Molecular Epidemiology of Escherichia coli with Resistance against Third-Generation Cephalosporines Isolated from Deployed German Soldiers—A Retrospective Assessment after Deployments to the African Sahel Region and Other Sites between 2007 and 2016
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain Collection
2.2. Mass-Spectrometry Based Species Identification and Phenotypic Resistance Testing
2.3. Whole-Genome Sequencing and Bioinformatics
2.4. Ethics
3. Results
3.1. Details on the Origin of the Assessed E. coli Strains
3.2. Phenotypic Resistance Patterns of the E. coli Isolates with Resistance against Third-Generation Cephalosporines against Carbapenems, Gentamicin, Ciprofloxacin, Moxifloxacin, and Trimethoprim–Sulfamethoxazole
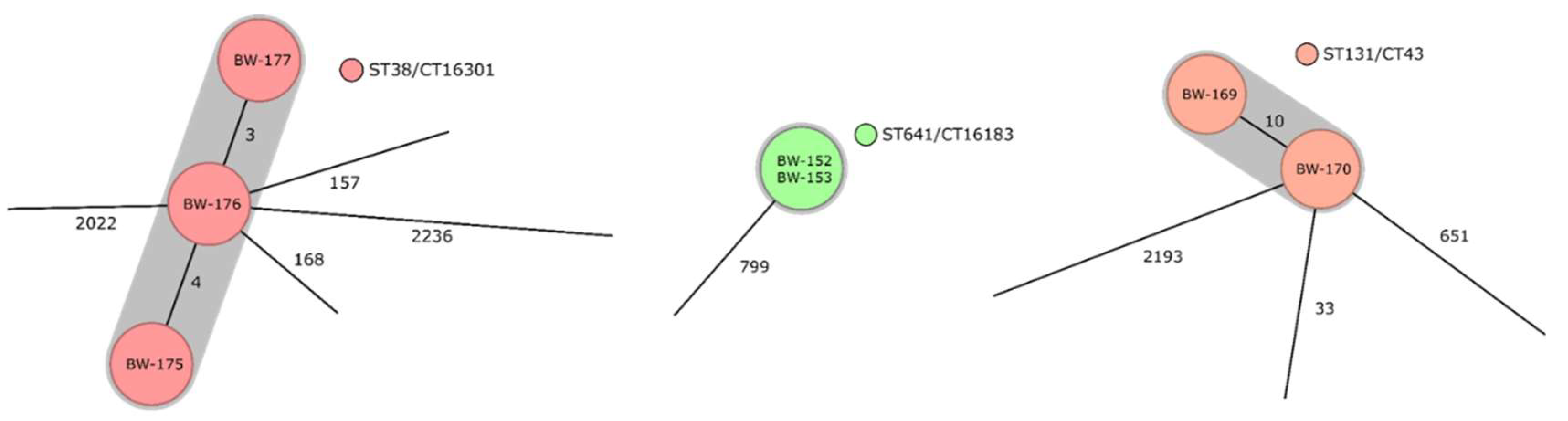
3.3. Identified Clonal Lineages, Resistance Determinants, as Well as Virulence Determinants, and Their Distribution on the Deployment Sites in the Sahel Regions and in Other Deployment Settings
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Anonymized Strain I.D. | Year of Isolation | Deployment Site | ASU | AZT | CTX | CPD | CTZ | CIP | FEP | FOF | GEN | IMI | MER | MOX | TZP | CTM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BW-151 | 2007 | Uzbekistan | ≥32 | ≥64 | ≥64 | ≥8 | 16 | ≥4 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≥8 | 8 | ≥320 |
| BW-152 | 2007 | Democratic Republic of the Congo | ≥32 | ≥64 | ≥64 | ≥8 | 4 | ≤0.25 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≤0.25 | ≤4 | ≤20 |
| BW-153 | 2007 | Lebanon | ≥32 | 16 | ≥64 | ≥8 | 4 | ≤0.25 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≤0.25 | ≤4 | ≤20 |
| BW-154 | 2007 | Uzbekistan | ≥32 | ≥64 | ≥64 | ≥8 | 16 | ≥4 | ≥64 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | ≥8 | 64 | ≥320 |
| BW-155 | 2007 | Afghanistan | 16 | 16 | ≥64 | ≥8 | 8 | ≤0.25 | 2 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | ≤0.25 | ≤4 | ≥320 |
| BW-156 | 2007 | Democratic Republic of the Congo | 16 | 16 | ≥64 | ≥8 | 16 | ≤0.25 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≤0.25 | ≤4 | ≥320 |
| BW-157 | 2007 | Uzbekistan | ≥32 | ≤64 | ≤64 | ≥8 | 16 | ≥4 | 8 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≥8 | 8 | ≥320 |
| BW-158 | 2007 | Sudan | ≥32 | ≤1 | 4 | ≥8 | ≤1 | ≥4 | ≤1 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≥8 | 8 | ≥320 |
| BW-159 | 2008 | Sudan | 16 | ≥64 | ≥64 | ≥8 | 16 | ≤0.25 | ≥64 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≤0.25 | ≤4 | ≥320 |
| BW-160 | 2009 | Sudan | ≥32 | ≥64 | ≥64 | ≥8 | 16 | ≥4 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≥8 | ≥128 | ≥320 |
| BW-161 | 2009 | Ghana | 16 | ≥64 | ≥64 | ≥8 | ≤1 | ≤0.25 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≤0.25 | ≤4 | ≤20 |
| BW-162 | 2009 | Sudan | ≥32 | ≥64 | ≥64 | ≥8 | ≥64 | ≥4 | ≥64 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≥8 | 16 | ≤20 |
| BW-163 | 2010 | Sudan | ≥32 | 16 | ≥64 | ≥8 | 16 | ≥4 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≥8 | 8 | ≤20 |
| BW-164 | 2010 | Afghanistan | 16 | 2 | ≥64 | ≥8 | ≤1 | ≥4 | 2 | ≤16 | ≤2 | ≤0.25 | ≤0.25 | ≥8 | ≤4 | ≥320 |
| BW-165 | 2010 | Sudan | 16 | 16 | ≥64 | ≥8 | 4 | ≤.25 | 2 | ≤16 | 4 | ≤0.25 | ≤0.25 | ≤0.25 | ≤4 | ≤20 |
| BW-166 | 2010 | Thailand | ≥2 | 2 | ≥64 | ≥8 | 4 | ≤0.25 | ≤1 | ≤16 | ≤1 | ≤0.25 | ≥0.25 | ≤0.25 | ≤4 | ≤20 |
| BW-167 | 2011 | Afghanistan | 16 | 16 | ≥64 | ≥8 | 4 | ≤0.25 | ≤1 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | 0.5 | ≤4 | ≥320 |
| BW-168 | 2011 | Sudan | 16 | 4 | ≥64 | ≥8 | 16 | 0.5 | 4 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | 0.5 | ≤4 | ≥320 |
| BW-169 | 2011 | Sudan | ≤2 | 4 | ≥64 | ≥8 | 4 | ≥4 | 2 | ≤16 | ≤1 | ≤0.25 | ≥0.25 | ≥8 | ≤4 | ≤20 |
| BW-170 | 2013 | Sudan | ≤2 | 2 | ≥64 | ≥8 | 4 | ≥4 | ≤1 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≥8 | ≤4 | ≥320 |
| BW-171 | 2013 | Djibouti | ≥32 | 16 | ≥64 | 4 | ≥64 | ≥4 | ≤1 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | ≥8 | ≤4 | ≤20 |
| BW-172 | 2013 | Tanzania | ≥32 | 16 | ≥64 | ≥8 | 16 | 1 | 4 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | 1 | 8 | ≥320 |
| BW-173 | 2014 | Uganda | 16 | 16 | ≥64 | ≥8 | 4 | 0.5 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | 2 | ≤4 | ≥320 |
| BW-174 | 2014 | Uganda | ≥32 | ≥64 | ≥64 | ≥8 | ≥64 | ≥4 | ≥64 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | ≥8 | 64 | ≥320 |
| BW-175 | 2014 | South Sudan | ≥32 | 4 | ≥64 | ≥8 | 16 | 1 | ≤1 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | 2 | ≤4 | ≥320 |
| BW-176 | 2014 | South Sudan | 16 | 16 | ≥64 | ≥8 | ≤1 | ≤0.25 | ≤1 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | 0.5 | 16 | ≤20 |
| BW-177 | 2014 | South Sudan | 16 | 16 | ≤64 | ≥8 | ≤0.25 | 2 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | 0.5 | ≤4 | ≥320 | |
| BW-178 | 2014 | South Sudan | ≥32 | 16 | ≥64 | ≥8 | 16 | ≤0.25 | 4 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | 0.5 | 64 | ≥320 |
| BW-179 | 2014 | Uganda | ≥32 | ≥64 | ≥64 | ≥8 | 16 | ≤0.25 | 32 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | 1 | 24 | ≤20 |
| BW-180 | 2013 | South Sudan | ≤2 | ≤1 | ≥64 | ≥8 | ≤1 | 1 | ≤1 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | 4 | ≤4 | ≤20 |
