Dietary Nutritional Level Affects Intestinal Microbiota and Health of Goats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Study Design
2.2. Experimental Diet and Feeding Management
2.3. Sample Collection
2.4. Microbial gDNA Extraction and Illumina-MiSeq Sequencing
2.5. Determination of Functional Bacteria Copy Number
2.6. Determination of pH, VFAs, and Endotoxin (LPS) Content
2.7. Statistical Analysis
3. Results
3.1. Comparison of Growth Performance under Different Nutritional Levels
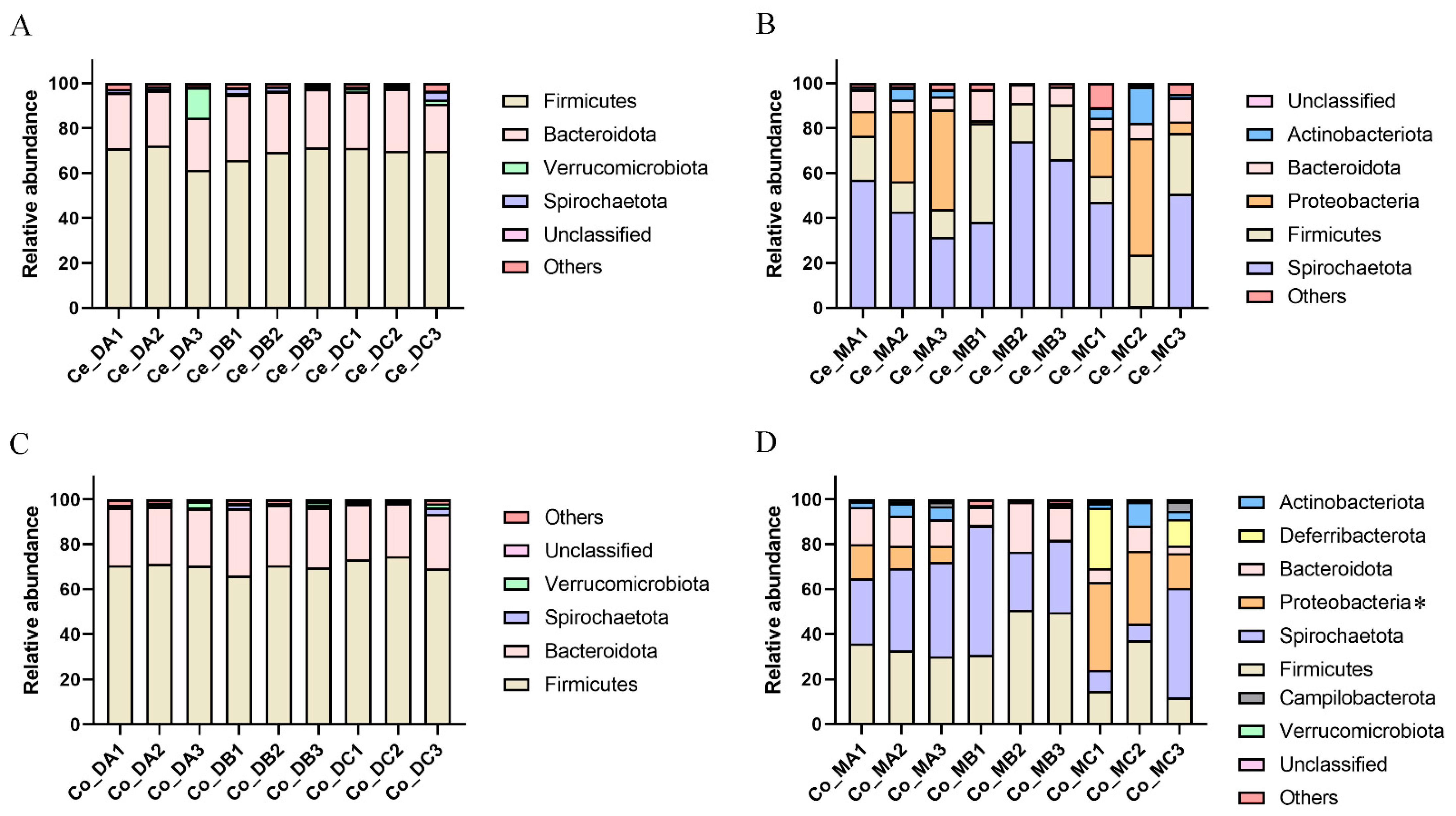
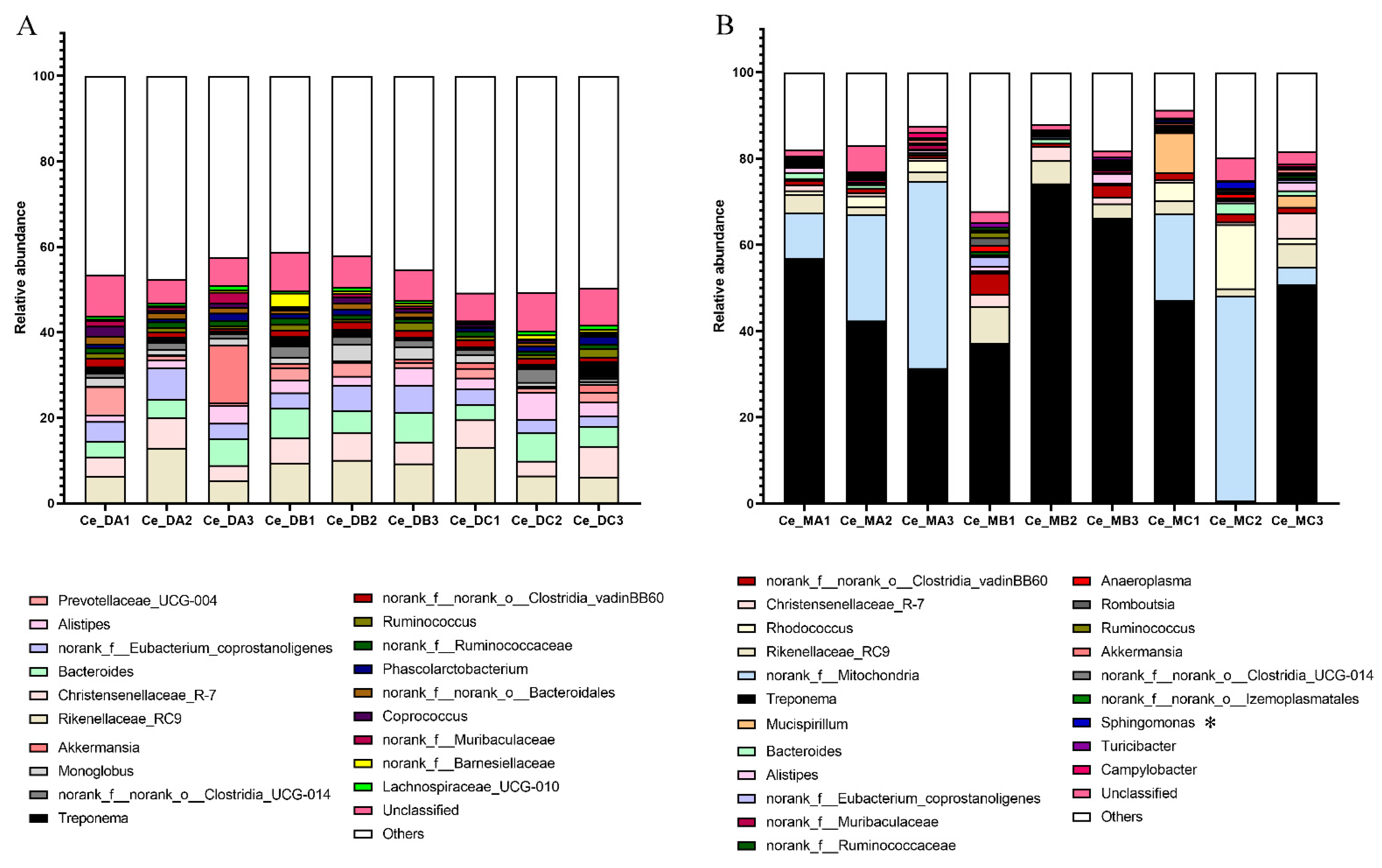
3.2. Comparison of Microbiota in the Cecum Digest under Different Nutritional Levels
3.3. Comparison of Microbiota in the Cecum Mucosa under Different Nutritional Levels
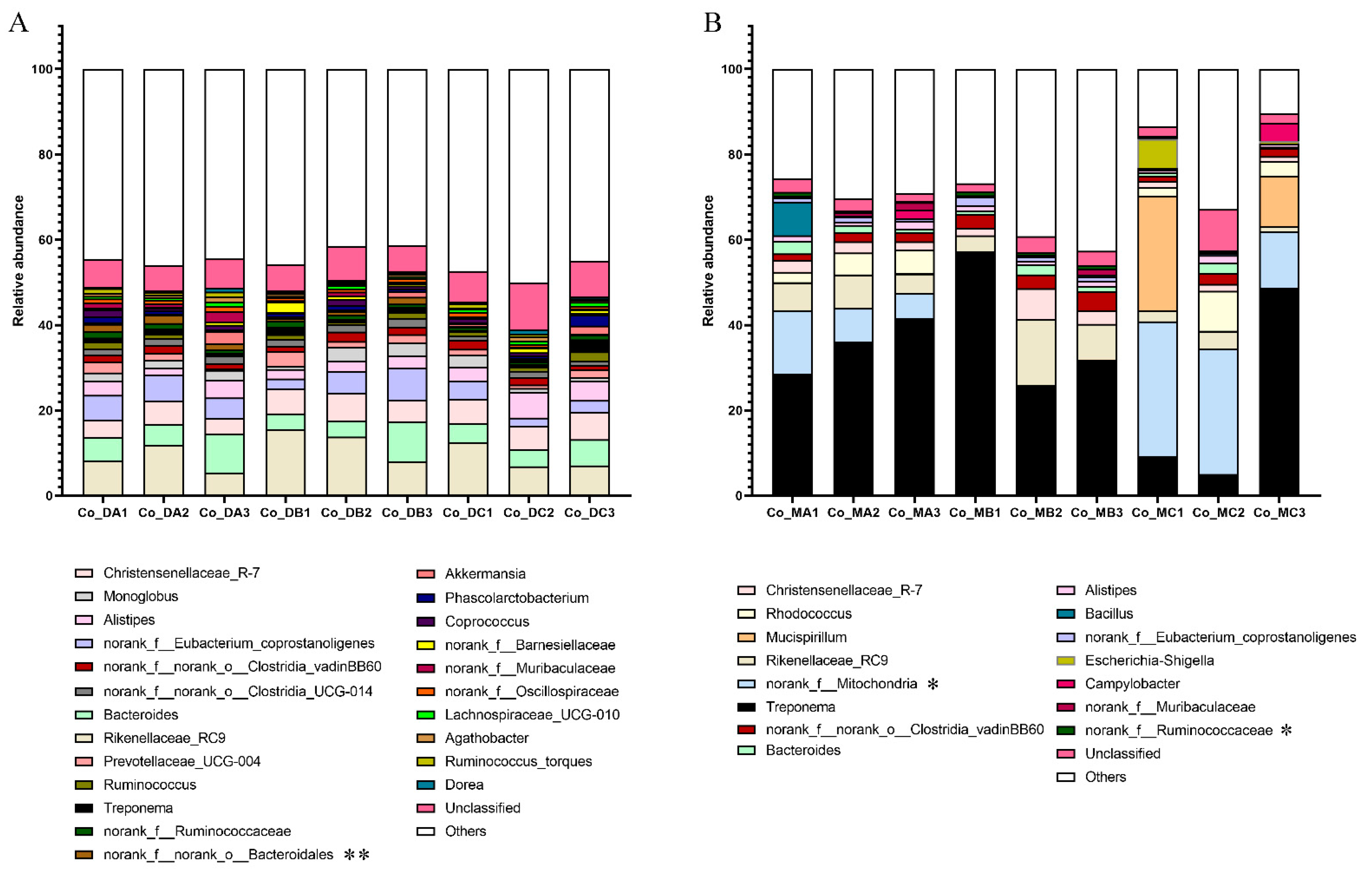
3.4. Comparison of Microbiota in the Colon Digest under Different Nutritional Levels
3.5. Comparison of Microbiota in the Colon Mucosa under Different Nutritional Levels
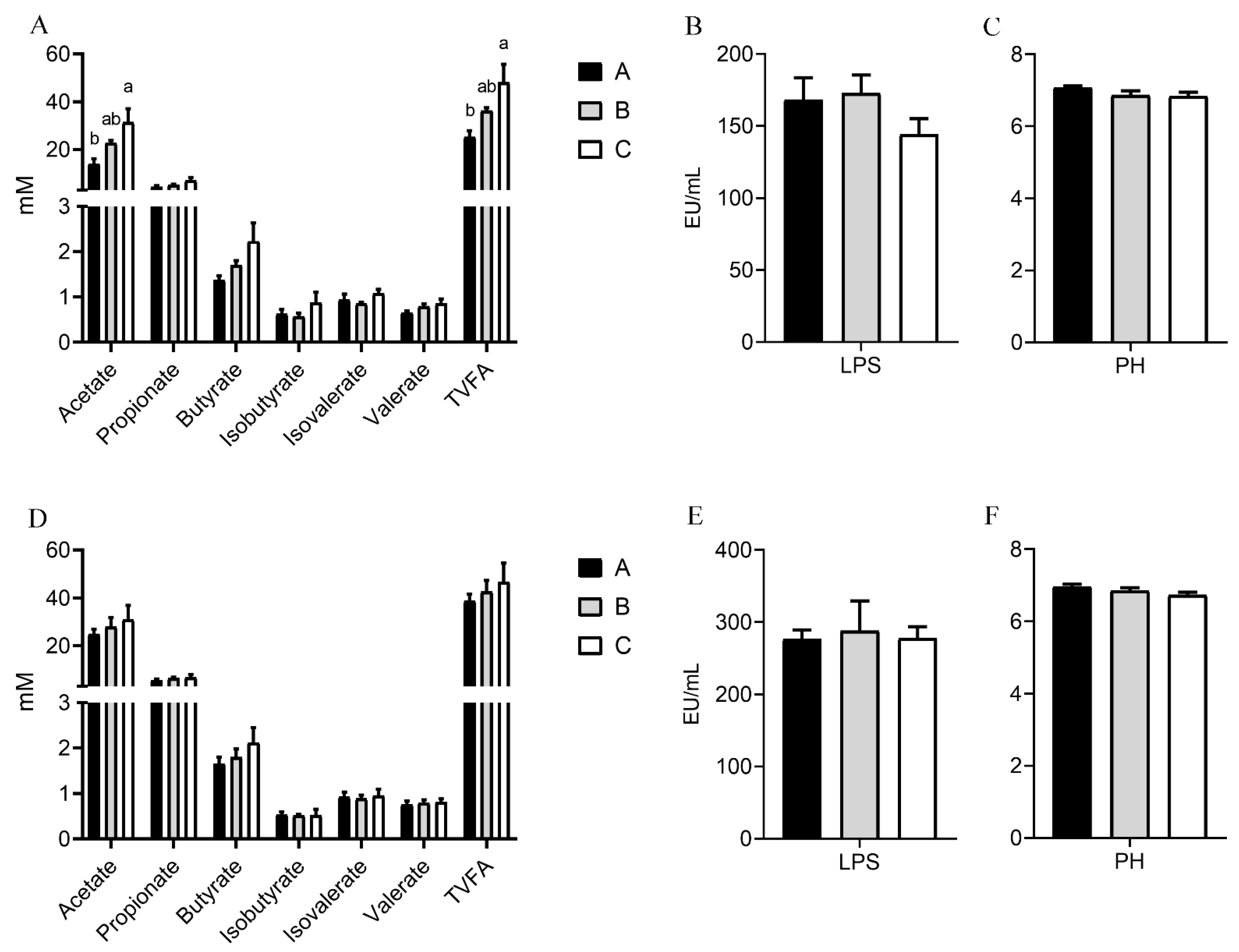
3.6. Effects of Dietary Nutritional Level on Intestinal pH, VFAs, and LPS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macfarlane, G.T.; Macfarlane, S. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012, 95, 50–60. [Google Scholar] [CrossRef]
- Chen, F.; Stappenbeck, T.S. Microbiome control of innate reactivity. Curr. Opin. Immunol. 2019, 56, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Wang, G.; Yu, L.; Tian, F.; Chen, W.; Zhai, Q. The effects of diet and gut microbiota on the regulation of intestinal mucin glycosylation. Carbohydr. Polym. 2021, 258, 117651. [Google Scholar] [CrossRef] [PubMed]
