Functional Microbial Communities in Hybrid Linear Flow Channel Reactors for Desulfurization of Tannery Effluent
Abstract
:1. Introduction
2. Materials and Methods
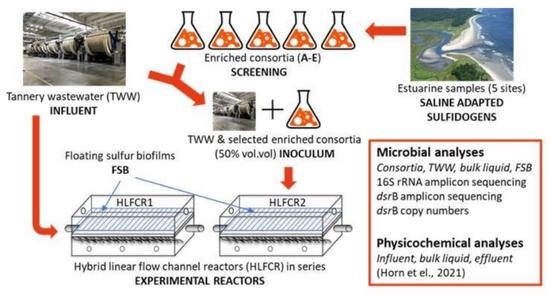
2.1. Tannery Wastewater and Inoculum
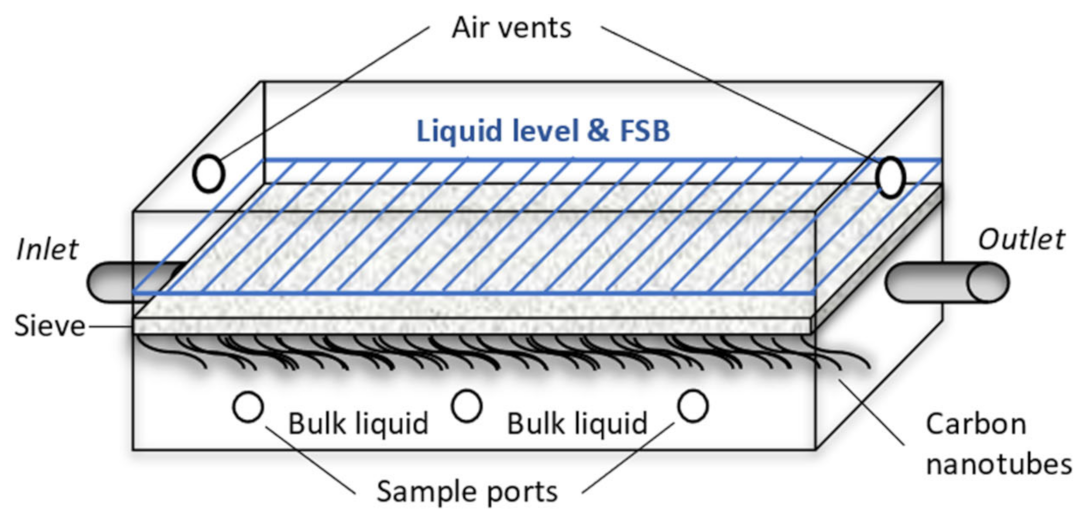
2.2. Set-Up and Operation, and Sampling Regime of Hybrid Linear Flow Channel Reactors
2.3. Physicochemical Analyses
2.4. Microbial Analyses
2.4.1. Extraction of Deoxyribonucleic Acid
2.4.2. Amplicon Sequencing
2.4.3. Quantitative Polymerase Chain Reaction
2.4.4. Statistical Analyses
3. Results and Discussion
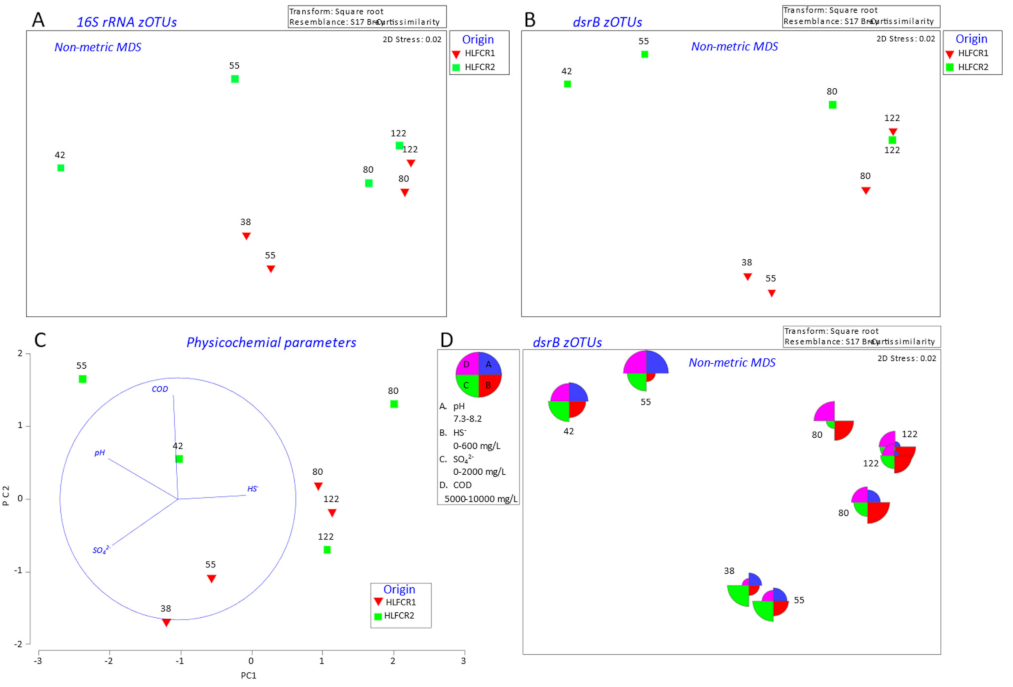
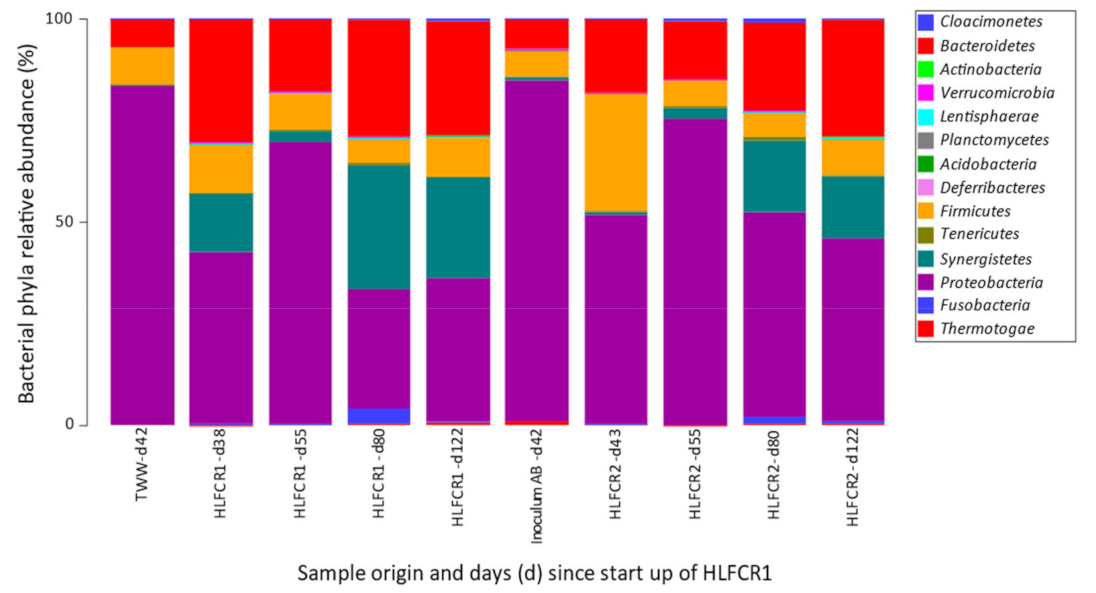
3.1. Influence of Endogenous and Exogenous Bacterial Communities on Bacterial Community Composition in Hybrid Linear Flow Channel Reactor Pre-Treating Tannery Wastewater
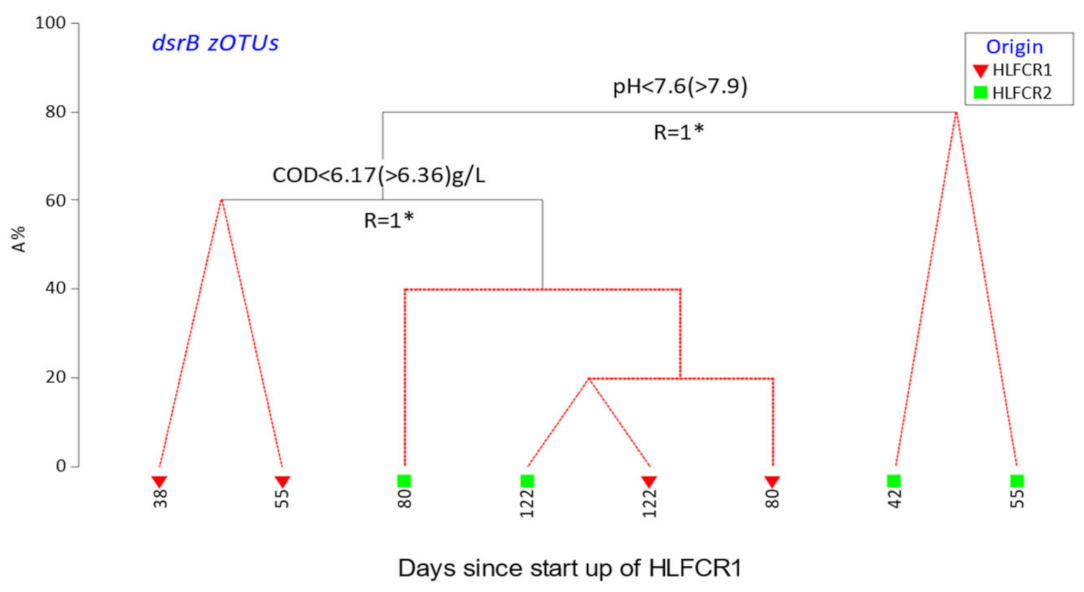
3.2. Influence of Physicochemical Parameters on Community Selection
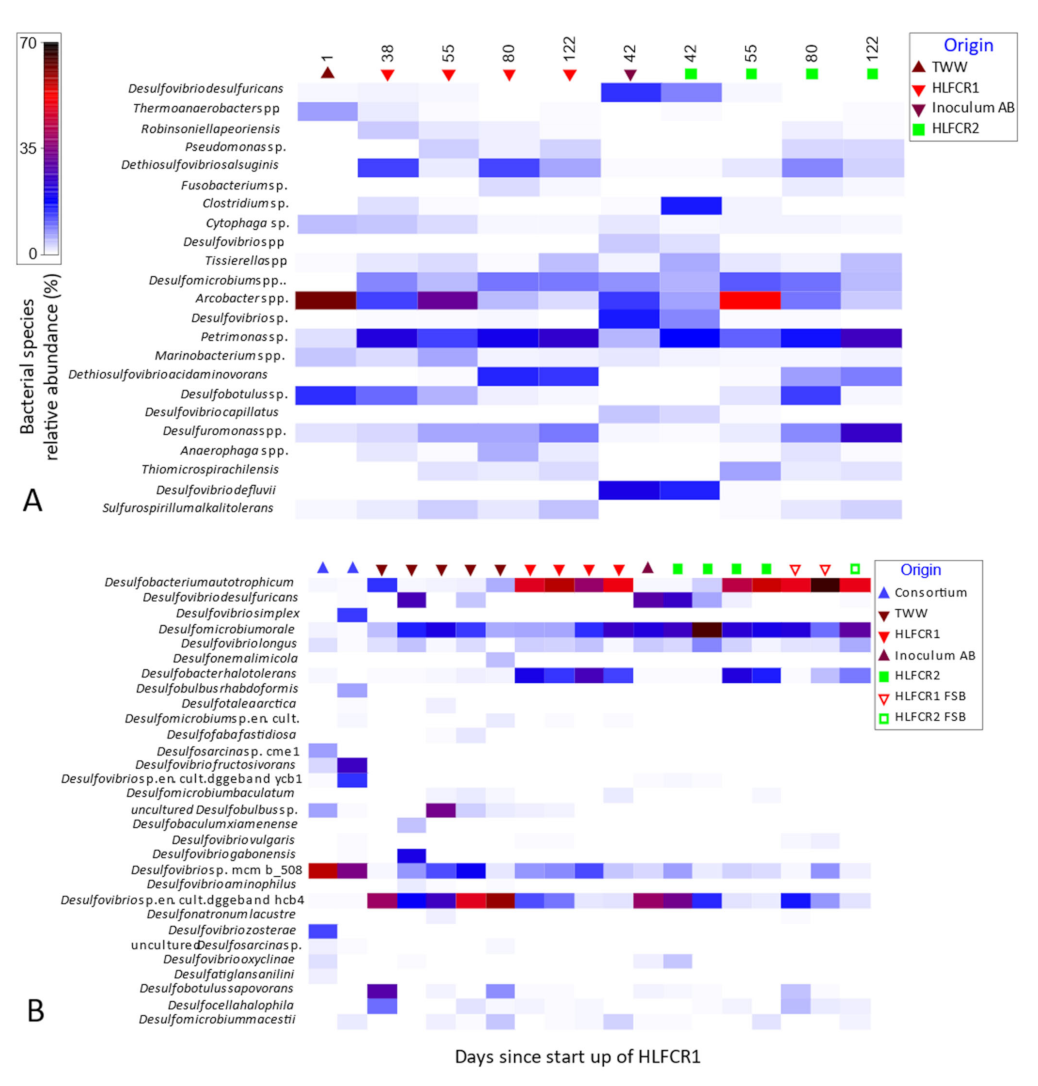
3.3. Selection of Bacterial Taxa in Hybrid Linear Flow Channel Reactors
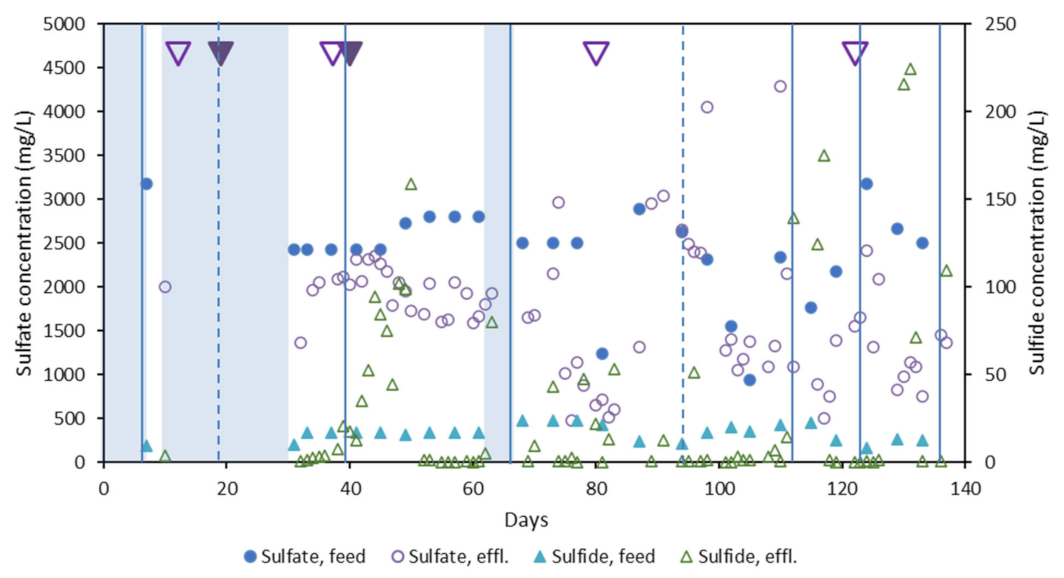
3.4. Dissimilatory Sulfite Reducing Gene Abundance and Sulfate Reduction Rates
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Mpofu, A.B.; Oyekola, O.O.; Welz, P.J. Anaerobic treatment of tannery wastewater in the context of a circular bioeconomy for developing countries. J. Clean. Prod. 2021, 296, 126490. [Google Scholar] [CrossRef]
- Swartz, C.D.; Jackson-Moss, C.; Rowswell, R.; Mpofu, A.B.; Welz, P.J. Water and Wastewater Management in the Tanning and Leather Finishing Industry; Water Research Commission: Pretoria, South Africa, 2017; ISBN 978-1-4312-0881-4. [Google Scholar] [CrossRef]
- Oliveira, C.A.; Fuess, L.T.; Soares, L.A.; Damianovic, M.H.R.Z. Increasing salinity concentrations determine the long-term participation of methanogenesis and sulfidogenesis in the biodigestion of sulfate-rich wastewater. J. Environ. Manag. 2021, 296, 113254. [Google Scholar] [CrossRef] [PubMed]
- Mpofu, A.; Kibangou, V.; Kaira, W.; Oyekola, O.; Welz, P. Anaerobic Co-Digestion of Tannery and Slaughterhouse Wastewater for Solids Reduction and Resource Recovery: Effect of Sulfate Concentration and Inoculum to Substrate Ratio. Energies 2021, 14, 2491. [Google Scholar] [CrossRef]
