Microbiome in Male Genital Mucosa (Prepuce, Glans, and Coronal Sulcus): A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Inclusion and Exclusion Criteria
2.3. Study Selection
2.4. Data Extraction
2.5. Study Quality Assessment
3. Results
3.1. Number of Retrieved Papers
3.2. Pediatric Foreskin Microbiome
3.3. Circumcision and Coronal Sulcus Microbiome
3.4. Male Genital Mucosa Microbiome and Bacterial Vaginosis
3.5. The Microbiome in Genital Mucosa Inflammation
3.6. Influence of Probiotic Supplementation on Glans Microbiome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waldman, A.J.; Balskus, E.P. The human microbiota, infectious disease, and global health: Challenges and opportunities. ACS Infect. Dis. 2018, 4, 14–26. [Google Scholar] [CrossRef]
- Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koedooder, R.; Mackens, S.; Budding, A.; Fares, D.; Blockeel, C.; Laven, J.; Schoenmakers, S. Identification and evaluation of the microbiome in the female and male reproductive tracts. Hum. Reprod. Update 2019, 25, 298–325. [Google Scholar] [CrossRef]
- Slingerland, A.E.; Schwabkey, Z.; Wiesnoski, D.H.; Jenq, R.R. Clinical evidence for the microbiome in inflammatory diseases. Front. Immunol. 2017, 8, 400. [Google Scholar] [CrossRef] [Green Version]
- Dreno, B.; Araviiskaia, E.; Berardesca, E.; Gontijo, G.; Sanchez Viera, M.; Xiang, L.F.; Martin, R.; Bieber, T. Microbiome in healthy skin, update for dermatologists. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 2038–2047. [Google Scholar] [CrossRef] [PubMed]
- Saus, E.; Iraola-Guzman, S.; Willis, J.R.; Brunet-Vega, A.; Gabaldon, T. Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Mol. Asp. Med. 2019, 69, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Katongole, P.; Sande, O.J.; Joloba, M.; Reynolds, S.J.; Niyonzima, N. The human microbiome and its link in prostate cancer risk and pathogenesis. Infect. Agent Cancer 2020, 15, 53. [Google Scholar] [CrossRef]
- Pagan, L.; Ederveen, R.A.M.; Huisman, B.W.; Schoones, J.W.; Zwittink, R.D.; Schuren, F.H.J.; Rissmann, R.; Piek, J.M.J.; Van Poelgeest, M.I.E. The human vulvar microbiome: A systematic review. Microorganisms 2021, 9, 2568. [Google Scholar] [CrossRef]
- Altmae, S.; Franasiak, J.M.; Mandar, R. The seminal microbiome in health and disease. Nat. Rev. Urol. 2019, 16, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Pattaroni, C.; Vulliemoz, N.; Castella, V.; Marsland, B.J.; Stojanov, M. Sperm microbiota and its impact on semen parameters. Front. Microbiol. 2019, 10, 234. [Google Scholar] [CrossRef]
- Tuddenham, S.; Ravel, J.; Marrazzo, J.M. Protection and risk: Male and female genital microbiota and sexually transmitted infections. J. Infect. Dis. 2021, 223, S222–S235. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.V.; Kafka, J.K.; Ferreira, V.H.; Roth, K.; Kaushic, C. Innate and adaptive immune responses in male and female reproductive tracts in homeostasis and following HIV infection. Cell. Mol. Immunol. 2014, 11, 410–427. [Google Scholar] [CrossRef] [Green Version]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [Green Version]
- National Heart Lung and Blood Institute. Study Quality Assessment Tools; NHLBI: Bethesda, MD, USA, 2019.