- Shan, K.; Qu, H.; Zhou, K.; Wang, L.; Zhu, C.; Chen, H.; Gu, Z.; Cui, J.; Fu, G.; Li, J.; et al. Distinct Gut Microbiota Induced by Different Fat-to-Sugar-Ratio High-Energy Diets Share Similar Pro-obesity Genetic and Metabolite Profiles in Prediabetic Mice. mSystems 2019, 4, e00219-19. [Google Scholar] [CrossRef] [Green Version]
- Tao, S.; Tian, P.; Luo, Y.; Tian, J.; Hua, C.; Geng, Y.; Cong, R.; Ni, Y.; Zhao, R. Microbiome-Metabolome Responses to a High-Grain Diet Associated with the Hind-Gut Health of Goats. Front. Microbiol. 2017, 8, 1764. [Google Scholar] [CrossRef]
- Jia, J.; Liang, C.; Wu, X.; Xiong, L.; Bao, P.; Chen, Q.; Yan, P. Effect of high proportion concentrate dietary on Ashdan Yak jejunal barrier and microbial function in cold season. Res. Veter. Sci. 2021, 140, 259–267. [Google Scholar] [CrossRef]
- Steele, M.A.; AlZahal, O.; Hook, S.E.; Croom, J.; McBride, B.W. Ruminal acidosis and the rapid onset of ruminal parakeratosis in a mature dairy cow: A case report. Acta Veter. Scand. 2009, 51, 39. [Google Scholar] [CrossRef] [Green Version]
- Caruso, R.; Lo, B.C.; Núñez, G. Host–microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020, 20, 411–426. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Duanmu, Y.; Dong, H.; Ni, Y.; Chen, J.; Shen, X.; Zhao, R. High Concentrate Diet Induced Mucosal Injuries by Enhancing Epithelial Apoptosis and Inflammatory Response in the Hindgut of Goats. PLoS ONE 2014, 9, 111596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tovaglieri, A.; Sontheimer-Phelps, A.; Geirnaert, A.; Prantil-Baun, R.; Camacho, D.M.; Chou, D.B.; Jalili-Firoozinezhad, S.; de Wouters, T.; Kasendra, M.; Super, M.; et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome 2019, 7, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Roewe, J.; Stavrides, G.; Strueve, M.; Sharma, A.; Marini, F.; Mann, A.; Smith, S.A.; Kaya, Z.; Strobl, B.; Mueller, M.; et al. Bacterial polyphosphates interfere with the innate host defense to infection. Nat. Commun. 