| BW-181 | 2014 | South Sudan | 16 | 4 | ≥64 | ≥8 | 16 | ≤0.25 | 4 | ≤16 | ≤1 | ≤0.25 | ≤0.26 | ≤0.25 | ≤4 | ≤20 |
| BW-182 | 2014 | Nigeria | ≥32 | 16 | ≥64 | ≥8 | 4 | 1 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | 1 | ≤4 | ≥320 |
| BW-183 | 2014 | Democratic Republic of the Congo | ≥32 | ≥64 | ≥64 | ≥8 | ≥64 | ≥4 | ≥64 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | ≥8 | 8 | ≥320 |
| BW-184 | 2014 | Mali | ≤2 | 16 | ≥64 | ≥8 | 4 | ≤0.25 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≤0.25 | ≤4 | ≥321 |
| BW-185 | 2014 | Mali | ≥32 | 16 | ≥64 | ≥8 | 4 | ≤0.25 | 4 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≤0.25 | ≤4 | ≥320 |
| BW-186 | 2014 | South Sudan | ≥32 | ≥64 | ≥64 | ≥8 | 64 | ≥4 | 4 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | ≥8 | 8 | ≥320 |
| BW-187 | 2015 | Mali | ≤2 | 16 | ≥64 | ≥8 | 4 | ≤0.25 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≤0.25 | ≤4 | ≥320 |
| BW-188 | 2015 | Not further defined African destinations | ≥32 | 4 | 8 | ≥8 | 2 | ≤0.25 | ≤1 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | 0.5 | ≤4 | ≥320 |
| BW-189 | 2014 | Unknown or multiple deployment settings | ≥32 | 16 | ≥64 | ≥8 | 16 | 1 | 2 | 64 | ≥16 | ≤0.25 | ≤0.25 | 2 | 24 | ≥320 |
| BW-192 | 2016 | Sudan and Uganda | ≥32 | 2 | ≥64 | ≥8 | 4 | ≤0.25 | ≤1 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | 1 | 16 | ≥320 |
| BW-193 | 2016 | Nigeria | 16 | 2 | ≥64 | ≥8 | ≤1 | ≥4 | 2 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | ≥8 | ≤4 | ≥320 |
| BW-194 | 2016 | Mali | 8 | 16 | ≥64 | ≥8 | 16 | ≥4 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≥8 | ≤4 | ≤20 |
| BW-195 | 2016 | Mali | ≤2 | ≥64 | ≥64 | ≥8 | 4 | ≤0.25 | 8 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | 0.5 | ≤4 | ≤20 |
| BW-198 | 2016 | Ethiopia | ≥32 | 32 | ≤64 | ≥8 | ≥64 | ≥4 | 8 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | ≥8 | 24 | ≥320 |
| BW-199 | 2016 | Mali | 16 | 16 | ≥64 | ≥8 | 4 | ≥4 | 2 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≥8 | ≤4 | ≥320 |
| BW-201 | 2016 | India and Nepal | 16 | 4 | ≥64 | ≥8 | 4 | ≤0.25 | ≤1 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | ≤0.25 | ≤4 | ≥320 |
| BW-203 | 2016 | Sudan and Uganda | 16 | 2 | ≥64 | ≥8 | ≤1 | ≤0.25 | ≤1 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | 1 | ≤4 | ≥320 |
| BW-206 | 2016 | Mali | 8 | 16 | ≥64 | ≥8 | 16 | ≥4 | ≥64 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≥8 | ≤4 | ≥320 |
| BW-207 | 2016 | Iraq | 16 | 16 | ≥64 | ≥8 | 16 | 0.5 | 2 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | 1 | ≤4 | ≥320 |
| BW-208 | 2016 | Mali | ≥32 | 16 | ≥64 | ≥8 | 16 | ≥4 | ≥64 | ≤16 | ≤1 | ≤0.25 | ≤0.25 | ≥8 | 8 | ≥320 |
| BW-209 | 2016 | Iraq | 4 | 16 | ≥64 | ≥8 | 4 | 1 | 4 | ≤16 | ≥16 | ≤0.25 | ≤0.25 | 4 | ≤4 | ≥320 |
References
- Tribble, D.R.; Murray, C.K.; Lloyd, B.A.; Ganesan, A.; Mende, K.; Blyth, D.M.; Petfield, J.L.; McDonald, J. After the Battlefield: Infectious Complications among Wounded Warriors in the Trauma Infectious Disease Outcomes Study. Mil. Med. 2019, 184 (Suppl. S2), 18–25. [Google Scholar] [CrossRef] [PubMed]
- Nawfal Dagher, T.; Al-Bayssari, C.; Diene, S.M.; Azar, E.; Rolain, J.M. Bacterial infection during wars, conflicts and post-natural disasters in Asia and the Middle East: A narrative review. Expert Rev. Anti. Infect. Ther. 2020, 18, 511–529. [Google Scholar] [CrossRef] [PubMed]
- Collignon, P.; Athukorala, P.C.; Senanayake, S.; Khan, F. Antimicrobial resistance: The major contribution of poor governance and corruption to this growing problem. PLoS ONE 2015, 10, e0116746.0. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collignon, P.; Beggs, J.J.; Walsh, T.R.; Gandra, S.; Laxminarayan, R. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: A univariate and multivariable analysis. Lancet Planet Health 2018, 2, e398–e405. [Google Scholar] [CrossRef] [PubMed]
- Kaba, H.E.J.; Kuhlmann, E.; Scheithauer, S. Thinking outside the box: Association of antimicrobial resistance with climate warming in Europe—A 30 country observational study. Int. J. Hyg. Environ. Health 2020, 223, 151–158. [Google Scholar] [CrossRef]
- Kaba, H.E.J.; Scheithauer, S. Estimating the effect of practicing nursing professionals density on cumulative carbapenem-resistance prevalence in gram-negative invasive Isolates: A 30 European country observational modeling study. Antimicrob. Resist. Infect. Control 2022, 11, 41. [Google Scholar] [CrossRef]
- Frickmann, H.; Podbielski, A.; Kreikemeyer, B. Resistant Gram-Negative Bacteria and Diagnostic Point-of-Care Options for the Field Setting during Military Operations. Biomed. Res. Int. 2018, 2018, 9395420. [Google Scholar] [CrossRef] [Green Version]
- Frickmann, H.; Wiemer, D.; Frey, C.; Hagen, R.M.; Hinz, R.; Podbielski, A.; Köller, T.; Warnke, P. Low Enteric Colonization with Multidrug-Resistant Pathogens in Soldiers Returning from Deployments- Experience from the Years 2007–2015. PLoS ONE 2016, 11, e0162129. [Google Scholar] [CrossRef]
- Tompkins, K.; Juliano, J.J.; van Duin, D. Antimicrobial Resistance in Enterobacterales and Its Contribution to Sepsis in Sub-saharan Africa. Front. Med. (Lausanne) 2021, 26, 615649. [Google Scholar] [CrossRef]
