Effect of the Type VI Secretion System Secreted Protein Hcp on the Virulence of Aeromonas salmonicida
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Sequence Alignment Analysis, Phylogenetic Tree Construction, and Protein Structure Prediction
2.3. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.4. Construction of hcp Silent Strains
2.5. Growth Curve Measurement
2.6. Biofilm Formation Measurement
2.7. Preparation of Mucus
2.8. Adhesion Test
2.9. Hemolysis Test
2.10. Artificial Infection and Sample Collection
2.11. Preparation of Extracellular Products
2.12. SDS-PAGE Electrophoresis
2.13. Effect of hcp Gene on the Enzyme Activity of A. salmonicida Extracellular Products
2.14. Preparation and Determination of Extracellular Polysaccharide and Protein in EPSs
2.15. Statistical Analysis
3. Results
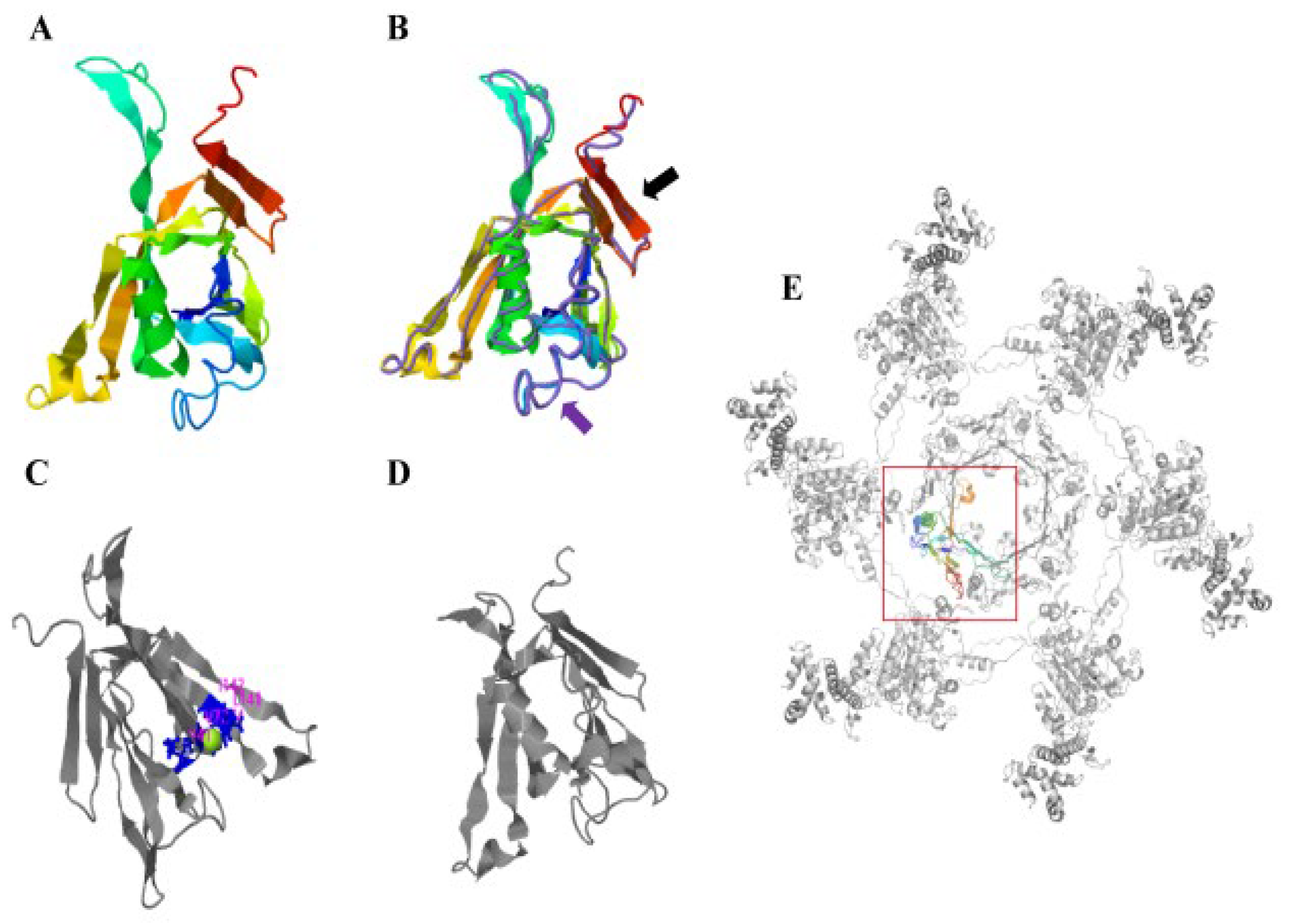
3.1. Phylogenetic Tree and Protein Structure Analysis of Hcp from A. salmonicida
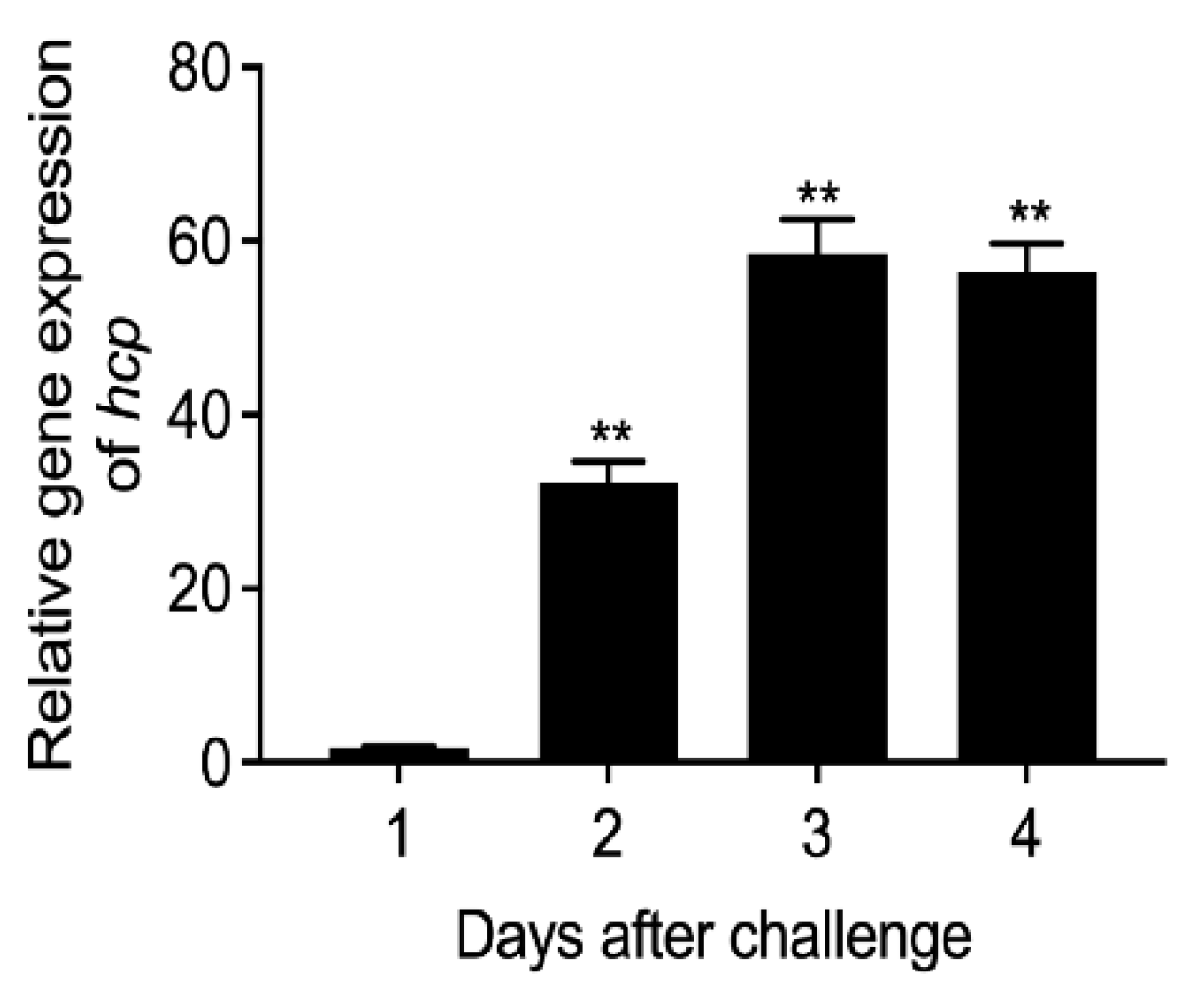
3.2. Expression Level of Hcp Gene during Infection of E. coioides
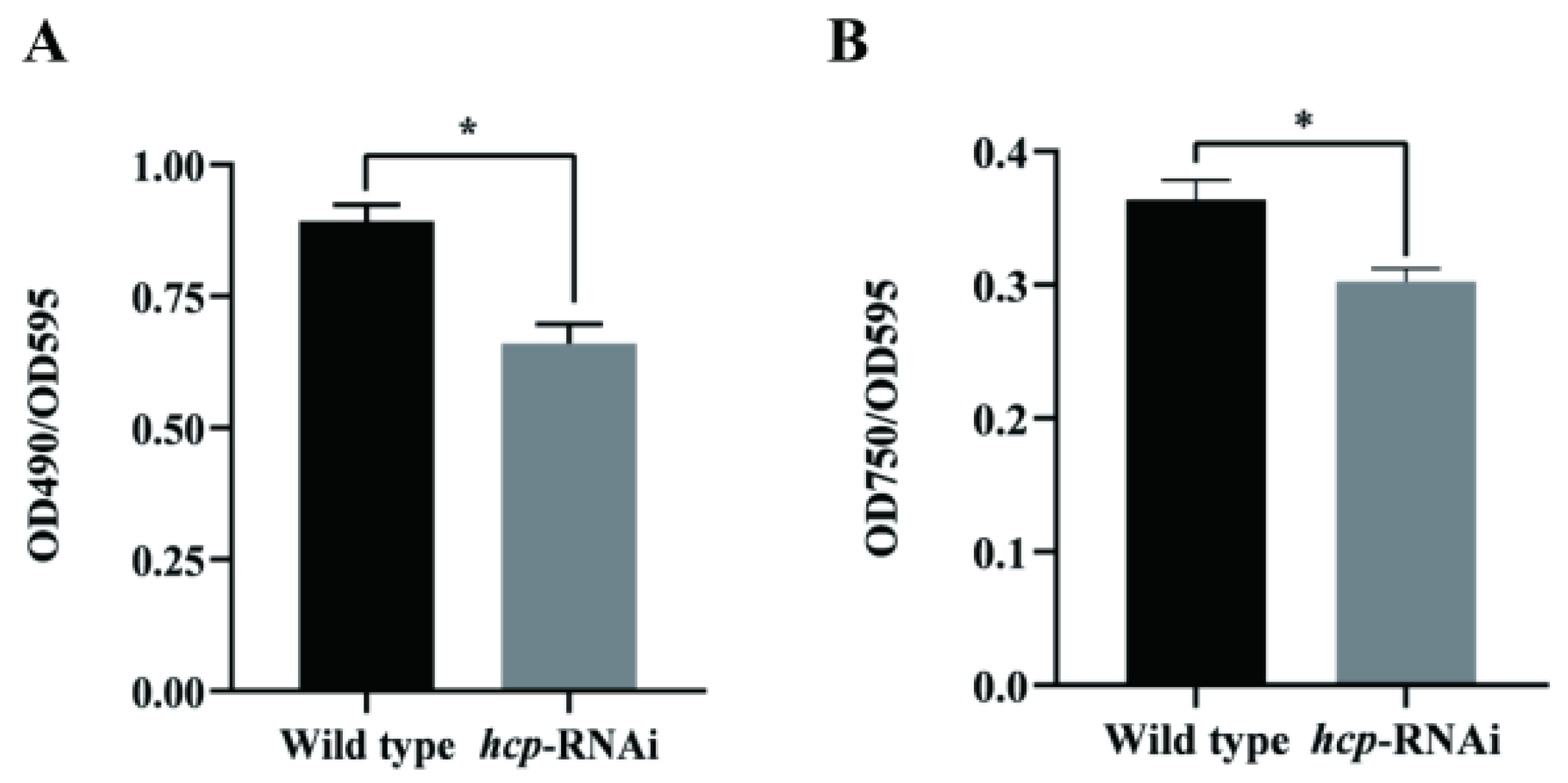
3.3. Effects of Hcp Gene Silencing on the Virulence of A. salmonicida
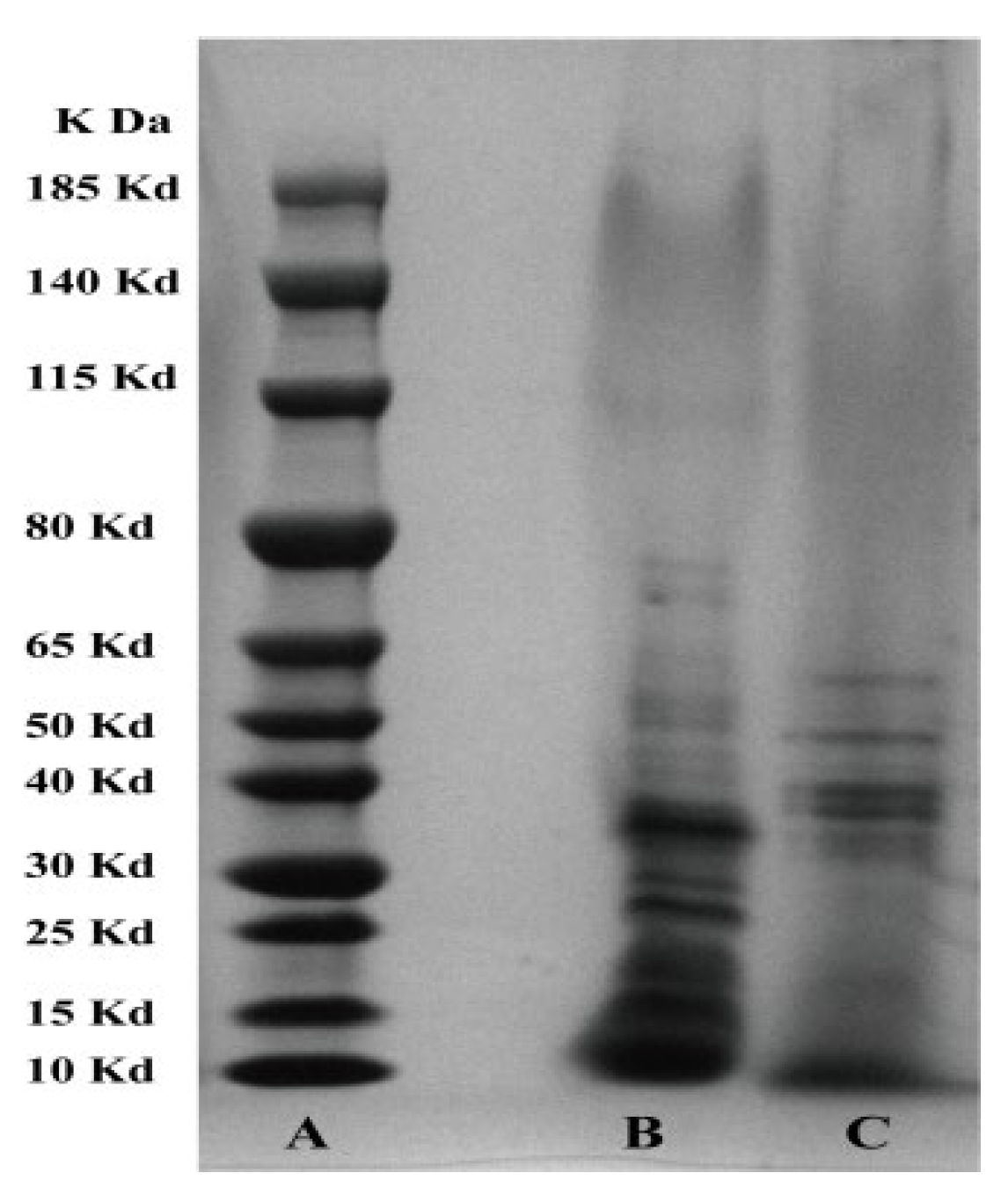
3.4. SDS-PAGE Analysis of the Effect of Hcp on the Protein Composition of Extracellular Products
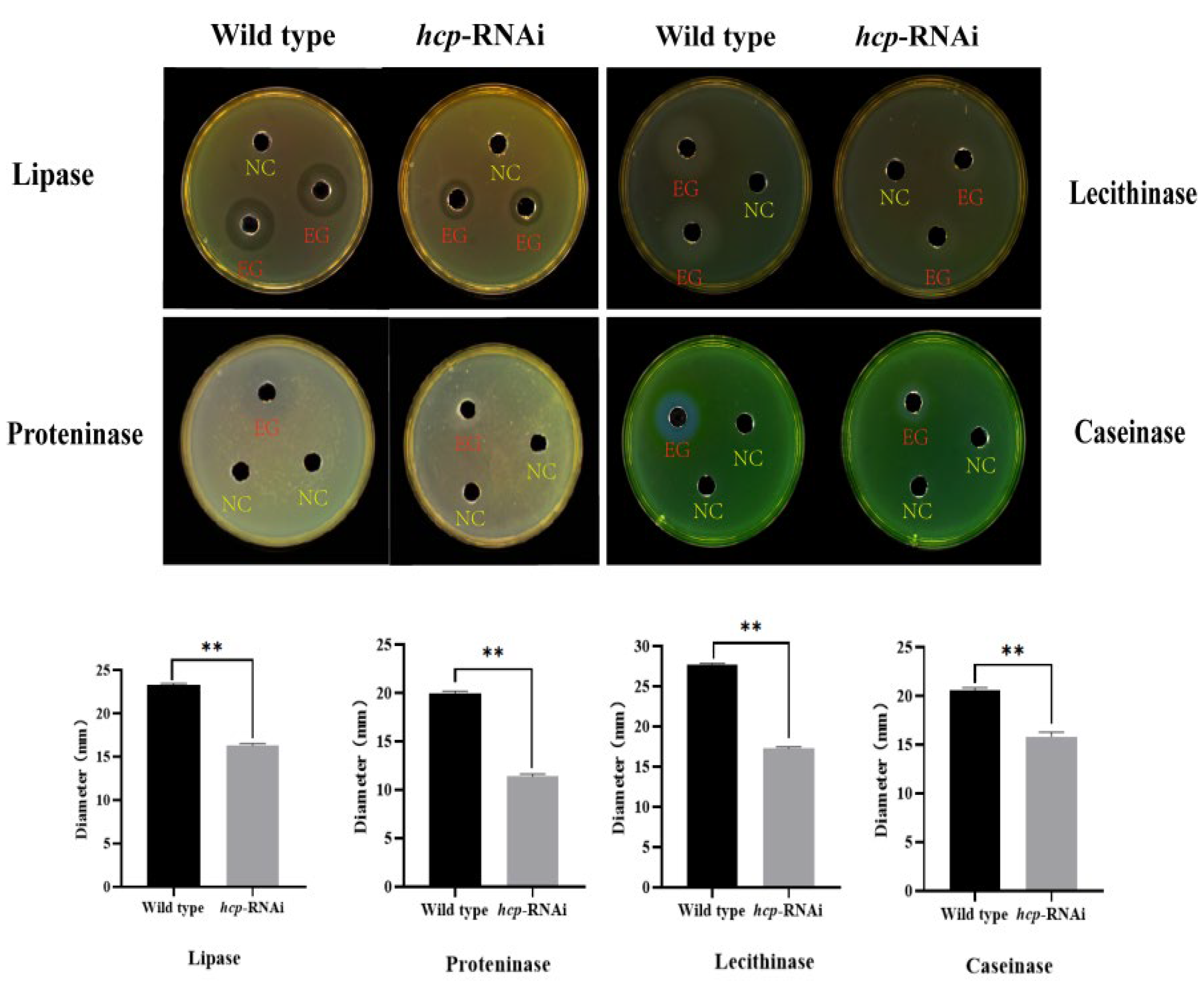
3.5. Effect of Hcp Gene on the Enzyme Activity of A. salmonicida EPS
3.6. Effects of Hcp Silencing on EPS Content of A. salmonicida
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jin, S.; Fu, S. Identification and histopathological and pathogenicity analysis of Aeromonas salmonicida salmonicida from goldfish (Carassius auratus) in North China. Aquac. Fish. 2020, 5, 36–41. [Google Scholar] [CrossRef]
- Van Banning, P. Bacterial Fish Pathogens: Disease in Farmed and Wild Fish: B. Austin and DA Austin; Ellis Horwood Limited: Chichester, UK, 1987; 364p. [Google Scholar]