- Tran, T.; Kannoorpatti, K.; Padovan, A.; Thennadil, S. Sulphate-Reducing Bacteria’s Response to Extreme pH Environments and the Effect of Their Activities on Microbial Corrosion. Appl. Sci. 2021, 11, 2201. [Google Scholar] [CrossRef]
- Yang, J.H. Hydrogen sulfide removal technology: A focused review on adsorption and catalytic oxidation. Korean J. Chem. Eng. 2021, 38, 674–691. [Google Scholar] [CrossRef]
- Midha, V.; Dey, A. Biological Treatment of Tannery Wastewater for Sulfide Removal. Int. J. Chem. Sci. 2008, 6, 472–486. [Google Scholar]
- Boshoff, G.; Duncan, J.; Rose, P. Tannery effluent as a carbon source for biological sulphate reduction. Water Res. 2004, 38, 2651–2658. [Google Scholar] [CrossRef]
- Zhao, C.; Yang, Q.; Chen, W.; Li, H.; Zhang, H. Isolation of a sulfate reducing bacterium and its application in sulfate removal from tannery wastewater. Afr. J. Biotechnol. 2011, 10, 11966–11971. [Google Scholar]
- Hao, T.-W.; Xiang, P.-Y.; Mackey, H.R.; Chi, K.; Lu, H.; Chui, H.-K.; van Loosdrecht, M.C.; Chen, G.-H. A review of biological sulfate conversions in wastewater treatment. Water Res. 2014, 65, 1–21. [Google Scholar] [CrossRef]
- Marais, T.; Huddy, R.; Harrison, S.; van Hille, R. Demonstration of simultaneous biological sulphate reduction and partial sulphide oxidation in a hybrid linear flow channel reactor. J. Water Process Eng. 2020, 34, 101143. [Google Scholar] [CrossRef]
- Mpofu, A.; Kaira, W.; Oyekola, O.; Welz, P. Anaerobic co-digestion of tannery effluents: Process optimisation for resource recovery, recycling and reuse in a circular bioeconomy. Process Saf. Environ. Prot. 2022, 158, 547–559. [Google Scholar] [CrossRef]
- Marais, T.; Huddy, R.; Harrison, S.; van Hille, R. Effect of hydraulic residence time on biological sulphate reduction and elemental sulphur recovery in a single-stage hybrid linear flow channel reactor. Biochem. Eng. J. 2020, 162, 107717. [Google Scholar] [CrossRef]
- Horn, E.J.; Oyekola, O.O.; Welz, P.J.; van Hille, R.P. Biological Desulfurization of Tannery Effluent Using Hybrid Linear Flow Channel Reactors. Water 2021, 14, 32. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater, 22nd ed.; Rice, E.W., Baird, R.B., Eaton, A.D., Clesceri, L.S., Eds.; American Public Health Association (APHA): Washington, DC, USA; American WaterWorks Association (AWWA): Denver, CO, USA; Water Environment Federation (WEF): Alexandria, VA, USA, 2012. [Google Scholar]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Geets, J.; Borremans, B.; Diels, L.; Springael, D.; Vangronsveld, J.; van der Lelie, D.; Vanbroekhoven, K. DsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. J. Microbiol. Methods 2006, 66, 194–205. [Google Scholar] [CrossRef]
- Wagner, M.; Roger, A.J.; Flax, J.L.; Brusseau, G.A.; Stahl, D.A. Phylogeny of Dissimilatory Sulfite Reductases Supports an Early Origin of Sulfate Respiration. J. Bacteriol. 1998, 180, 2975–2982. [Google Scholar] [CrossRef] [Green Version]
- Willis, A. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef] [Green Version]
- Travanty, N.V.; Ponnusamy, L.; Kakumanu, M.L.; Nicholson, W.L.; Apperson, C.S. Diversity and structure of the bacterial microbiome of the American dog tick, Dermacentor variabilis, is dominated by the endosymbiont Francisella. Symbiosis 2019, 79, 239–250. [Google Scholar] [CrossRef]
- Pramanick, R.; Nathani, N.; Warke, H.; Mayadeo, N.; Aranha, C. Vaginal dysbiotic microbiome in women with no symptoms of genital infections. Front. Cell. Infect. Microbiol. 2022, 11, 760459. [Google Scholar] [CrossRef]
- McCurdie, P.J.; Holmes, S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef]
- Kibangou, V.A.; Lilly, M.; Mpofu, A.B.; de Jonge, N.; Oyekola, O.O.; Welz, P.J. Sulfate-reducing and methanogenic microbial community responses during anaerobic digestion of tannery effluent. Bioresour. Technol. 2021, 347, 126308. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Chon, K.; Ren, X.; Kou, Y.; Hwang, M.-H.; Chae, K.-J. Bioaugmentation treatment of a novel microbial consortium for degradation of organic pollutants in tannery wastewater under a full-scale oxic process. Biochem. Eng. J. 2021, 175, 108131. [Google Scholar] [CrossRef]
- Grabowski, A.; Tindall, B.J.; Bardin, V.; Blanchet, D.; Jeanthon, C. Petrimonas sulfuriphila gen. nov., sp. nov., a mesophilic fermentative bacterium isolated from a biodegraded oil reservoir. Int. J. Syst. Evol. Microbiol. 2005, 55, 1113–1121. [Google Scholar] [CrossRef]
- Hahnke, S.; Langer, T.; Koeck, D.E.; Klocke, M. Description of Proteiniphilum saccharofermentans sp. nov., Petrimonas mucosa sp. nov. and Fermentimonas caenicola gen. nov., sp. nov., isolated from mesophilic laboratory-scale biogas reactors, and emended description of the genus Proteiniphilum. Int. J. Syst. Evol. Microbiol. 2016, 66, 1466–1475. [Google Scholar] [CrossRef]