2020, 11, 4035. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, F.; Wu, W.; Sun, M.; Bilotta, A.J.; Yao, S.; Xiao, Y.; Huang, X.; Eaves-Pyles, T.D.; Golovko, G.; et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018, 11, 752–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fachi, J.L.; Sécca, C.; Rodrigues, P.B.; De Mato, F.C.P.; Di Luccia, B.; Felipe, J.D.S.; Pral, L.P.; Rungue, M.; Rocha, V.D.M.; Sato, F.T.; et al. Acetate coordinates neutrophil and ILC3 responses against C. difficile through FFAR2. J. Exp. Med. 2019, 217, 20190489. [Google Scholar] [CrossRef] [PubMed]
- Tulkens, J.; Vergauwen, G.; Van Deun, J.; Geeurickx, E.; Dhondt, B.; Lippens, L.; De Scheerder, M.-A.; Miinalainen, I.; Rappu, P.; De Geest, B.G.; et al. Increased levels of systemic LPS-positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut 2020, 69, 191–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Zhang, K.; Li, C.; Wang, X.; Chen, Y.; Yang, Y. Characterization and Comparison of Microbiota in the Gastrointestinal Tracts of the Goat (Capra hircus) during Preweaning Development. Front. Microbiol. 2019, 10, 2125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, H.; Zhou, G.; Tian, G.; Liu, Y.; Dong, N.; Li, L.; Zhang, S.; Chai, H.; Chen, Y.; Yang, Y. Changes in Rumen Microbiota Affect Metabolites, Immune Responses and Antioxidant Enzyme Activities of Sheep under Cold Stimulation. Animals 2021, 11, 712. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Q.; Cao, Y.; Gao, Y.; Yu, C.; Du, L.; Wang, X.; Li, J. Effects of dietary energy and protein levels on growing-fattening performance and blood biochemical indicators of holstein bulls. Chin. J. Anim. Nutr. 2015, 27, 1252–1261. [Google Scholar]