- Bricknell, I.R.; Bowden, T.J. Susceptibility of Atlantic halibut, Hippoglossus hippoglossus (L.) to infection with typical and atypical Aeromonas salmonicida. Aquaculture 1999, 175, 1–13. [Google Scholar] [CrossRef]
- Magariños, B.; Devesa, S. Furunculosis in Senegalese sole (Solea senegalensis) cultured in a recirculation system. Vet. Rec.-Engl. Ed. 2011, 168, 431. [Google Scholar] [CrossRef] [PubMed]
- Lago, E.P.; Nieto, T.P. Virulence factors of Aeromonas salmonicida subsp. salmonicida strains associated with infections in turbot Psetta maxima. Dis. Aquat. Org. 2012, 99, 145–151. [Google Scholar] [CrossRef] [Green Version]
- Briones, V.; Fernandez, A. Haemorrhagic Septicaemia by Aeromonas salmonicida subsp. salmonicida in a Black-tip Reef Shark (Carcharhinus melanopterus). J. Vet. Med. Ser. B 1998, 45, 443–445. [Google Scholar] [CrossRef]
- Burr, S.E.; Pugovkin, D. Attenuated virulence of an Aeromonas salmonicida subsp. salmonicida type III secretion mutant in a rainbow trout model. Microbiology 2005, 151, 2111–2118. [Google Scholar] [CrossRef] [Green Version]
- Orozova, P.; Barker, M. Identification and pathogenicity to rainbow trout, Oncorhynchus mykiss (Walbaum), of some aeromonads. J. Fish Dis. 2009, 32, 865–871. [Google Scholar] [CrossRef]
- Marcoux, P.É.; Vincent, A.T. Systematic analysis of the stress-induced genomic instability of Type Three Secretion System in Aeromonas salmonicida subsp. salmonicida. Microorganisms 2020, 9, 85. [Google Scholar] [CrossRef]
- Balado, M.; Souto, A. Two catechol siderophores, acinetobactin and amonabactin, are simultaneously produced by Aeromonas salmonicida subsp. salmonicida sharing part of the biosynthetic pathway. ACS Chem. Biol. 2015, 10, 2850–2860. [Google Scholar] [CrossRef]
- Schwenteit, J.; Gram, L. Quorum sensing in Aeromonas salmonicida subsp. achromogenes and the effect of the autoinducer synthase AsaI on bacterial virulence. Vet. Microbiol. 2011, 147, 389–397. [Google Scholar] [CrossRef]
- Garduño, R.A.; Moore, A.R. Host cell invasion and intracellular residence by Aeromonas salmonicida: Role of the S-layer. Can. J. Microbiol. 2000, 46, 660–668. [Google Scholar] [CrossRef]