- Maus, I.; Tubbesing, T.; Wibberg, D.; Heyer, R.; Hassa, J.; Tomazetto, G.; Huang, L.; Bunk, B.; Spröer, C.; Benndorf, D.; et al. The Role of Petrimonas mucosa ING2-E5AT in Mesophilic Biogas Reactor Systems as Deduced from Multiomics Analyses. Microorganisms 2020, 8, 2024. [Google Scholar] [CrossRef]
- Qian, Y.; Xu, M.; Deng, T.; Hu, W.; He, Z.; Yang, X.; Wang, B.; Song, D.; Chen, L.; Huang, Y.; et al. Synergistic interactions of Desulfovibrio and Petrimonas for sulfate-reduction coupling polycyclic aromatic hydrocarbon degradation. J. Hazard. Mater. 2020, 407, 124385. [Google Scholar] [CrossRef]
- Magot, M.; Ravot, G.; Campaignolle, X.; Ollivier, B.; Patel, B.K.C.; Fardeau, M.-L.; Thomas, P.; Crolet, J.-L.; Garcia, J.-L. Dethiosulfovibrio peptidovorans gen. nov., sp. nov., a New Anaerobic, Slightly Halophilic, Thiosulfate-Reducing Bacterium from Corroding Offshore Oil Wells. Int. J. Syst. Bacteriol. 1997, 47, 818–824. [Google Scholar] [CrossRef] [Green Version]
- Surkov, A.V.; Dubinina, G.A.; Lysenko, A.M.; Glőckner, F.O.; Kuever, J. Dethiosulfovibrio russensis sp. nov., Dethisulfovibrio marinus sp. nov. and Dethiosulfovibrio acidaminovorans sp. nov., novel anaerobic thiosulfate- and sulfur-reducing bacteria isolated from “Thiodendron” sulfur mats in different saline environments. Int. J. Syst. Evol. Microbiol. 2001, 51, 327–337. [Google Scholar] [CrossRef] [Green Version]
- Díaz-Cárdenas, C.; López, G.; Patel, B.; Baena, S. Dethiosulfovibrio salsuginis sp. nov., an anaerobic, slightly halophilic bacterium isolated from a saline spring. Int. J. Syst. Evol. Microbiol. 2010, 60, 850–853. [Google Scholar] [CrossRef]
- Surkov, A.V.; Böttcher, M.E.; Kuever, J. Stable sulfur isotope fractionation during the reduction of thiosulfate by Dethiosulfovibrio russensis. Arch. Microbiol. 2000, 174, 448–451. [Google Scholar] [CrossRef] [PubMed]