- Le Bellego, L.; van Milgen, J.; Noblet, J. Effect of high temperature and low-protein diets on the performance of growing-finishing pigs. J. Anim. Sci. 2002, 80, 691–701. [Google Scholar] [CrossRef]
- Pu, G.; Li, P.; Du, T.; Niu, Q.; Fan, L.; Wang, H.; Liu, H.; Li, K.; Niu, P.; Wu, C.; et al. Adding Appropriate Fiber in Diet Increases Diversity and Metabolic Capacity of Distal Gut Microbiota without Altering Fiber Digestibility and Growth Rate of Finishing Pig. Front. Microbiol. 2020, 11, 533. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; He, X.; Zou, Y.; Ding, Y.; Li, H.; Chen, H. Altered gut microbiome promotes proteinuria in mice induced by Adriamycin. AMB Express 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailén, M.; Bressa, C.; Martínez-López, S.; González-Soltero, R.; Lominchar, M.G.M.; Juan, C.S.; Larrosa, M. Microbiota Features Associated with a High-Fat/Low-Fiber Diet in Healthy Adults. Front. Nutr. 2020, 7, 583608. [Google Scholar] [CrossRef] [PubMed]
- White, D.C.; Sutton, S.D.; Ringelberg, D.B. The genus Sphingomonas: Physiology and ecology. Curr. Opin. Biotechnol. 1996, 7, 301–306. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, S.; Nie, Q.; He, H.; Tan, H.; Geng, F.; Ji, H.; Hu, J.; Nie, S. Gut firmicutes: Relationship with dietary fiber and role in host homeostasis. Crit. Rev. Food Sci. Nutr. 2022, 12, 1–16. [Google Scholar] [CrossRef]