- Daher, R.K.; Filion, G. Alteration of virulence factors and rearrangement of pAsa5 plasmid caused by the growth of Aeromonas salmonicida in stressful conditions. Vet. Microbiol. 2011, 152, 353–360. [Google Scholar] [CrossRef]
- Carniel, E.; Guilvout, I. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J. Bacteriol. 1996, 178, 6743–6751. [Google Scholar] [CrossRef] [Green Version]
- Russell, A.B.; Peterson, S.B. Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 2014, 12, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Cherrak, Y.; Flaugnatti, N. Structure and activity of the type VI secretion system. Microbiol. Spectr. 2019, 7, 7.4.11. [Google Scholar] [CrossRef] [Green Version]
- Bröms, J.E.; Meyer, L. DotU and VgrG, core components of type VI secretion systems, are essential for Francisella LVS pathogenicity. PLoS ONE 2012, 7, e34639. [Google Scholar] [CrossRef] [Green Version]
- Boyer, F.; Fichant, G. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: What can be learned from available microbial genomic resources? BMC Genom. 2009, 10, 104. [Google Scholar] [CrossRef] [Green Version]
- Silverman, J.M.; Agnello, D.M. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol. Cell 2013, 51, 584–593. [Google Scholar] [CrossRef] [Green Version]
- Hachani, A.; Wood, T.E. Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 2016, 29, 81–93. [Google Scholar] [CrossRef]
- Sha, J.; Rosenzweig, J.A. Evaluation of the roles played by Hcp and VgrG type 6 secretion system effectors in Aeromonas hydrophila SSU pathogenesis. Microbiology 2013, 159 Pt 6, 1120–1135. [Google Scholar] [CrossRef]
- Manera, K.; Caro, F. Sensing of intracellular Hcp levels controls T6SS expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2021, 118, e2104813118. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tao, J. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infect. Immun. 2012, 80, 1243–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carruthers, D.; Nicholson, A. Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS ONE 2013, 8, e59388. [Google Scholar] [CrossRef]