- Surkov, A.V.; Böttcher, M.E.; Kuever, J. Sulphur isotope fractionation during the reduction of elemental sulphur and thiosulphate by Dethiosulfovibrio spp. Isot. Environ. Health Stud. 2012, 48, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Jurado, V.; D’Angeli, I.; Martin-Pozas, T.; Cappelletti, M.; Ghezzi, D.; Gonzalez-Pimentel, J.L.; Cuezva, S.; Miller, A.Z.; Fernandez-Cortes, A.; De Waele, J.; et al. Dominance of Arcobacter in the white filaments from the thermal sulfidic spring of Fetida Cave (Apulia, southern Italy). Sci. Total Environ. 2021, 800, 149465. [Google Scholar] [CrossRef] [PubMed]
- Ramond, J.-B.; Welz, P.J.; le Roes-Hill, M.; Tuffin, M.; Burton, S.G.; Cowan, D.A. Selection of Clostridium sp. in biological sand filters neutralizing acid mine drainage. FEMS Microb. Ecol. 2014, 87, 678–690. [Google Scholar] [CrossRef] [Green Version]
- Bukhtiyarova, P.A.; Antsiferov, D.V.; Brasseur, G.; Avakyan, M.R.; Frank, Y.A.; Ikkert, O.P.; Pimenov, N.V.; Tuovinen, O.H.; Karnachuk, O.V. Isolation, characterization, and genome insights into an anaerobic sulfidogenic Tissierella bacterium from Cu-bearing coins. Anaerobe 2019, 56, 66–77. [Google Scholar] [CrossRef]
- Jørgensen, B.B.; Findlay, A.; Pellerin, A. The Biogeochemical Sulfur Cycle of Marine Sediments. Front. Microbiol. 2019, 10, 849. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Dashti, M.; Prange, A.; Rainey, F.A.; Rohde, M.; Whitman, W.; Wiegel, J. Thermoanaerobacter sulfurigignens sp. nov., an anaerobic thermophilic bacterium that reduces 1 M thiosulfate to elemental sulfur and tolerates 90 mM sulfite. Int. J. Syst. Evol. Microbiol. 2007, 57, 1429–1434. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.-J.; Chen, C.; Guo, H.-L.; Wang, A.-J.; Ren, N.-Q.; Lee, D.-J. Characterization of a newly isolated strain Pseudomonas sp. C27 for sulfide oxidation: Reaction kinetics and stoichiometry. Sci. Rep. 2016, 6, 21032. [Google Scholar] [CrossRef]
- Fadeev, E.; Cardozo-Mino, M.G.; Rapp, J.Z.; Bienhold, C.; Salter, I.; Salman-Carvalho, V.; Molari, M.; Tegetmeyer, H.E.; Buttigieg, P.L.; Boetius, A. Comparison of Two 16S rRNA Primers (V3–V4 and V4–V5) for Studies of Arctic Microbial Communities. Front. Microbiol. 2021, 12, 637526. [Google Scholar] [CrossRef]
- Amann, J.; Lange, D.; Schüler, M.; Rabus, R. Substrate-Dependent Regulation of Carbon Catabolism in Marine Sulfate-Reducing Desulfobacterium autotrophicum HRM2. J. Mol. Microbiol. Biotechnol. 2010, 18, 74–84. [Google Scholar] [CrossRef]
