipids. Anal. Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Hammel, A.; Zimmer, D.; Sommer, F.; Mühlhaus, T.; Schroda, M. Absolute Quantification of Major Photosynthetic Protein Complexes in Chlamydomonas Reinhardtii Using Quantification Concatamers (QconCATs). Front. Plant Sci. 2018, 9, 1265. [Google Scholar] [CrossRef] [Green Version]
- Rütgers, M.; Muranaka, L.S.; Mühlhaus, T.; Sommer, F.; Thoms, S.; Schurig, J.; Willmund, F.; Schulz-Raffelt, M.; Schroda, M. Substrates of the Chloroplast Small Heat Shock Proteins 22E/F Point to Thermolability as a Regulative Switch for Heat Acclimation in Chlamydomonas Reinhardtii. Plant Mol. Biol. 2017, 95, 579–591. [Google Scholar] [CrossRef] [Green Version]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Dolan, K.T.; Duguid, E.M.; He, C. Crystal Structures of SlyA Protein, a Master Virulence Regulator of Salmonella, in Free and DNA-Bound States. J. Biol. Chem. 2011, 286, 22178–22185. [Google Scholar] [CrossRef] [Green Version]
- Schroedinger, L.L.C. The PyMOL Molecular Graphics System, Version 2.4; Schroedinger: New York, NY, USA, 2015. [Google Scholar]
- Wu, X.; Haakonsen, D.L.; Sanderlin, A.G.; Liu, Y.J.; Shen, L.; Zhuang, N.; Laub, M.T.; Zhang, Y. Structural Insights into the Unique Mechanism of Transcription Activation by Caulobacter Crescentus GcrA. Nucleic Acids Res. 2018, 46, 3245–3256. [Google Scholar] [CrossRef]
- Munson, G.P.; Scott, J.R. Rns, a Virulence Regulator within the AraC Family, Requires Binding Sites Upstream and Downstream of Its Own Promoter to Function as an Activator: Rns Requires Upstream and Downstream Binding Sites. Mol. Microbiol. 2002, 36, 1391–1402. [Google Scholar] [CrossRef]
- Midgett, C.R.; Talbot, K.M.; Day, J.L.; Munson, G.P.; Kull, F.J. Structure of the Master Regulator Rns Reveals an Inhibitor of Enterotoxigenic Escherichia Coli Virulence Regulons. Sci. Rep. 2021, 11, 15663. [Google Scholar] [CrossRef]
- Ishihama, A. Role of the RNA Polymerase Alpha Subunit in Transcription Activation. Mol. Microbiol. 1992, 6, 3283–3288. [Google Scholar] [CrossRef]
- Cvirkaitė-Krupovič, V.; Krupovič, M.; Daugelavičius, R.; Bamford, D.H. Calcium Ion-Dependent Entry of the Membrane-Containing Bacteriophage PM2 into Its Pseudoalteromonas Host. Virology 2010, 405, 120–128. [Google Scholar] [CrossRef]
- Nagaraja, V.; Gopinathan, K.P. Requirement for Calcium Ions in Mycobacteriophage I3 DNA Injection and Propagation. Arch. Microbiol. 1980, 124, 249–254. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]

| Protein IDs | Intensity WT | Intensity G106A (Normalized) | Difference WT-G106A |
|---|---|---|---|
| rpoC2 (PMM1483) | 587,230 | 894 | 586,336 |
| rpoB (PMM1485) | 477,070 | 3316 | 473,754 |
| rps19 (PMM1554) | 904,660 | 462,906 | 441,754 |
| eno (PMM0208) | 332,800 | 2638 | 330,162 |
| PMM0855 | 339,410 | 15,255 | 324,155 |
| dapL (PMM1500) | 326,460 | 34,569 | 291,891 |
| pdxJ (PMM1107) | 237,270 | 0 | 237,270 |
| tufA (PMM1508) | 387,360 | 244,715 | 142,645 |
| rps17 (PMM1549) | 160,920 | 36,521 | 124,399 |
| rpoA (PMM1535) | 97,779 | 0 | 97,779 |
| rbcL (PMM0549) | 97,015 | 393 | 96,622 |
| rpoC1 (PMM1484) | 93,962 | 0 | 93,962 |
| rpl27 (PMM1345) | 119,740 | 30,858 | 88,882 |
| PMM0599 | 88,057 | 0 | 88,057 |
| bait protein | 2,051,700 | 2,051,700 | 0 |
| No 3-AT, +His | +3-AT, No His | |||||
|---|---|---|---|---|---|---|
| Dilution Factor | 1 | 10−1 | 10−2 | 1 | 10−1 | 10−2 |
| WT_R67 | WT_R67 | |||||
| WT_R67 | 3 | 3 | 3 | 3 | 2 | 1 |
| S66_R67 | 3 | 3 | 3 | 3 | 2 | 0 |
| G106_R67 | 3 | 3 | 2 | 0 | 0 | 0 |
| rpoC2 (PMM1483) | 3 | 3 | 2 | 0 | 0 | 0 |
| rpoC1 (PMM1484) | 3 | 3 | 2 | 0 | 0 | 0 |
| rpoB (PMM1485) | 3 | 3 | 2 | 0 | 0 | 0 |
| rpoA (PMM1535) | 3 | 3 | 3 | 3 | 2 | 0 |
| PMM0486 | 3 | 3 | 3 | 0 | 0 | 0 |
| PMM0599 | 3 | 3 | 2 | 0 | 0 | 0 |
| pdxJ (PMM1107) | 3 | 3 | 3 | 0 | 0 | 0 |
| tufA (PMM1508) | 3 | 3 | 2 | 0 | 0 | 0 |
| gp1, RNAP (PSSP7_013) | 3 | 3 | 2 | 0 | 0 | 0 |
| S66_R67 | S66_R67 | |||||
| WT_R67 | 3 | 3 | 2 | 2 | 1 | 0 |
| rpoA (PMM1535) | 3 | 3 | 1 | 3 | 1 | 0 |
| G106_R67 | G106_R67 | |||||
| WT_R67 | 3 | 3 | 3 | 0 | 0 | 0 |
| rpoA (PMM1535) | 3 | 3 | 2 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lambrecht, S.J.; Stappert, N.; Sommer, F.; Schroda, M.; Steglich, C. A Cyanophage MarR-Type Transcription Factor Regulates Host RNase E Expression during Infection. Microorganisms 2022, 10, 2245. https://doi.org/10.3390/microorganisms10112245
Lambrecht SJ, Stappert N, Sommer F, Schroda M, Steglich C. A Cyanophage MarR-Type Transcription Factor Regulates Host RNase E Expression during Infection. Microorganisms. 2022; 10(11):2245. https://doi.org/10.3390/microorganisms10112245
Chicago/Turabian StyleLambrecht, S. Joke, Nils Stappert, Frederik Sommer, Michael Schroda, and Claudia Steglich. 2022. "A Cyanophage MarR-Type Transcription Factor Regulates Host RNase E Expression during Infection" Microorganisms 10, no. 11: 2245. https://doi.org/10.3390/microorganisms10112245
APA StyleLambrecht, S. J., Stappert, N., Sommer, F., Schroda, M., & Steglich, C. (2022). A Cyanophage MarR-Type Transcription Factor Regulates Host RNase E Expression during Infection. Microorganisms, 10(11), 2245. https://doi.org/10.3390/microorganisms10112245