- Huang, L.; Qi, W. The immune response of a warm water fish orange-spotted grouper (Epinephelus coioides) infected with a typical cold water bacterial pathogen Aeromonas salmonicida is AhR dependent. Dev. Comp. Immunol. 2020, 113, 103779. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Qi, W. Insights into mesophilic virulence, antibiotic resistant and human pathogenicity: A genomics study on the Aeromonas salmonicida SRW-OG1 newly isolated from the Asian fish Epinephelus coioides. Aquaculture 2021, 539, 736630. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Yang, J.; Roy, A. BioLiP: A semi-manually curated database for biologically relevant ligand–protein interactions. Nucleic Acids Res. 2012, 41, D1096–D1103. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Zhang, Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucleic Acids Res 2007, 35, 3375–3382. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Kihara, D. Local energy landscape flattening: Parallel hyperbolic Monte Carlo sampling of protein folding. Proteins 2002, 48, 192–201. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y. REMO: A new protocol to refine full atomic protein models from C-alpha traces by optimizing hydrogen-bonding networks. Proteins 2009, 76, 665–676. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Liang, Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure 2011, 19, 1784–1795. [Google Scholar] [CrossRef]
- Roy, A.; Xu, D. A protocol for computer-based protein structure and function prediction. J. Vis. Exp. 2011, 57, e3259. [Google Scholar] [CrossRef] [Green Version]
- Webb, E.C. Enzyme Nomenclature 1992: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes; Academic Press: Cambridge, MA, USA, 1992. [Google Scholar]