- Strittmatter, A.W.; Liesegang, H.; Rabus, R.; Decker, I.; Amann, J.; Andres, S.; Henne, A.; Fricke, W.F.; Martinez-Arias, R.; Bartels, D.; et al. Genome sequence of Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ. Microbiol. 2009, 11, 1038–1055. [Google Scholar] [CrossRef] [PubMed]
- Dworkin, M.; Falkow, S.; Rosenberg, E.; Schleifer, K.-H.; Stackebrandt, E. (Eds.) The Prokaryotes: Vol. 2: Ecophysiology and Biochemistry, 3rd ed.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Tarpgaard, I.H.; Jørgensen, B.B.; Kjeldsen, K.U.; Røy, H. The marine sulfate reducer Desulfobacterium autotrophicum HRM2 can switch between low and high apparent half-saturation constants for dissimilatory sulfate reduction. FEMS Microbiol. Ecol. 2017, 93, fix012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandt, K.K.; Ingvorsen, K. Desulfobacter halotolerans sp. nov., a Halotolerant Acetate-Oxidizing Sulfate-Reducing Bacterium Isolated from Sediments of Great Salt Lake, Utah. Syst. Appl. Microbiol. 1997, 20, 366–373. [Google Scholar] [CrossRef]
- Langendijk, P.S.; Kulik, E.M.; Sandmeier, H.; Meyer, J.; Van Der Hoeven, J.S. Isolation of Desulfomicrobium orale sp. nov. and Desulfovibrio strain NY682, oral sulfate-reducing bacteria involved in human periodontal disease. Int. J. Syst. Evol. Microbiol. 2001, 51, 1035–1044. [Google Scholar] [CrossRef] [Green Version]
- Blagodatskaya, E.V.; Blagodatskii, S.A.; Anderson, T.-H. Quantitative Isolation of Microbial DNA from Different Types of Soils of Natural and Agricultural Ecosystems. Microbiology 2003, 72, 744–749. [Google Scholar] [CrossRef]
- Joergensen, R.G.; Emmerling, C. Methods for evaluating human impact on soil microorganisms based on their activity, biomass, and diversity in agricultural soils. J. Plant Nutr. Soil Sci. 2006, 169, 295–309. [Google Scholar] [CrossRef]
- Marstorp, H.; Guan, X.; Gong, P. Relationship between dsDNA, chloroform labile C and ergosterol in soils of different organic matter contents and pH. Soil Biol. Biochem. 2000, 32, 879–882. [Google Scholar] [CrossRef]
- Quňoz, K.; Buchmann, C.; Meyer, M.; Schmidt-Heydt, M.; Steinmetz, Z.; Diehl, D. Physicochemical and microbial soil quality indictors as affected by the agricultural management system in strawberry cultivation using straw or black polyethylene mulching. Appl. Soil Ecol. 2017, 113, 36–46. [Google Scholar] [CrossRef]