- Akpan, M.R.; Isemin, N.U.; Udoh, A.E.; Ashiru-Oredope, D. Implementation of antimicrobial stewardship programmes in African countries: A systematic literature review. J. Glob. Antimicrob. Resist. 2020, 22, 317–324. [Google Scholar] [CrossRef]
- Timberlake, L. The Sahel: Drought, desertification and famine. Draper Fund Rep. 1985, 14, 17–19. [Google Scholar]
- Braima, O.A.; Ali, M.A.; Abdulla, E.M. Bacteriological profile and antibiotic resistance in newborn infants with possible community-acquired neonatal sepsis in Khartoum State, Sudan. Sudan. J. Paediatr. 2021, 21, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Azab, K.S.M.; Abdel-Rahman, M.A.; El-Sheikh, H.H.; Azab, E.; Gobouri, A.A.; Farag, M.M.S. Distribution of Extended-Spectrum β-Lactamase (ESBL)-Encoding Genes among Multidrug-Resistant Gram-Negative Pathogens Collected from Three Different Countries. Antibiotics (Basel) 2021, 10, 247. [Google Scholar] [CrossRef] [PubMed]
- Dirar, M.H.; Bilal, N.E.; Ibrahim, M.E.; Hamid, M.E. Prevalence of extended-spectrum β-lactamase (ESBL) and molecular detection of blaTEM, blaSHV and blaCTX-M genotypes among Enterobacteriaceae isolates from patients in Khartoum, Sudan. Pan. Afr. Med. J. 2020, 37, 213. [Google Scholar] [CrossRef] [PubMed]
- Moglad, E.H. Antibiotics Profile, Prevalence of Extended-Spectrum Beta-Lactamase (ESBL), and Multidrug-Resistant Enterobacteriaceae from Different Clinical Samples in Khartoum State, Sudan. Int. J. Microbiol. 2020, 2020, 8898430. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Wen, P.; Xu, H.; Chi, X.; Li, S.; Yu, X.; Lin, X.; Wu, S.; Zheng, B. Emergence and Comparative Genomics Analysis of Extended-Spectrum-β-Lactamase-Producing Escherichia coli Carrying mcr-1 in Fennec Fox Imported from Sudan to China. mSphere 2019, 4, e00732-19. [Google Scholar] [CrossRef] [Green Version]
- Toy, T.; Pak, G.D.; Duc, T.P.; Campbell, J.I.; El Tayeb, M.A.; Von Kalckreuth, V.; Im, J.; Panzner, U.; Cruz Espinoza, L.M.; Eibach, D.; et al. Multicountry Distribution and Characterization of Extended-spectrum β-Lactamase-associated Gram-negative Bacteria From Bloodstream Infections in Sub-Saharan Africa. Clin. Infect. Dis. 2019, 69 (Suppl. S6), S449–S458. [Google Scholar] [CrossRef] [Green Version]
- Saeed, A.; Hamid, S.A.; Bayoumi, M.; Shanan, S.; Alouffi, S.; Alharbi, S.A.; Alshammari, F.D.; Abd, H. Elevated antibiotic resistance of Sudanese urinary tract infection bacteria. EXCLI J. 2017, 16, 1073–1080. [Google Scholar]
- Hamdan, H.Z.; Kubbara, E.; Adam, A.M.; Hassan, O.S.; Suliman, S.O.; Adam, I. Urinary tract infections and antimicrobial sensitivity among diabetic patients at Khartoum, Sudan. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 26. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, M.E.; Bilal, N.E.; Hamid, M.E. Increased multi-drug resistant Escherichia coli from hospitals in Khartoum state, Sudan. Afr. Health Sci. 2012, 12, 368–375. [Google Scholar] [CrossRef] [Green Version]
- Hamdan, H.Z.; Ziad, A.H.; Ali, S.K.; Adam, I. Epidemiology of urinary tract infections and antibiotics sensitivity among pregnant women at Khartoum North Hospital. Ann. Clin. Microbiol. Antimicrob. 2011, 10, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmadiena, M.M.; El Hussein, A.A.; Muckle, C.A.; Cole, L.; Wilkie, E.; Mistry, K.; Perets, A. Antimicrobial susceptibility and multi-drug resistance of Salmonella enterica subspecies enterica serovars in Sudan. Trop. Anim. Health. Prod. 2013, 45, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Elshayeb, A.A.; Ahmed, A.A.; El Siddig, M.A.; El Hussien, A.A. Prevalence of current patterns and predictive trends of multidrug-resistant Salmonella Typhi in Sudan. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.E.; Pham, D.T.; Boinett, C.; Wong, V.K.; Pak, G.D.; Panzner, U.; Espinoza, L.M.C.; von Kalckreuth, V.; Im, J.; Schütt-Gerowitt, H.; et al. The phylogeography and incidence of multi-drug resistant typhoid fever in sub-Saharan Africa. Nat. Commun. 2018, 9, 5094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, H.S. Sensitivity of Salmonella and Shigella to antibiotics and chemotherapeutic agents in Sudan. J. Trop. Med. Hyg. 1985, 88, 243–247. [Google Scholar]
- Shears, P.; Hart, C.A.; Broadhead, R.L.; Coulter, J.B. A note on antibiotic resistance in Escherichia coli isolated from children with diarrhoea in the Sudan. Ann. Trop. Paediatr. 1987, 7, 38–41. [Google Scholar] [CrossRef]
- Musa, H.A.; Shears, P. Antibiotic resistant Escherichia coli in a Sudanese hospital. J. Hosp. Infect. 1998, 38, 148–150. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Osman, H.; Mansour, A.M.; Musa, H.A.; Ahmed, A.B.; Karrar, Z.; Hassan, H.S. Antimicrobial agent resistance in bacterial isolates from patients with diarrhea and urinary tract infection in the Sudan. Am. J. Trop. Med. Hyg. 2000, 63, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Doczeova, A.; Liskova, A.; Krcmery, V., Jr. Low antibiotic resistance in respiratory pathogens in a remote area in southern Sudan that was isolated by civil war for 18 years. Clin. Infect. Dis. 2003, 37, 1582. [Google Scholar] [CrossRef] [Green Version]