- Luo, G.; Huang, L. flrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerg. Microbes Infect. 2016, 5, e85. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Liu, Y.; Teng, X.; Luan, P.; Teng, X.; Yin, X. Immunosuppression participated in complement activation-mediated inflammatory injury caused by 4-octylphenol via TLR7/IκBα/NF-κB pathway in common carp (Cyprinus carpio) gills. Aquat. Toxicol. 2022, 249, 106211. [Google Scholar] [CrossRef]
- Cui, J.; Zhou, Q.; Yu, M.; Liu, Y.; Teng, X.; Gu, X. 4-tert-butylphenol triggers common carp hepatocytes ferroptosis via oxidative stress, iron overload, SLC7A11/GSH/GPX4 axis, and ATF4/HSPA5/GPX4 axis. Ecotoxicol. Environ. Saf. 2022, 242, 113944. [Google Scholar] [CrossRef]
- Choi, K.H.; Schweizer, H.P. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat. Protoc. 2006, 1, 153–161. [Google Scholar] [CrossRef]
- Darsigny, M.; Babeu, J.-P. Hepatocyte nuclear factor-4α promotes gut neoplasia in mice and protects against the production of reactive oxygen species. Cancer Res. 2010, 70, 9423–9433. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Y.; Zhao, L. Mechanisms underlying the virulence regulation of new Vibrio alginolyticus ncRNA Vvrr1 with a comparative proteomic analysis. Emerg. Microbes Infect. 2019, 8, 1604–1618. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Zhao, L. The Effect of tonB Gene on the Virulence of Pseudomonas plecoglossicida and the Immune Response of Epinephelus coioides. Front. Microbiol. 2021, 12, 720967. [Google Scholar] [CrossRef]
- Mao, L.; Qin, Y. Role of LuxR-type regulators in fish pathogenic Aeromonas hydrophila. J. Fish Dis. 2020, 43, 215–225. [Google Scholar] [CrossRef]
- Huang, L.; Huang, L. The TCA pathway is an important player in the regulatory network governing Vibrio alginolyticus adhesion under adversity. Front. Microbiol. 2016, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Zuo, Y. Copper stress by nutritional immunity activates the CusS-CusR two-component system that contributes to Vibrio alginolyticus anti-host response but affects virulence-related properties. Aquaculture 2021, 532, 736012. [Google Scholar] [CrossRef]