- Sterne, J.A.C.; Savovic, J.; Page, M.J.; Elbers, R.G.; Blencowe, N.S.; Boutron, I.; Cates, C.J.; Cheng, H.Y.; Corbett, M.S.; Eldridge, S.M.; et al. RoB 2: A revised tool for assessing risk of bias in randomised trials. BMJ 2019, 366, l4898. [Google Scholar] [CrossRef] [Green Version]
- Iniesta, S.; Esteban, S.; Armijo, O.; Lobo, S.; Manzano, S.; Espinosa, I.; Cardenas, N.; Bartha, J.L.; Jimenez, E. Ligilactobacillus salivarius PS11610 exerts an effect on the microbial and immunological profile of couples suffering unknown infertility. Am. J. Reprod. Immunol. 2022, 88, e13552. [Google Scholar] [CrossRef]
- Li, M.; Mao, J.X.; Jiang, H.H.; Huang, C.M.; Gao, X.H.; Zhang, L. Microbiome profile in patients with adult balanoposthitis: Relationship with redundant prepuce, genital mucosa physical barrier status and inflammation. Acta Derm. Venereol. 2021, 101, adv00466. [Google Scholar] [CrossRef]
- Liu, C.M.; Hungate, B.A.; Tobian, A.A.; Serwadda, D.; Ravel, J.; Lester, R.; Kigozi, G.; Aziz, M.; Galiwango, R.M.; Nalugoda, F.; et al. Male circumcision significantly reduces prevalence and load of genital anaerobic bacteria. mBio 2013, 4, e00076. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.M.; Prodger, J.L.; Tobian, A.A.R.; Abraham, A.G.; Kigozi, G.; Hungate, B.A.; Aziz, M.; Nalugoda, F.; Sariya, S.; Serwadda, D.; et al. Penile anaerobic dysbiosis as a risk factor for HIV infection. mBio 2017, 8, e00996-17. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.E.; Dong, Q.; Van der Pol, B.; Toh, E.; Fan, B.; Katz, B.P.; Mi, D.; Rong, R.; Weinstock, G.M.; Sodergren, E.; et al. Bacterial communities of the coronal sulcus and distal urethra of adolescent males. PLoS ONE 2012, 7, e36298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plummer, E.L.; Vodstrcil, L.A.; Doyle, M.; Danielewski, J.A.; Murray, G.L.; Fehler, G.; Fairley, C.K.; Bulach, D.M.; Garland, S.M.; Chow, E.P.F.; et al. A prospective, open-label pilot study of concurrent male partner treatment for bacterial vaginosis. mBio 2021, 12, e0232321. [Google Scholar] [CrossRef]
- Price, L.B.; Liu, C.M.; Johnson, K.E.; Aziz, M.; Lau, M.K.; Bowers, J.; Ravel, J.; Keim, P.S.; Serwadda, D.; Wawer, M.J.; et al. The effects of circumcision on the penis microbiome. PLoS ONE 2010, 5, e8422. [Google Scholar] [CrossRef] [Green Version]
- Storm, D.W.; Copp, H.L.; Halverson, T.M.; Du, J.; Juhr, D.; Wolfe, A.J. A Child’s urine is not sterile: A pilot study evaluating the Pediatric Urinary Microbiome. J. Pediatr. Urol. 2022, 18, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Watchorn, R.E.; Van den Munckhof, E.H.A.; Quint, K.D.; Eliahoo, J.; de Koning, M.N.C.; Quint, W.G.V.; Bunker, C.B. Balanopreputial sac and urine microbiota in patients with male genital lichen sclerosus. Int. J. Dermatol. 2021, 60, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Zozaya, M.; Ferris, M.J.; Siren, J.D.; Lillis, R.; Myers, L.; Nsuami, M.J.; Eren, A.M.; Brown, J.; Taylor, C.M.; Martin, D.H. Bacterial communities in penile skin, male urethra, and vaginas of heterosexual couples with and without bacterial vaginosis. Microbiome 2016, 4, 16. [Google Scholar] [CrossRef] [Green Version]