- Albasha, A.M.; Osman, E.H.; Abd-Alhalim, S.; Alshaib, E.F.; Al-Hassan, L.; Altayb, H.N. Detection of several carbapenems resistant and virulence genes in classical and hyper-virulent strains of Klebsiella pneumoniae isolated from hospitalized neonates and adults in Khartoum. BMC Res. Notes 2020, 13, 312. [Google Scholar] [CrossRef]
- Mahmoud, N.E.; Altayb, H.N.; Gurashi, R.M. Detection of Carbapenem-Resistant Genes in Escherichia coli Isolated from Drinking Water in Khartoum, Sudan. J. Environ. Public Health 2020, 2020, 2571293. [Google Scholar] [CrossRef] [PubMed]
- Osman, E.A.; El-Amin, N.E.; Al-Hassan, L.L.; Mukhtar, M. Multiclonal spread of Klebsiella pneumoniae across hospitals in Khartoum, Sudan. J. Glob. Antimicrob. Resist. 2021, 24, 241–245. [Google Scholar] [CrossRef]
- Ejaz, H.; Younas, S.; Abosalif, K.O.A.; Junaid, K.; Alzahrani, B.; Alsrhani, A.; Abdalla, A.E.; Ullah, M.I.; Qamar, M.U.; Hamam, S.S.M. Molecular analysis of blaSHV, blaTEM, and blaCTX-M in extended-spectrum β-lactamase producing Enterobacteriaceae recovered from fecal specimens of animals. PLoS ONE 2021, 16, e0245126. [Google Scholar] [CrossRef]
- Abd Alfadil, N.A.; Suliman Mohamed, M.; Ali, M.M.; El Nima, E.A.I. Characterization of Pathogenic Bacteria Isolated from Sudanese Banknotes and Determination of Their Resistance Profile. Int. J. Microbiol. 2018, 2018, 4375164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awad, A.; Eltayeb, I.; Matowe, L.; Thalib, L. Self-medication with antibiotics and antimalarials in the community of Khartoum State, Sudan. J. Pharm. Pharm. Sci. 2005, 8, 326–331. [Google Scholar] [PubMed]
- Rosenthal, V.D.; Belkebir, S.; Zand, F.; Afeef, M.; Tanzi, V.L.; Al-Abdely, H.M.; El-Kholy, A.; Aziz AlKhawaja, S.A.; Demiroz, A.P.; Sayed, A.F.; et al. Six-year multicenter study on short-term peripheral venous catheters-related bloodstream infection rates in 246 intensive units of 83 hospitals in 52 cities of 14 countries of Middle East: Bahrain, Egypt, Iran, Jordan, Kingdom of Saudi Arabia, Kuwait, Lebanon, Morocco, Pakistan, Palestine, Sudan, Tunisia, Turkey, and United Arab Emirates-International Nosocomial Infection Control Consortium (INICC) findings. J. Infect. Public. Health 2020, 13, 1134–1141. [Google Scholar] [PubMed]
- Shears, P.; Suliman, G.; Hart, C.A. Occurrence of multiple antibiotic resistance and R plasmids in Enterobacteriaceae isolated from children in the Sudan. Epidemiol. Infect. 1988, 100, 73–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musa, H.A.; Hassan, H.S.; Shears, P. Occurrence in Sudan of Shigella dysenteriae type 1 with transferable antimicrobial resistance. Ann. Trop. Med. Parasitol. 1997, 91, 669–671. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.E.; Magzoub, M.A.; Bilal, N.E.; Hamid, M.E. Distribution of Class I integrons and their effect on the prevalence of multi-drug resistant Escherichia coli clinical isolates from Sudan. Saudi Med. J. 2013, 34, 240–247. [Google Scholar]
- Abdelgader, S.A.; Shi, D.; Chen, M.; Zhang, L.; Hejair, H.M.A.; Muhammad, U.; Yao, H.; Zhang, W. Antibiotics Resistance Genes Screening and Comparative Genomics Analysis of Commensal Escherichia coli Isolated from Poultry Farms between China and Sudan. Biomed. Res. Int. 2018, 2018, 5327450. [Google Scholar] [CrossRef] [Green Version]
- Najjuka, C.F.; Kateete, D.P.; Lodiongo, D.K.; Mambo, O.; Mocktar, C.; Kayondo, W.; Baluku, H.; Kajumbula, H.M.; Essack, S.Y.; Joloba, M.L. Prevalence of plasmid-mediated AmpC beta-lactamases in Enterobacteria isolated from urban and rural folks in Uganda. AAS Open Res. 2020, 3, 62. [Google Scholar] [CrossRef] [PubMed]
- Sangare, S.A.; Maiga, A.I.; Guindo, I.; Maiga, A.; Camara, N.; Savadogo, S.; Diallo, S.; Bougoudogo, F.; Armand-Lefevre, L.; Andremont, A.; et al. Prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae isolated from blood cultures in Africa. Med. Mal. Infect. 2015, 45, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Nkansa-Gyamfi, N.A.; Kazibwe, J.; Traore, D.A.K.; Nji, E. Prevalence of multidrug-, extensive drug-, and pandrug-resistant commensal Escherichia coli isolated from healthy humans in community settings in low- and middle-income countries: A systematic review and meta-analysis. Glob. Health. Action 2019, 12 (Suppl. S1), 1815272. [Google Scholar] [CrossRef] [PubMed]
- Sangaré, S.A.; Maïga, A.I.; Maïga, A.; Diallo, S.; Camara, N.; Savadogo, S.; Guindo, I.; Bougoudogo, F.; Armand-Lefèvre, L.; Andremont, A.; et al. Prevalence of extended-spectrum beta-lactamase phenotypes in enterobacteria isolated from blood cultures of patients at admission to the University Hospital of Bamako. Med. Sante Trop. 2017, 27, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Sangare, S.A.; Rondinaud, E.; Maataoui, N.; Maiga, A.I.; Guindo, I.; Maiga, A.; Camara, N.; Dicko, O.A.; Dao, S.; Diallo, S.; et al. Very high prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in bacteriemic patients hospitalized in teaching hospitals in Bamako, Mali. PLoS ONE 2017, 12, e0172652. [Google Scholar] [CrossRef] [Green Version]