- Tsou, A.M.; Zhu, J. Quorum sensing negatively regulates hemolysin transcriptionally and posttranslationally in Vibrio cholerae. Infect. Immun. 2010, 78, 461–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Zhang, Y. Phenotypic characterization, virulence, and immunogenicity of Pseudomonas plecoglossicida rpoE knock-down strain. Fish Shellfish. Immunol. 2019, 87, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zuo, Y. The Zinc Nutritional Immunity of Epinephelus Coioides Contributes to the Importance of znuC During Pseudomonas plecoglossicida Infection. Front. Immunol. 2021, 12, 678699. [Google Scholar] [CrossRef]
- Lutwyche, P.; Exner, M.M. A conserved Aeromonas salmonicida porin provides protective immunity to rainbow trout. Infect. Immun. 1995, 63, 3137–3142. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Li, J. Extracellular products-mediated interspecific interaction between Pseudomonas aeruginosa and Escherichia coli. J. Microbiol. 2021, 59, 29–40. [Google Scholar] [CrossRef]
- Kim, H.S.; Park, H.D. Ginger extract inhibits biofilm formation by Pseudomonas aeruginosa PA14. PLoS ONE 2013, 8, e76106. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Zuo, Y. A metabolomic investigation into the temperature-dependent virulence of Pseudomonas plecoglossicida from large yellow croaker (Pseudosciaena crocea). J. Fish Dis. 2019, 42, 431–446. [Google Scholar] [CrossRef]
- Hayes, S.; Aoki, K. Bacterial contact-dependent delivery systems. Annu. Rev. Genet. 2010, 44, 71–90. [Google Scholar] [CrossRef]
- Williams, G.; Varcoe, T. Vibrio cholerae Hcp, a secreted protein coregulated with HlyA. Infect. Immun. 1996, 64, 283–289. [Google Scholar] [CrossRef]
- Pukatzki, S.; Ma, T. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 2006, 103, 1528–1533. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Leung, Y. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 2007, 66, 1192–1206. [Google Scholar] [CrossRef]