- Matheron, C.; Delort, A.-M.; Gaudet, G.; Forano, E. Re-investigation of glucose metabolism in Fibrobacter succinogenes, using NMR spectroscopy and enzymatic assays.: Evidence for pentose phosphates phosphoketolase and pyruvate formate lyase activities. Biochim. et Biophys. Acta 1997, 1355, 50–60. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Liu, K.; Wang, Z.; Bai, X.; Peng, Q.; Jin, L. Bacterial Community Diversity Associated with Different Utilization Efficiencies of Nitrogen in the Gastrointestinal Tract of Goats. Front. Microbiol. 2019, 10, 239. [Google Scholar] [CrossRef]
- Christensen, L.; Vuholm, S.; Roager, H.M.; Nielsen, D.S.; Krych, L.; Kristensen, M.; Astrup, A.; Hjorth, M.F. Prevotella Abundance Predicts Weight Loss Success in Healthy, Overweight Adults Consuming a Whole-Grain Diet Ad Libitum: A Post Hoc Analysis of a 6-Wk Randomized Controlled Trial. J. Nutr. 2019, 149, 2174–2181. [Google Scholar] [CrossRef]
- Fettig, N.M.; Robinson, H.G.; Allanach, J.R.; Davis, K.M.; Simister, R.L.; Wang, E.J.; Sharon, A.J.; Ye, J.; Popple, S.J.; Seo, J.H.; et al. Inhibition of Th1 activation and differentiation by dietary guar gum ameliorates experimental autoimmune encephalomyelitis. Cell Rep. 2022, 40, 111328. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.; Chen, X.; Chen, C.; Sun, J.; Bai, X.; Yuan, Y. Effect of copper ions on glucose fermentation pathways in bioelectrochemical system. Chemosphere 2021, 272, 129627. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, X.; Tao, S.; Pi, Y.; Han, D.; Ye, H.; Feng, C.; Zhao, J.; Chen, L.; Wang, J. Maternal supplementation with combined galactooligosaccharides and casein glycomacropeptides modulated microbial colonization and intestinal development of neonatal piglets. J. Funct. Foods 2020, 74, 104170. [Google Scholar] [CrossRef]
- Bordoni, L.; Gabbianelli, R.; Fedeli, D.; Fiorini, D.; Bergheim, I.; Jin, C.J.; Marinelli, L.; Di Stefano, A.; Nasuti, C. Positive effect of an electrolyzed reduced water on gut permeability, fecal microbiota and liver in an animal model of Parkinson’s disease. PLoS ONE 2019, 14, e0223238. [Google Scholar] [CrossRef]