- Sha, B.E.; Chen, H.Y.; Wang, Q.J.; Zariffard, M.R.; Cohen, M.H.; Spear, G.T. Utility of Amsel criteria, Nugent score, and quantitative PCR for Gardnerella vaginalis, Mycoplasma hominis, and Lactobacillus spp. for diagnosis of bacterial vaginosis in human immunodeficiency virus-infected women. J. Clin. Microbiol. 2005, 43, 4607–4612. [Google Scholar] [CrossRef] [Green Version]
- Karaduta, O.; Dvanajscak, Z.; Zybailov, B. Metaproteomics—An advantageous option in studies of host-microbiota interaction. Microorganisms 2021, 9, 980. [Google Scholar] [CrossRef]
- Buffet-Bataillon, S.; Rizk, G.; Cattoir, V.; Sassi, M.; Thibault, V.; Del Giudice, J.; Gangneux, J.P. Efficient and quality-optimized metagenomic pipeline designed for taxonomic classification in routine microbiological clinical tests. Microorganisms 2022, 10, 711. [Google Scholar] [CrossRef]
- Pérez-Cobas, A.E.; Gosalbes, M.J.; Friedrichs, A.; Knecht, H.; Artacho, A.; Eismann, K.; Otto, W.; Rojo, D.; Bargiela, R.; Von Bergen, M.; et al. Gut microbiota disturbance during antibiotic therapy: A multi-omic approach. Gut 2013, 62, 1591–1601. [Google Scholar] [CrossRef]
- Wu, I.W.; Gao, S.S.; Chou, H.C.; Yang, H.Y.; Chang, L.C.; Kuo, Y.L.; Vinh Dinh, M.C.; Chung, W.H.; Yang, C.W.; Lai, H.C.; et al. Integrative metagenomic and metabolomic analyses reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics 2020, 10, 5398. [Google Scholar] [CrossRef]
- Feng, Y.; Ramnarine, V.R.; Bell, R.; Volik, S.; Davicioni, E.; Hayes, V.M.; Ren, S.; Collins, C.C. Metagenomic and metatranscriptomic analysis of human prostate microbiota from patients with prostate cancer. BMC Genom. 2019, 20, 146. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Tseng, C.H.; Pei, Z.; Blaser, M.J. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. USA 2007, 104, 2927–2932. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E. Prevotella in the gut: Choose carefully. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 69–70. [Google Scholar] [CrossRef] [PubMed]
- Campisciano, G.; Iebba, V.; Zito, G.; Luppi, S.; Martinelli, M.; Fischer, L.; De Seta, F.; Basile, G.; Ricci, G.; Comar, M. Lactobacillus iners and gasseri, Prevotella bivia and HPV belong to the microbiological signature negatively affecting human reproduction. Microorganisms 2020, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.C.; Frick, I.M. Gram-positive anaerobic cocci-commensals and opportunistic pathogens. FEMS Microbiol. Rev. 2013, 37, 520–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandar, R. Microbiota of male genital tract: Impact on the health of man and his partner. Pharmacol. Res. 2013, 69, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Sengupta, P.; Izuka, E.; Menuba, I.; Jegasothy, R.; Nwagha, U. Staphylococcal infections and infertility: Mechanisms and management. Mol. Cell. Biochem. 2020, 474, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.L. Antimicrobial activity of host-derived lipids. Antibiotics 2020, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Baurecht, H.; Ruhlemann, M.C.; Rodriguez, E.; Thielking, F.; Harder, I.; Erkens, A.S.; Stolzl, D.; Ellinghaus, E.; Hotze, M.; Lieb, W.; et al. Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J. Allergy Clin. Immunol. 2018, 141, 1668–1676. [Google Scholar] [CrossRef] [Green Version]