- Mougous, J.D.; Cuff, M.E. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 2006, 312, 1526–1530. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Kucukural, A. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Russel, B.; Hood, D. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 2011, 475, 343–347. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Brackmann, M. Cryo-EM structure of the extended type VI secretion system sheath–tube complex. Nat. Microbiol. 2017, 2, 1507–1512. [Google Scholar] [CrossRef]
- Gallique, M.; Bouteiller, M. The Type VI Secretion System: A Dynamic System for Bacterial Communication? Front. Microbiol. 2017, 8, 1454. [Google Scholar] [CrossRef]
- Pukatzki, S.; McAuley, S.B. The type VI secretion system: Translocation of effectors and effector-domains. Curr. Opin. Microbiol. 2009, 12, 11–17. [Google Scholar] [CrossRef]
- Kuhlman, B.; Bradley, P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. [Google Scholar] [CrossRef]
- Aubert, F.; Flannagan, S. A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect. Immun. 2008, 76, 1979–1991. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Hao, S. The Type VI Secretion System Modulates Flagellar Gene Expression and Secretion in Citrobacter freundii and Contributes to Adhesion and Cytotoxicity to Host Cells. Infect. Immun. 2015, 83, 2596–2604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Liu, J. Diverse roles of Hcp family proteins in the environmental fitness and pathogenicity of Aeromonas hydrophila Chinese epidemic strain NJ-35. Appl. Microbiol. Biotechnol. 2018, 102, 7083–7095. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Fang, L. VgrG2 of type VI secretion system 2 of Vibrio parahaemolyticus induces autophagy in macrophages. Front. Microbiol. 2015, 6, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bleumink-Pluym, N.M.; van Alphen, L.B. Identification of a functional type VI secretion system in Campylobacter jejuni conferring capsule polysaccharide sensitive cytotoxicity. PLoS Pathog. 2013, 9, e1003393. [Google Scholar] [CrossRef] [Green Version]