- Ogunade, I.; Schweickart, H.; McCoun, M.; Cannon, K.; McManus, C. Integrating 16S rRNA Sequencing and LC–MS-Based Metabolomics to Evaluate the Effects of Live Yeast on Rumen Function in Beef Cattle. Animals 2019, 9, 28. [Google Scholar] [CrossRef]
- Li, X.; Li, Z.; He, Y.; Li, P.; Zhou, H.; Zeng, N. Regional distribution of Christensenellaceae and its associations with metabolic syndrome based on a population-level analysis. PeerJ 2020, 8, 9591. [Google Scholar] [CrossRef]
- Rincon, M.T.; Dassa, B.; Flint, H.J.; Travis, A.J.; Jindou, S.; Borovok, I.; Lamed, R.; Bayer, E.A.; Henrissat, B.; Coutinho, P.M.; et al. Abundance and Diversity of Dockerin-Containing Proteins in the Fiber-Degrading Rumen Bacterium, Ruminococcus flavefaciens FD-1. PLoS ONE 2010, 5, 12476. [Google Scholar] [CrossRef] [Green Version]
- Trocino, A.; Fragkiadakis, M.; Majolini, D.; Tazzoli, M.; Radaelli, G.; Xiccato, G. Soluble fibre, starch and protein level in diets for growing rabbits: Effects on digestive efficiency and productive traits. Anim. Feed Sci. Technol. 2013, 180, 73–82. [Google Scholar] [CrossRef]
- Manzanilla, E.G.; Pérez, J.F.; Martin, M.; Blandón, J.C.; Baucells, F.; Kamel, C.; Gasa, J. Dietary protein modifies effect of plant extracts in the intestinal ecosystem of the pig at weaning1. J. Anim. Sci. 2009, 87, 2029–2037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daïen, C.; Tan, J.; Audo, R.; Mielle, J.; Quek, L.; Krycer, J.; Angelatos, A.; Duraes, M.; Pinget, G.; Ni, D.; et al. Gut-derived acetate promotes B10 cells with antiinflammatory effects. JCI Insight 2021, 6, 144156. [Google Scholar] [CrossRef]