- Sangare, S.A.; Maiga, A.I.; Guindo, I.; Maiga, A.; Camara, N.; Dicko, O.A.; Diallo, S.; Bougoudogo, F.; Armand-Lefevre, L.; Andremont, A.; et al. Prevalence of ESBL-producing Enterobacteriaceae isolated from blood cultures in Mali. J. Infect. Dev. Ctries 2016, 10, 1059–1064. [Google Scholar] [CrossRef] [Green Version]
- Tandé, D.; Jallot, N.; Bougoudogo, F.; Montagnon, T.; Gouriou, S.; Sizun, J. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in a Malian orphanage. Emerg. Infect. Dis. 2009, 15, 472–474. [Google Scholar] [CrossRef]
- Tandé, D.; Boisramé-Gastrin, S.; Münck, M.R.; Héry-Arnaud, G.; Gouriou, S.; Jallot, N.; Nordmann, P.; Naas, T. Intrafamilial transmission of extended-spectrum-beta-lactamase-producing Escherichia coli and Salmonella enterica Babelsberg among the families of internationally adopted children. J. Antimicrob. Chemother. 2010, 65, 859–865. [Google Scholar] [CrossRef] [Green Version]
- Boisramé-Gastrin, S.; Tandé, D.; Münck, M.R.; Gouriou, S.; Nordmann, P.; Naas, T. Salmonella carriage in adopted children from Mali: 2001-08. J. Antimicrob. Chemother. 2011, 66, 2271–2276. [Google Scholar] [CrossRef]
- Meli, H.; Cissoko, Y.; Konaté, I.; Soumaré, M.; Fofana, A.; Dembélé, J.P.; Kaboré, M.; Cissé, M.A.; Zaré, A.; Dao, S. Tuberculosis and HIV coinfection complicated by nosocomial infection caused by Klebsiella pneumoniae: About 4 cases in a Department of Infectious diseases in Mali. Pan. Afr. Med. J. 2020, 37, 141. [Google Scholar]
- Frickmann, H.; Warnke, P.; Frey, C.; Schmidt, S.; Janke, C.; Erkens, K.; Schotte, U.; Köller, T.; Maaßen, W.; Podbielski, A.; et al. Surveillance of Food- and Smear-Transmitted Pathogens in European Soldiers with Diarrhea on Deployment in the Tropics: Experience from the European Union Training Mission (EUTM) Mali. Biomed. Res. Int. 2015, 2015, 573904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagen, R.M.; Hinz, R.; Frickmann, H. β-Lactamases Encoded by blaCTX-M Group I Genes as Determinants of Resistance of Esbl-Positive Enterobacteriaceae in European Soldiers in Tropical Mali. Eur. J. Microbiol. Immunol. (Bp) 2015, 5, 281–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frickmann, H.; Hagen, R.M.; Geiselbrechtinger, F.; Hoysal, N. Infectious diseases during the European Union training mission Mali (EUTM MLI)—A four-year experience. Mil. Med. Res. 2018, 5, 19. [Google Scholar] [CrossRef]
- Mikhail, I.A.; Fox, E.; Haberberger, R.L., Jr.; Ahmed, M.H.; Abbatte, E.A. Epidemiology of bacterial pathogens associated with infectious diarrhea in Djibouti. J. Clin. Microbiol. 1990, 28, 956–961. [Google Scholar] [CrossRef] [Green Version]
- Cavallo, J.D.; Bercion, R.; Baudet, J.M.; Samson, T.; France, M.; Meyran, M. Etude de la sensibilité aux antibiotiques de 140 souches de shigelles isolées à Djibouti [Antibiotic sensitivity of 140 strains of Shigella isolated in Djibouti]. Bull. Soc. Pathol. Exot. 1993, 86, 35–40. [Google Scholar] [PubMed]
- Masseron, T.; Hovette, P. Evolution de l’antibiorésistance des shigelles à Djibouti [Evolution of Shigella antibiotic resistance in Djibouti]. Med. Trop. (Mars) 1999, 59, 205–206. [Google Scholar] [PubMed]
- Plantamura, J.; Bousquet, A.; Védy, S.; Larréché, S.; Bigaillon, C.; Delacour, H.; Mérens, A. Molecular epidemiological of extended-spectrum β-lactamase producing Escherichia coli isolated in Djibouti. J. Infect. Dev. Ctries. 2019, 13, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Riddle, M.S.; Connor, P.; Fraser, J.; Porter, C.K.; Swierczewski, B.; Hutley, E.J.; Danboise, B.; Simons, M.P.; Hulseberg, C.; Lalani, T.; et al. Trial Evaluating Ambulatory Therapy of Travelers’ Diarrhea (TrEAT TD) Study: A Randomized Controlled Trial Comparing 3 Single-Dose Antibiotic Regimens With Loperamide. Clin. Infect. Dis. 2017, 65, 2008–2017. [Google Scholar] [CrossRef] [Green Version]
- Johnson, R.C.; Van Nostrand, J.D.; Tisdale, M.; Swierczewski, B.; Simons, M.P.; Connor, P.; Fraser, J.; Melton-Celsa, A.R.; Tribble, D.R.; Riddle, M.S. Fecal Microbiota Functional Gene Effects Related to Single-Dose Antibiotic Treatment of Travelers’ Diarrhea. Open Forum Infect. Dis. 2021, 8, ofab271. [Google Scholar] [CrossRef]
- Lo, S.; Robin, F.; Ba-Diallo, A.; Diallo, O.F.; Dia, M.L.; Beyrouthy, R.; Gaye-Diallo, A.; Sow, A.I.; Bonnet, R. Fortuitous Detection of cmy-2 and dha-1 from ESBL-producing Escherichia coli in Senegal. Bull. Soc. Pathol. Exot. 2017, 110, 221–223. [Google Scholar] [CrossRef]