- Labella, A.M.; Rosado, J.J. Virulence properties of three new Photobacterium species affecting cultured fish. J. Appl. Microbiol. 2020, 129, 37–50. [Google Scholar] [CrossRef]
- Santos, Y.; Bandin, I. Comparison of the extracellular biological activities of Vibrio anguillarum and Aeromonas hydrophila. Aquaculture 1992, 107, 259–270. [Google Scholar] [CrossRef]
- Li, J.; Ni, X. Detection of three virulence genes alt, ahp and aerA in Aeromonas hydrophila and their relationship with actual virulence to zebrafish. J. Appl. Microbiol. 2011, 110, 823–830. [Google Scholar] [CrossRef]
- Zhou, Q.L.; Wang, Y. Distribution and virulence gene comparison of Aeromonas strains isolated from diseased fish and water environment. Pol. J. Microbiol. 2013, 62, 299–302. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, X. Roles of Hcp family proteins in the pathogenesis of the porcine extraintestinal pathogenic Escherichia coli type VI secretion system. Sci. Rep. 2016, 6, 26816. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Caro, F. Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 2018, 359, 210–213. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, H.; Yu, J.; Qiao, Y.; Ma, Y.; Zheng, J.; Lin, M.; Yan, Q.; Huang, L. Effect of the Type VI Secretion System Secreted Protein Hcp on the Virulence of Aeromonas salmonicida. Microorganisms 2022, 10, 2307. https://doi.org/10.3390/microorganisms10122307
Cai H, Yu J, Qiao Y, Ma Y, Zheng J, Lin M, Yan Q, Huang L. Effect of the Type VI Secretion System Secreted Protein Hcp on the Virulence of Aeromonas salmonicida. Microorganisms. 2022; 10(12):2307. https://doi.org/10.3390/microorganisms10122307
Chicago/Turabian StyleCai, Hongyan, Jiaying Yu, Ying Qiao, Ying Ma, Jiang Zheng, Mao Lin, Qingpi Yan, and Lixing Huang. 2022. "Effect of the Type VI Secretion System Secreted Protein Hcp on the Virulence of Aeromonas salmonicida" Microorganisms 10, no. 12: 2307. https://doi.org/10.3390/microorganisms10122307