- Iacob, S.I.; Feinn, R.S.; Sardi, L. Systematic review of complications arising from male circumcision. BJUI Compass 2022, 3, 99–123. [Google Scholar] [CrossRef]
- Dong, Q.; Nelson, D.E.; Toh, E.; Diao, L.; Gao, X.; Fortenberry, J.D.; Van der Pol, B. The microbial communities in male first catch urine are highly similar to those in paired urethral swab specimens. PLoS ONE 2011, 6, e19709. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.E.; Van Der Pol, B.; Dong, Q.; Revanna, K.V.; Fan, B.; Easwaran, S.; Sodergren, E.; Weinstock, G.M.; Diao, L.; Fortenberry, J.D. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS ONE 2010, 5, e14116. [Google Scholar] [CrossRef]
- Yang, X.; Da, M.; Zhang, W.; Qi, Q.; Zhang, C.; Han, S. Role of Lactobacillus in cervical cancer. Cancer Manag. Res. 2018, 10, 1219–1229. [Google Scholar] [CrossRef] [Green Version]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef] [Green Version]
- Heidrich, V.; Inoue, L.T.; Asprino, P.F.; Bettoni, F.; Mariotti, A.C.H.; Bastos, D.A.; Jardim, D.L.F.; Arap, M.A.; Camargo, A.A. Choice of 16S ribosomal RNA primers impacts male urinary microbiota profiling. Front. Cell. Infect. Microbiol. 2022, 12, 862338. [Google Scholar] [CrossRef]
- Kameoka, S.; Motooka, D.; Watanabe, S.; Kubo, R.; Jung, N.; Midorikawa, Y.; Shinozaki, N.O.; Sawai, Y.; Takeda, A.K.; Nakamura, S. Benchmark of 16S rRNA gene amplicon sequencing using Japanese gut microbiome data from the V1–V2 and V3–V4 primer sets. BMC Genom. 2021, 22, 527. [Google Scholar] [CrossRef]
- Hoffman, C.; Siddiqui, N.Y.; Fields, I.; Gregory, W.T.; Simon, H.M.; Mooney, M.A.; Wolfe, A.J.; Karstens, L. Species-level resolution of female bladder microbiota from 16S rRNA amplicon sequencing. mSystems 2021, 6, e00518–e00521. [Google Scholar] [CrossRef]
- Graspeuntner, S.; Loeper, N.; Kunzel, S.; Baines, J.F.; Rupp, J. Selection of validated hypervariable regions is crucial in 16S-based microbiota studies of the female genital tract. Sci. Rep. 2018, 8, 9678. [Google Scholar] [CrossRef]

| Author | Study Design | Field of Study | Participants | Age (Years) | Ethnicity, Country | Anatomical Site | Sample Type | Microbial Analysis | Key Findings | Limitations | Quality Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
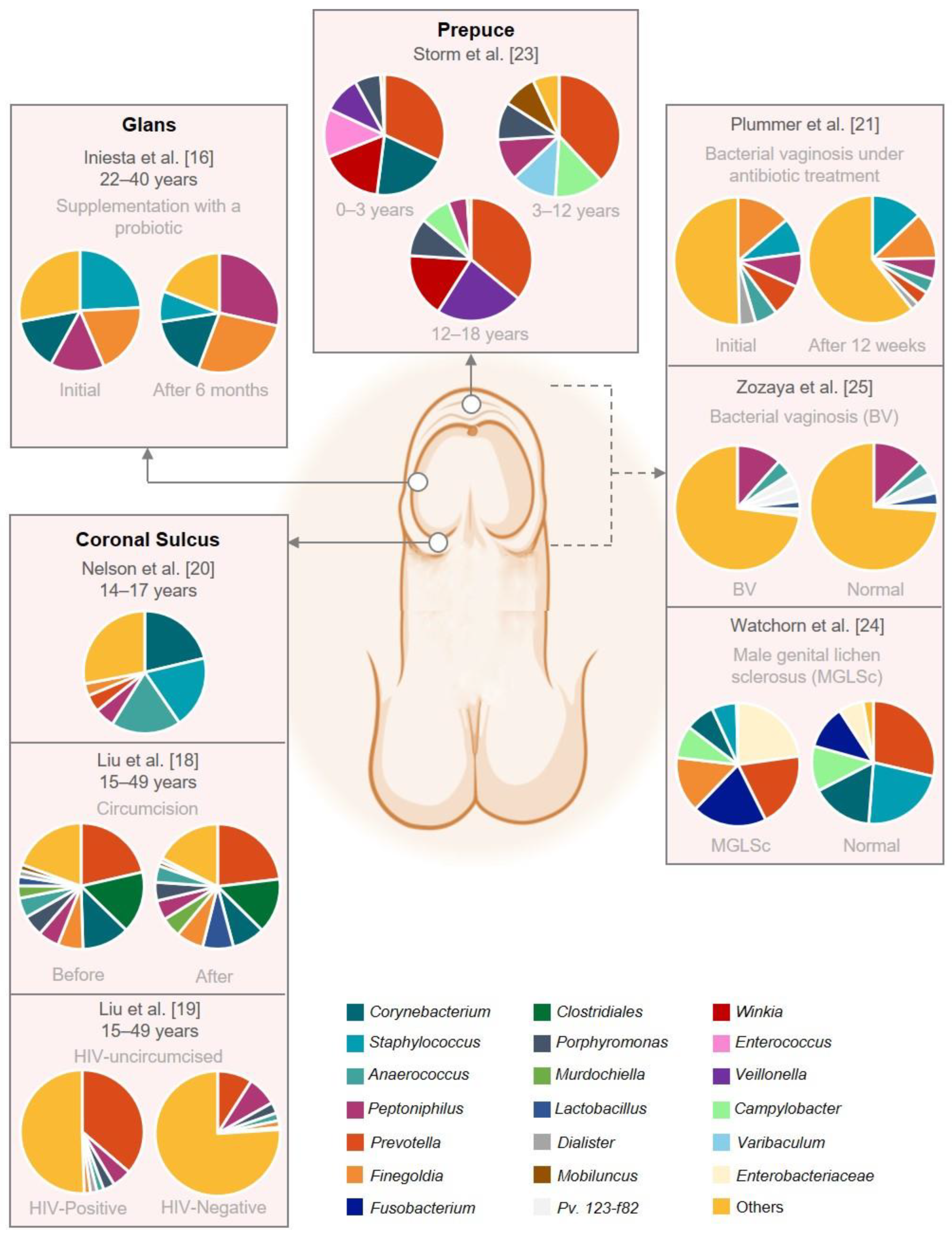
| Iniesta et al. [16] | Control intervention | Supplementation with a probiotic | 17 couples under artificial reproductive treatment | Couples: 22–40 Male (mean 36) Female (mean 35) | Caucasian, Spain | Glans | Glans, semen, and blood | 16S rRNA amplification of the V3–V4 region | Treatment with Lactobacillus salivarius PS11610 modified the microbiota composition improving the urogenital tract microbiome, solving the dysbiosis in 88.9% of the couples. Male samples showed higher bacterial diversity at genus level than female samples. Most prevalent genera in glans: Initial: Peptoniphilus, Staphylococcus, Fineglodia, and Corynebacterium; 3 and 6 months: Peptonophilus, Finegoldia, Corynebacterium (↓ Staphylococcus) | Small sample size. No control group treated with placebo. | Fair |