- Ndir, A.; Diop, A.; Ka, R.; Faye, P.M.; Dia-Badiane, N.M.; Ndoye, B.; Astagneau, P. Infections caused by extended-spectrum beta-lactamases producing Enterobacteriaceae: Clinical and economic impact in patients hospitalized in 2 teaching hospitals in Dakar, Senegal. Antimicrob. Resist. Infect. Control 2016, 5, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vounba, P.; Arsenault, J.; Bada-Alambédji, R.; Fairbrother, J.M. Prevalence of antimicrobial resistance and potential pathogenicity, and possible spread of third generation cephalosporin resistance, in Escherichia coli isolated from healthy chicken farms in the region of Dakar, Senegal. PLoS ONE 2019, 14, e0214304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndir, A.; Diop, A.; Faye, P.M.; Cissé, M.F.; Ndoye, B.; Astagneau, P. Epidemiology and Burden of Bloodstream Infections Caused by Extended-Spectrum Beta-Lactamase Producing Enterobacteriaceae in a Pediatric Hospital in Senegal. PLoS ONE 2016, 11, e0143729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dia, M.L.; Ngom, B.; Diagne, R.; Ka, R.; Lo, S.; Cisse, M.F.; Arlet, G.; Sow, A.I. Molecular detection of CTX-M-15-type β-lactamases in Escherichia coli strains from Senegal. New Microbes New Infect 2015, 9, 45–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrois, D.; Breurec, S.; Seck, A.; Delauné, A.; Le Hello, S.; Pardos de la Gándara, M.; Sontag, L.; Perrier-Gros-Claude, J.D.; Sire, J.M.; Garin, B.; et al. Prevalence and characterization of extended-spectrum β-lactamase-producing clinical Salmonella enterica isolates in Dakar, Senegal, from 1999 to 2009. Clin. Microbiol. Infect. 2014, 20, O109-16. [Google Scholar] [CrossRef]
- Page, A.L.; de Rekeneire, N.; Sayadi, S.; Aberrane, S.; Janssens, A.C.; Rieux, C.; Djibo, A.; Manuguerra, J.C.; Ducou-le-Pointe, H.; Grais, R.F.; et al. Infections in children admitted with complicated severe acute malnutrition in Niger. PLoS ONE 2013, 8, e68699. [Google Scholar] [CrossRef] [Green Version]
- Langendorf, C.; Le Hello, S.; Moumouni, A.; Gouali, M.; Mamaty, A.A.; Grais, R.F.; Weill, F.X.; Page, A.L. Enteric bacterial pathogens in children with diarrhea in Niger: Diversity and antimicrobial resistance. PLoS ONE 2015, 10, e0120275. [Google Scholar] [CrossRef] [Green Version]
- Dembélé, R.; Konaté, A.; Traoré, O.; Kaboré, W.A.D.; Soulama, I.; Kagambèga, A.; Traoré, A.S.; Guessennd, N.K.; Aidara-Kane, A.; Gassama-Sow, A.; et al. Extended spectrum beta-lactamase and fluoroquinolone resistance genes among Escherichia coli and Salmonella isolates from children with diarrhea, Burkina Faso. BMC Pediatr. 2020, 20, 459. [Google Scholar] [CrossRef]
- Cardinale, M.; Bourbotte-Salmon, F.; Scheiwe, C.; Boulezaz, S.; Ridet, M.; Laitselart, P. Antimicrobial resistance in N’Djamena (Chad): Four-year experience of the French Forward Medical and Surgical Team engaged in the “Barkhane Operation”. Med. Mal. Infect. 2020, 50, 665–669. [Google Scholar] [CrossRef]
- Baron, S.A.; Mediannikov, O.; Abdallah, R.; Kuete Yimagou, E.; Medkour, H.; Dubourg, G.; Elamire, Y.; Afouda, P.; Ngom, I.I.; Angelakis, E.; et al. Multidrug-Resistant Klebsiella pneumoniae Clones from Wild Chimpanzees and Termites in Senegal. Antimicrob. Agents Chemother. 2021, 65, e0255720. [Google Scholar] [CrossRef]
- Ruppé, E.; Armand-Lefèvre, L.; Estellat, C.; Consigny, P.H.; El Mniai, A.; Boussadia, Y.; Goujon, C.; Ralaimazava, P.; Campa, P.; Girard, P.M.; et al. High Rate of Acquisition but Short Duration of Carriage of Multidrug-Resistant Enterobacteriaceae After Travel to the Tropics. Clin. Infect. Dis. 2015, 61, 593–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arcilla, M.S.; van Hattem, J.M.; Haverkate, M.R.; Bootsma, M.C.J.; van Genderen, P.J.J.; Goorhuis, A.; Grobusch, M.P.; Lashof, A.M.O.; Molhoek, N.; Schultsz, C.; et al. Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): A prospective, multicentre cohort study. Lancet Infect. Dis. 2017, 17, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Frickmann, H.; Schwarz, N.G.; Wiemer, D.F.; Fischer, M.; Tannich, E.; Scheid, P.L.; Müller, M.; Schotte, U.; Bock, W.; Hagen, R.M. Food and drinking water hygiene and intestinal protozoa in deployed German soldiers. Eur. J. Microbiol. Immunol. (Bp) 2013, 3, 53–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schawaller, M.; Wiemer, D.; Hagen, R.M.; Frickmann, H. Infectious diseases in German military personnel after predominantly tropical deployments: A retrospective assessment over 13 years. BMJ Mil. Health 2020. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Kantele, A.; Lääveri, T.; Mero, S.; Vilkman, K.; Pakkanen, S.H.; Ollgren, J.; Antikainen, J.; Kirveskari, J. Antimicrobials increase travelers’ risk of colonization by extended-spectrum betalactamase-producing Enterobacteriaceae. Clin. Infect. Dis. 2015, 60, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Denamur, E.; Clermont, O.; Bonacorsi, S.; Gordon, D. The population genetics of pathogenic Escherichia coli. Nat. Rev. Microbiol. 2021, 19, 37–54. [Google Scholar] [CrossRef]
- Dekker, D.; Pankok, F.; Thye, T.; Taudien, S.; Oppong, K.; Akenten, C.W.; Lamshöft, M.; Jaeger, A.; Kaase, M.; Scheithauer, S.; et al. Clonal Clusters, Molecular Resistance Mechanisms and Virulence Factors of Gram-Negative Bacteria Isolated from Chronic Wounds in Ghana. Antibiotics (Basel) 2021, 10, 339. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 11 November 2022).
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Jünemann, S.; Sedlazeck, F.J.; Prior, K.; Albersmeier, A.; John, U.; Kalinowski, J.; Mellmann, A.; Goesmann, A.; von Haeseler, A.; Stoye, J.; et al. Updating benchtop sequencing performance comparison. Nature Biotechnol. 2013, 31, 294–296. [Google Scholar] [CrossRef] [Green Version]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Abricate, Github. Available online: https://github.com/tseemann/abricate (accessed on 22 March 2021).