- Di Lorenzo, F.; De Castro, C.; Silipo, A.; Molinaro, A. Lipopolysaccharide structures of Gram-negative populations in the gut microbiota and effects on host interactions. FEMS Microbiol. Rev. 2019, 43, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Ananthakrishnan, A.N. New approaches along the IBD course: Diet, tight control and stem cells. Nat. Rev. Gastroenterol. Hepatol. 2018, 16, 82–84. [Google Scholar] [CrossRef]
- Kim, J.D.; Yoon, N.A.; Jin, S.; Diano, S. Microglial UCP2 Mediates Inflammation and Obesity Induced by High-Fat Feeding. Cell Metab. 2019, 30, 952–962. [Google Scholar] [CrossRef]
- Tian, P.; Zhu, H.; Qian, X.; Chen, Y.; Wang, Z.; Zhao, J.; Zhang, H.; Wang, G.; Chen, W. Consumption of Butylated Starch Alleviates the Chronic Restraint Stress-Induced Neurobehavioral and Gut Barrier Deficits through Reshaping the Gut Microbiota. Front. Immunol. 2021, 12, 755481. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, H.; Li, B.; Gao, M.; Li, Q.; Gao, Y.; Dong, N.; Liu, G.; Wang, Z.; Gao, W.; Chen, Y.; et al. Dietary Nutritional Level Affects Intestinal Microbiota and Health of Goats. Microorganisms 2022, 10, 2322. https://doi.org/10.3390/microorganisms10122322
Guo H, Li B, Gao M, Li Q, Gao Y, Dong N, Liu G, Wang Z, Gao W, Chen Y, et al. Dietary Nutritional Level Affects Intestinal Microbiota and Health of Goats. Microorganisms. 2022; 10(12):2322. https://doi.org/10.3390/microorganisms10122322
Chicago/Turabian StyleGuo, Hongran, Bibo Li, Meiqi Gao, Qian Li, Yawei Gao, Ning Dong, Gongwei Liu, Zhichao Wang, Wenrui Gao, Yulin Chen, and et al. 2022. "Dietary Nutritional Level Affects Intestinal Microbiota and Health of Goats" Microorganisms 10, no. 12: 2322. https://doi.org/10.3390/microorganisms10122322