| Li et al. [17] | Case control | Balanoposthitis (BP) | 26 BP uncircumcised 29 HV uncircumcised | 18–65 | Unknown, China | Glans, penis, and prepuce | Swabs | 16S rRNA amplification of the V4 region | Microbiome BP ∼ HV, but ≠ HV with redundant prepuce. Most prevalent species BP: Staphylococcus warneri (with condom use) and Prevotella bivia (without sexual activity). Most prevalent HV: Ezakiella (redundant prepuce), Porphyromonas somerae (normal prepuce). | Small sample size. No ethnicity records. V4 region is considered a relatively low informative region for taxonomic assignment. | Fair |
| Liu et al. [18] | Randomized controlled trial | Circumcision | 77 HV uncircumcised 79 HIV-negative pre- and post-circumcision | 15–49 | Unknown, USA | Coronal sulcus (CS) | Swabs | 16S rRNA amplification of the V3-V6 region | Male circumcision reduced the prevalence and the absolute abundance of CS bacteria and the diversity of microbiota. Day 0: Prevalent but low abundance: Prevotella sp., Clostridiales and Corynebacterium sp. At 5%: Peptoniphilus sp., Anaerococcus sp., Fenigoldia sp., Murdochiella sp., Porphyromonas sp., and Lactobacillus sp. Year 1: Reduction in bacterial load on post-circumcision: Porphymonas sp., Prevotella sp., Negativicoccus sp., Dialister sp., Mobiluncus sp., and 6 genera from Clostridiales family XI. No reduction: Atopolium sp., Sneathia sp., and Megasphaera. More prevalent: Kocuria sp. and Facklamia sp. | No ethnicity records. | Good |
| Liu et al. [19] | Case control | Circumcision and Human immunodeficiency virus (HIV) | 46 HIV-positive uncircumcised 136 HIV-negative uncircumcised | 15–49 | Unknown, Uganda | Coronal sulcus | Swabs | 16S rRNA amplification of the V3–V4 region | HIV-positive uncircumcised: ↑ penile anaerobes. Most prevalent genera HIV-positive uncircumcised: Prevotella, Dialister, Mobiluncus, Murdochiella, and Peptostreptococcus. | No ethnicity records. | Good |
| Nelson et al. [20] | Observational cohort | Circumcision | 18 HV (6 uncircumcised and 12 circumcised) | 14–17 | Mixed Black, Caucasian, and Latin, USA | Coronal sulcus | Swabs and first catch urine | 16S rRNA amplification of the V1–V3, V3–V5, and V6–V9 regions | Microbiome CS ≠ urine. Most prevalent genera CS: Corynebacteria, Staphylococcus, Anaerococcus, Peptoniphilus, Prevotella, Finegoldia, Porphyromonas, Propionibacterium, and Delftia. Uncircumcised: ↑ Prevotella and Porphyromonas. Most prevalent genera urine: Streptococcus, Lactobacillus, Gardnerella, and Veillonella. | Small sample size. | Fair |
| Plummer et al. [21] | Randomized controlled trial | Bacterial vaginosis under antibiotic treatment | 27 couples | >18 | Unknown, Australia | Penis | Swab and first catch urine | 16S rRNA amplification of the V3–V4 region | Day 0: Male specimens were heterogeneous in composition. Most abundant in penile swab: Corynebacterium, Staphylococcus, Peptoniphilus, and Prevotella. Day 8: Decreased on penile swab taxa—Anaerococcus, Finegoldia, Peptoniphilus, Prevotela spp., and Dialister. (↑ Staphyloccocus) | Small sample size. Self-collected swab. | Fair |
| Price et al. [22] | Randomized controlled trial | Circumcision | 12 HIV-negative pre- and post-circumcision | 15–49 | Unknown, Uganda | Coronal sulcus | Swabs | 16S rRNA amplification of the V4 region | Microbiome post-circumcision: ↓ penile anaerobes. Most prevalent genera pre-circumcision: Anaerococcus, Peptoniphilus, Finegoldia, and Prevotella. Most prevalent genera post-circumcision: Staphylococcus and Corynebacterium. | Small sample size. No ethnicity records. | Poor |