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Prah, I.; Ayibieke, A.; Nguyen, T.T.H.; Iguchi, A.; Mahazu, S.; Sato, W.; Hayashi, T.; Yamaoka, S.; Suzuki, T.; Iwanaga, S.; et al. Virulence Profiles of Diarrheagenic Escherichia coli Isolated from the Western Region of Ghana. Jpn. J. Infect. Dis. 2021, 74, 115–121. [Google Scholar] [CrossRef]
- Seni, J.; Moremi, N.; Matee, M.; van der Meer, F.; DeVinney, R.; Mshana, S.E.D.; Pitout, J.D. Preliminary insights into the occurrence of similar clones of extended-spectrum beta-lactamase-producing bacteria in humans, animals and the environment in Tanzania: A systematic review and meta-analysis between 2005 and 2016. Zoonoses Public Health 2018, 65, 1–10. [Google Scholar] [CrossRef]
- Guiral, E.; Gonçalves Quiles, M.; Muñoz, L.; Moreno-Morales, J.; Alejo-Cancho, I.; Salvador, P.; Alvarez-Martinez, M.J.; Marco, F.; Vila, J. Emergence of Resistance to Quinolones and β-Lactam Antibiotics in Enteroaggregative and Enterotoxigenic Escherichia coli Causing Traveler’s Diarrhea. Antimicrob. Agents Chemother. 2019, 63, e01745-18. [Google Scholar] [CrossRef] [Green Version]
- Seni, J.; Peirano, G.; Mshana, S.E.; Pitout, J.D.D.; DeVinney, R. The importance of Escherichia coli clonal complex 10 and ST131 among Tanzanian patients on antimicrobial resistance surveillance programs. Eur. J. Clin. Microbiol. Infect. Dis. 2021. Epub ahead of print. [Google Scholar] [CrossRef]
- Yehouenou, C.L.; Bogaerts, B.; De Keersmaecker, S.C.J.; Roosens, N.H.C.; Marchal, K.; Tchiakpe, E.; Affolabi, D.; Simon, A.; Dossou, F.M.; Vanneste, K.; et al. Whole-Genome Sequencing-Based Antimicrobial Resistance Characterization and Phylogenomic Investigation of 19 Multidrug-Resistant and Extended-Spectrum Beta-Lactamase-Positive Escherichia coli Strains Collected From Hospital Patients in Benin in 2019. Front. Microbiol. 2021, 12, 752883. [Google Scholar] [CrossRef]
- Mshana, S.E.; Imirzalioglu, C.; Hain, T.; Domann, E.; Lyamuya, E.F.; Chakraborty, T. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin. Microbiol. Infect. 2011, 17, 1279–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mbelle, N.M.; Feldman, C.; Osei Sekyere, J.; Maningi, N.E.; Modipane, L.; Essack, S.Y. The Resistome, Mobilome, Virulome and Phylogenomics of Multidrug-Resistant Escherichia coli Clinical Isolates from Pretoria, South Africa. Sci. Rep. 2019, 9, 16457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moser, A.I.; Kuenzli, E.; Büdel, T.; Campos-Madueno, E.I.; Bernasconi, O.J.; DeCrom-Beer, S.; Jakopp, B.; Mohammed, A.H.; Hassan, N.K.; Fehr, J.; et al. Travellers returning from the island of Zanzibar colonized with MDR Escherichia coli strains: Assessing the impact of local people and other sources. J. Antimicrob. Chemother. 2021, 76, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Musila, L.; Kyany’a, C.; Maybank, R.; Stam, J.; Oundo, V.; Sang, W. Detection of diverse carbapenem and multidrug resistance genes and high-risk strain types among carbapenem non-susceptible clinical isolates of target gram-negative bacteria in Kenya. PLoS ONE 2021, 16, e0246937. [Google Scholar] [CrossRef]
- Eger, E.; Heiden, S.E.; Korolew, K.; Bayingana, C.; Ndoli, J.M.; Sendegeya, A.; Gahutu, J.B.; Kurz, M.S.E.; Mockenhaupt, F.P.; Müller, J.; et al. Circulation of Extended-Spectrum Beta-Lactamase-Producing Escherichia coli of Pandemic Sequence Types 131, 648, and 410 Among Hospitalized Patients, Caregivers, and the Community in Rwanda. Front. Microbiol. 2021, 12, 662575. [Google Scholar] [CrossRef] [PubMed]
- Musicha, P.; Feasey, N.A.; Cain, A.K.; Kallonen, T.; Chaguza, C.; Peno, C.; Khonga, M.; Thompson, S.; Gray, K.J.; Mather, A.E.; et al. Genomic landscape of extended-spectrum β-lactamase resistance in Escherichia coli from an urban African setting. J. Antimicrob. Chemother. 2017, 72, 1602–1609. [Google Scholar] [CrossRef] [Green Version]
- Medugu, N.; Aworh, M.K.; Iregbu, K.; Nwajiobi-Princewill, P.; Abdulraheem, K.; Hull, D.M.; Harden, L.; Singh, P.; Obaro, S.; Egwuenu, A.; et al. Molecular characterization of multi drug resistant Escherichia coli isolates at a tertiary hospital in Abuja, Nigeria. Sci. Rep. 2022, 12, 14822. [Google Scholar] [CrossRef]
- Piarroux, R.; Moore, S.; Rebaudet, S. Cholera in Haiti. Presse Med. 2022, 51, 104136. [Google Scholar] [CrossRef]
| Beta- Lactamase (BL) | No. | Deployment Sites | MIC Value | MIC (mg/L) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | CTZ | CPD | FEP | ERT | MER | TZP | CTM | CIP | GEN | FOF | AZT | ||||
| blaCTX-M-15, blaTEM-1B | n = 12 | Afghanistan, Congo, India/Nepal, Iraq, Mali, Sudan, Uganda | Median | ≥64 | 4 | ≥8 | 2 | ≤0.5 | ≤0.25 | ≤4 | ≥320 | ≤0.25 | ≤1 | ≤16 | 16 |
| Range | ≥64 to ≥64 | ≤1 to 16 | ≥8 to ≥8 | ≤1 to ≥64 | ≤0.5 to ≤0.5 | ≤0.25 to ≤0.25 | ≤4 to 16 | ≤20 to >320 | ≤0.25 to ≥4 | ≤1 to ≥16 | ≤16 to 64 | ≤1 to >64 | |||
| blaCTX-M-15, blaTEM-1B, blaOXA-1 | n = 9 | Congo, Mali, Nigeria, Sudan, Uganda, Uzbekistan | Median | ≥64 | ≥64 | ≥8 | 8 | ≤0.5 | ≤0.25 | 8 | ≥320 | ≥ 4 | ≤1 | ≤16 | ≥64 |
| Range | ≥64 to ≥64 | 4 to ≥64 | ≥8 to ≥8 | 2 to ≥64 | ≤0.5 to ≤0.5 | ≤0.25 to ≤0.25 | ≤4 to >128 | ≤20 to >320 | ≤0.25 to ≥4 | ≤1 to ≥16 | ≤16 to 64 | 16 to ≥64 | |||
| blaCTX-M-15, blaOXA-1 | n = 7 | Ethiopia, Tanzania, Sudan, Uzbekistan | Median | ≥64 | 16 | ≥8 | 4 | ≤0.5 | ≤0.25 | 8 | ≥320 | ≥ 4 | ≤1 | ≤16 | 32 |