- Welz, P.J.; Khan, N.; Prins, A. The effect of biogenic and chemically synthesised silver nanoparticles on the benthic bacterial communities in river sediments. Sci. Total Environ. 2018, 644C, 1380–1390. [Google Scholar] [CrossRef]


| Parameter | H-TOC TWW | L-TOC TWW | ||
|---|---|---|---|---|
| Average | SD | Average | SD | |
| pH | 10.24 | 1.6 | 7.81 | 0.52 |
| EC (mS/cm) | 32.01 | 2.2 | 31.5 | 1.59 |
| TOC (mg/L) | 6116 | 1875 | 886 | 253 |
| COD (mg/L) | 28,169 | 3665 | 4968 | 2940 |
| BOD (mg/L) | 6200 | 812 | 1539 | 520 |
| VOAt (mg/L AAE) | 2920 | 718 | 1041 | 647 |
| Protein (mg/L) | 2875 | 1024 | 310.6 | 83.9 |
| TN (mg/L) | 1258 | 215 | 679 | 138 |
| TAN (mg/L NH3-N) | 301 | 286 | 350 | 235 |
| NO3− (mg/L) | 70.8 | 25.0 | 40.5 | 23.6 |
| NO2− (mg/L) | 5.1 | 5.2 | 2.35 | 2.63 |
| PO43− mg/L | 0 | 0 | 1.21 | 1.96 |
| SO42− (mg/L) | 1951 | 574 | 3687 | 383 |
| HS− (mg/L) | 699 | 114 | 83 | 76 |
| Cl (mg/L) | 7744 | 460 | 7713 | 325 |
| TS (g/L) | 36.1 | 5.8 | 17.96 | 3.23 |
| TVS (g/L) | 13.1 | 3.3 | 1.93 | 0.47 |
| K (mg/L) | 95.7 | 31.1 | 100.3 | 19.5 |
| Na (mg/L) | 6412 | 571 | 6225 | 276 |
| Fe (mg/L) | 0.11 | 0.08 | 0.19 | 0.12 |
| Ca (mg/L) | 692 | 482 | 230.9 | 58.5 |
| Mg (mg/L) | 120 | 138 | 220.7 | 27.6 |
| Mn (mg/L) | 0.50 | 0.41 | 15.14 | 6.92 |
| Zn (mg/L) | 0.50 | 0.33 | 0.19 | 0.12 |
| Cr (mg/L) | 0.09 | 0.05 | 0.20 | 0.08 |
| Alk (mg/L CaCO3) | 3256 | 907 | 1999 | 385 |
| COD:SO42− | 15.4 | 3.5 | 1.4 | 0.76 |
| TVS:TS | 0.36 | 0.03 | 0.11 | 0.02 |
| BOD:COD | 0.23 | 0.05 | 0.37 | 0.14 |
| C:N | 5.1 | 2.2 | 1.3 | 0.21 |
| VOA:Alk | 0.94 | 0.27 | 0.48 | 0.24 |
| COD:TVS | 2.2 | 0.52 | 2.5 | 0.93 |
| HLFCR2 (n = 4) | HLFCR1 (n = 4) | TWW (n = 5) | |
|---|---|---|---|
| HLFCR1 (n = 4) | NS | - | - |
| TWW (n = 5) | NS | 0.75 * | - |
| Consortia (n = 5) | 0.956 ** | 1 ** | 0.988 ** |
| Inoc. AB | TWW * | HLFCR * | Comments | |
|---|---|---|---|---|
| Selected species | ||||
| Desulfobacterium autotrophicum | + | ++ | ++++ | - |
| Desulfobacter halotolerans | + | + | +++ | - |
| Desulfomicrobium macestii | + | + to ++ | + to ++ | Temporal increase in RA |
| Species outcompeted | ||||
| Desulfobacterium desulfuricans | +++ | ++ | + | - |
| Desulfovibrio longus | ++ | ++ | ++ to + | Temporal decrease in RA |
| Uncultured Desulfobulbus sp. | + | +++ | + | Temporal decrease in HLFCR1 |