| Storm et al. [23] | Observational cohort | Healthy | 48 HV males 18 HV females | 0–18 | Unknown, USA | Males: urethra, perineum, and foreskin Females: urethra, perineum, and vagina | Swabs | 16S rRNA amplification of the V4 region | Perineal microbiomes differed significantly by age; urethral and foreskin microbiomes did not. Most common genera foreskin: Prevotella, Staphylococcus, Corynebacterium, Peptoniphilus, Mobiluncus, and Winkia. | Small sample size. No record of the pubertal status of the older cohort. No ethnicity records. V4 region is considered a relatively low informative region for taxonomic assignment. | Fair |
| Watchorn et al. [24] | Case control | Male genital lichen sclerosus (MGLSc) | 20 MGLSc uncircumcised 20 HV uncircumcised | MGLSs:26–73 HV:19–63 | Unknown, UK | Balanopreputial sac (BS) (Glans + inner prepuce) | Swabs and first catch urine | 16S rRNA amplification of the V3–V4 region | Microbiome BS (MGLSc) ∼ urine (MGLSc). Microbiome BS (MGLSc) ≠ balanopreputial sac (HV). Most prevalent genera BS (MGLSc): Enterobacteriaceae, Prevotella, Fusobacterium, and Finegoldia. Most prevalent genera BS (HV): ↑ Finegoldia, Staphyloccocus, Corynebacterium. | Small sample size. No ethnicity records. | Poor |
| Zozaya et al. [25] | Cross-sectional | Bacterial vaginosis (BV) | 65 HV-males (23 circumcised) 65 BV-males (35 circumcised) | Mean 30.7 | African American, USA | Glans, coronal sulcus, penis | Swab | 16S rRNA amplification of the V4-V6 region | More penile skin diversity of BV-males than normal-males, but urethral diversity did not differ between groups. BV-associated bacteria were more abundant in penile and urethral specimens on BV-males. Most abundant on penile skin of BV-males: Peptoniphilus, Anaerococcus, Pv. 123-f-82, Pv. 123-b-46, Lactobacillus iners, and Pv.123-f-110. Most abundant on penile skin of HV: Peptoniphilus, Anaerococcus, Pv 123-f-82, L. iners, Porphyromonas, and Prevotela disiens. | No records of recruitment duration. | Good |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, M.F.M.; Fernandes, Â.R.; Rodrigues, A.G.; Lisboa, C. Microbiome in Male Genital Mucosa (Prepuce, Glans, and Coronal Sulcus): A Systematic Review. Microorganisms 2022, 10, 2312. https://doi.org/10.3390/microorganisms10122312
Gonçalves MFM, Fernandes ÂR, Rodrigues AG, Lisboa C. Microbiome in Male Genital Mucosa (Prepuce, Glans, and Coronal Sulcus): A Systematic Review. Microorganisms. 2022; 10(12):2312. https://doi.org/10.3390/microorganisms10122312
Chicago/Turabian StyleGonçalves, Micael F. M., Ângela Rita Fernandes, Acácio Gonçalves Rodrigues, and Carmen Lisboa. 2022. "Microbiome in Male Genital Mucosa (Prepuce, Glans, and Coronal Sulcus): A Systematic Review" Microorganisms 10, no. 12: 2312. https://doi.org/10.3390/microorganisms10122312
APA StyleGonçalves, M. F. M., Fernandes, Â. R., Rodrigues, A. G., & Lisboa, C. (2022). Microbiome in Male Genital Mucosa (Prepuce, Glans, and Coronal Sulcus): A Systematic Review. Microorganisms, 10(12), 2312. https://doi.org/10.3390/microorganisms10122312