| Desulfobaculum xiamenense | ND | +/++ | ND | TWW batches 2 & 5 only |
| Desulfovibrio gabonensis | + | +/+++ | + | Only +++ in TWW batch 2 |
| Desulfobacterium sapovorans | + | +/++ | + | Temporal decrease in RA |
| Desulfocella halophila | ND | +/++ | + | Temporal decrease in RA |
| Desulfovibrio sp. mcm b_508 | ++ | ++/+++ | ++ to + | Slow temporal decrease in RA |
| Desulfovibrio enrichment culture dgge band hcb4 | +++ | +++/++++ | ++ to + | Temporal decrease in RA |
| Resilient species | ||||
| Desulfomicrobium orale | +++ | +++ | +++ | - |
| Sample | Batch or Time (Days) | DNA Conc. (ng/µL) | dsrB Copy Numbers (Number/ng DNA) | SR Rate (mg/L·day) |
|---|---|---|---|---|
| TWW | Batch 1 | 25 | 2.11 × 105 | NA |
| Batch 2 | 3 | 8.62 × 103 | NA | |
| Batch 3 | 6 | 2.10 × 104 | NA | |
| Batch 4 | 5 | 1.24 × 104 | NA | |
| Batch 5 | 18 | 1.56 × 104 | NA | |
| HLFCR1 | 12 | 25 | 2.11 × 105 | 198 |
| 38 | 96 | 3.64 × 105 | 674 | |
| 55 | 87 | 4.49 × 105 | 537 | |
| 80 | 64 | 3.85 × 105 | 643 | |
| 112 | 186 | 3.65 × 105 | 568 | |
| Inoculum AB | 42 | 237 | 9.25 × 105 | NA |
| HLFCR2 | 42 | 137 | 6.31 × 105 | NA |
| 55 | 184 | 4.11 × 105 | 439 | |
| 80 | 113 | 3.46 × 105 | 182 | |
| 123 | 230 | 1.49 × 105 | 281 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horn, E.J.; van Hille, R.P.; Oyekola, O.O.; Welz, P.J. Functional Microbial Communities in Hybrid Linear Flow Channel Reactors for Desulfurization of Tannery Effluent. Microorganisms 2022, 10, 2305. https://doi.org/10.3390/microorganisms10112305
Horn EJ, van Hille RP, Oyekola OO, Welz PJ. Functional Microbial Communities in Hybrid Linear Flow Channel Reactors for Desulfurization of Tannery Effluent. Microorganisms. 2022; 10(11):2305. https://doi.org/10.3390/microorganisms10112305
Chicago/Turabian StyleHorn, Emma J., Rob P. van Hille, Oluwaseun O. Oyekola, and Pamela J. Welz. 2022. "Functional Microbial Communities in Hybrid Linear Flow Channel Reactors for Desulfurization of Tannery Effluent" Microorganisms 10, no. 11: 2305. https://doi.org/10.3390/microorganisms10112305
APA StyleHorn, E. J., van Hille, R. P., Oyekola, O. O., & Welz, P. J. (2022). Functional Microbial Communities in Hybrid Linear Flow Channel Reactors for Desulfurization of Tannery Effluent. Microorganisms, 10(11), 2305. https://doi.org/10.3390/microorganisms10112305