| Range | 4 to ≥64 | ≤1 to ≥64 | ≥8 to ≥8 | ≤1 to ≥64 | ≤0.5 to ≤0.5 | ≤0.25 to ≤0.25 | 8 to ≥64 | ≤20 to >320 | 1 to ≥4 | ≤1 to ≥16 | ≤16 to 64 | ≤1 to ≥64 | |||
| blaCTX-M-15 with or without other BL | n = 7 | Mali, Sudan, Uganda | Median | ≥64 | 16 | ≥8 | 4 | ≤0.5 | ≤0.25 | ≤4 | ≥320 | 0.5 | ≤1 | ≤16 | 16 |
| Range | ≥64 to ≥64 | ≤1 to 16 | ≥8 to ≥8 | ≤1 to ≥64 | ≤0.5 to ≤0.5 | ≤0.25 to ≤0.25 | ≤4 to 64 | ≤20 to >320 | ≤0.25 to ≥4 | ≤1 to ≤1 | ≤16 to 64 | 2 to 16 | |||
| blaCTX-M-1 with or without other BL | n = 6 | Congo, Ghana, Lebanon, Mali, Nigeria, Sudan | Median | ≥64 | 4 | ≥8 | 2 | ≤0.5 | ≤0.25 | ≤4 | ≤20 | ≤0.25 | ≤1 | ≤16 | 16 |
| Range | ≥64 to ≥64 | ≤1 to 4 | ≥8 to ≥8 | 2 to 4 | ≤0.5 to ≤0.5 | ≤0.25 to ≤0.25 | ≤4 to ≤4 | ≤20 to >320 | ≤0.25 to ≥4 | ≤1 to ≥16 | ≤16 to 64 | 2 to ≥64 | |||
| blaCTX-M-27 | n = 5 | Mali, Sudan, Thailand | Median | ≥64 | 4 | ≥8 | 2 | ≤0.5 | ≤0.25 | ≤4 | ≥320 | ≤0.25 | ≤1 | ≤16 | 4 |
| Range | ≥64 to ≥64 | 4 to 4 | ≥8 to ≥8 | ≤1 to 2 | ≤0.5 to ≤0.5 | ≤0.25 to ≤0.25 | ≤4 to ≤4 | ≤20 to >320 | ≤0.25 to ≥4 | ≤1 to ≤1 | ≤16 to ≤16 | 2 to 16 | |||
| Other BL | n = 5 | Afghanistan, Djibouti, Sudan | Median | ≥64 | ≤1 | ≥8 | ≤1 | ≤0.5 | ≤0.25 | ≤4 | ≥320 | 1 | ≥ 16 | ≤16 | 16 |
| Range | ≥64 to ≥64 | ≤1 to ≥64 | 4 to ≥8 | ≤1 to 2 | ≤0.5 to ≤0.5 | ≤0.25 to ≤0.25 | ≤4 to 16 | ≤20 to >320 | ≤0.25 to ≥4 | ≤1 to ≥16 | ≤16 to 64 | 2 to 16 | |||
| Anonymized Strain Identity | Year of Isolation | Deployment Site | Sequence Type (ST) According to the Warwick University Scheme | Beta-Lactamase Genes (ESBL-Associated Underlined) | Genes Mediating Aminoglycoside Resistance | Genes Mediating Chloramphenicol Resistance | Genes Mediating Resistance against Trimethoprim or Sulfonamides | Genes Mediating Resistance against Macrolides or Lincosamides or Streptogramins | Genes Mediating Resistance against Fluoroquinolones | Genes Mediating Resistance against Tetracyclines | Genes Mediating Tolerance against Disinfectants |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BW-151 | 2007 | Uzbekistan | 617 | blaCTX-M-15, blaOXA-1 | aac(62019)-Ib-cr, aadA5, aph(3″)-Ib, aph(6)-Id | - | dfrA17, sul1, sul2 | - | aac(6′)-Ib-cr | tet(B) | sitABCD |
| BW-152 | 2007 | Democratic Republic of the Congo | 641 | blaCTX-M-1, blaTEM-1B | aadA1 | cmlA1 | sul3 | mph(A) | - | tet(B) | - |
| BW-153 | 2007 | Lebanon | 641 | blaCTX-M-1, blaTEM-1B | aadA1 | cmlA1 | sul3 | mph(A) | - | tet(B) | - |
| BW-154 | 2007 | Uzbekistan | 167 | blaCTX-M-15, blaOXA-1 | aac(3)-IIa, aac(6′)-Ib-cr, aadA5, aph(3″)-Ib, aph(6)-Id | - | dfrA17, sul2 | erm(B), mph(A) | aac(6′)-Ib-cr | tet(A) | sitABCD |
| BW-155 | 2007 | Afghanistan | 1312 | blaCTX-M-15, blaTEM-1B | aac(3)-IId, aadA2, aph(3″)-Ib, aph(6)-Id | catA1 | dfrA12, sul1, sul2 | mph(A) | - | tet(B) | sitABCD |
| BW-156 | 2007 | Democratic Republic of the Congo | 70 | blaCTX-M-15, blaTEM-1B | aph(3″)-Ib, aph(6)-Id | catA1 | dfrA7, sul1, sul2 | - | - | tet(B) | - |
| BW-157 | 2007 | Uzbekistan | 43 | blaCTX-M-15, blaOXA-1, blaTEM-1B | aac(6′)-Ib-cr, aadA1, aph(3″)-Ib, aph(6)-Id | - | dfrA1, sul2 | - | aac(6′)-Ib-cr | tet(A), tet(B) | - |
| BW-158 | 2007 | Sudan | 131 | blaCTX-M-15, blaOXA-1 | aac(6′)-Ib-cr, aadA5 | - | dfrA17, sul1 | mph(A) | aac(6′)-Ib-cr | - | - |
| BW-159 | 2008 | Sudan | 38 | blaCTX-M-15, blaTEM-1B | aadA1, aph(6)-Id | - | dfrA1, sul2 | - | - | - | - |
| BW-160 | 2009 | Sudan | 410 | blaCTX-M-15, blaOXA-1, blaTEM-1B | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id | - | dfrA14, sul2 | mph(A) | aac(6′)-Ib-cr | tet(A) | sitABCD |
| BW-161 | 2009 | Ghana | 446 | blaCTX-M-1 | - | - | - | - | - | - | - |
| BW-162 | 2009 | Sudan | 405 | blaCTX-M-15, blaOXA-1 | aac(6′)-Ib-cr | - | - | - | aac(6′)-Ib-cr | tet(B) | - |
| BW-163 | 2010 | Sudan | 10 | blaCTX-M-15, blaOXA-1 | aac(6′)-Ib-cr | - | - | - | aac(6′)-Ib-cr | tet(B) | - |
| BW-164 | 2010 | Afghanistan | 38 | blaCTX-M-14 | aadA2, aph(3″)-Ib, aph(6)-Id | - | dfrA12, sul2 | mph(A) | - | - | - |
| BW-165 | 2010 | Sudan | 767 | blaCTX-M-1 | aph(3″)-Ib, aph(6)-Id | - | sul2 | mph(A) | - | - | sitABCD |
| BW-166 | 2010 | Thailand | 3037 | blaCTX-M-27 | - | - | - | - | - | tet(A) | - |
| BW-167 | 2011 | Afghanistan | 10 | blaCTX-M-15, blaTEM-1B | aadA1, aph(3″)-Ib, aph(6)-Id | catA1 | dfrA1, sul2 | - | - | tet(A) | - |
| BW-168 | 2011 | Sudan | 940 | blaCTX-M-15 | aph(3″)-Ib, aph(6)-Id | - | dfrA1, sul2 | - | - | tet(B) | - |
| BW-169 | 2011 | Sudan | 131 | blaCTX-M-27 | aph(3″)-Ib, aph(6)-Id | - | sul1, sul2 | mph(A) | - | - | - |
| BW-170 | 2013 | Sudan | 131 | blaCTX-M-27 | aadA5, aph(3″)-Ib, aph(6)-Id | - | dfrA17, sul1, sul2 | mph(A) | - | - | - |
| BW-171 | 2013 | Djibouti | 5942 | blaSHV-12, blaTEM-1B | aac(3)-IV, aadA2, aph(3″)-Ib, aph(4)-Ia, aph(6)-Id | - | - | - | qnrS1 | tet(B) | -- |
| BW-172 | 2013 | Tanzania | 131 | blaCTX-M-15, blaOXA-1 | aac(6′)-Ib-cr, aadA5, aph(3″)-Ib, aph(6)-Id | - | dfrA17, sul2 | mph(A) | aac(6′)-Ib-cr | - | - |
| BW-173 | 2014 | Uganda | 656 | blaCTX-M-15, blaSHV-13, blaTEM-1B | aph(3″)-Ib, aph(6)-Id | - | dfrA14, sul2 | mph(A) | - | tet(B) | sitABCD |
| BW-174 | 2014 | Uganda | 448 | blaCTX-M-15, blaOXA-1, blaTEM-1B | aac(3)-IIa, aac(6′)-Ib-cr, aadA5, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id | - | dfrA17, sul2 | mph(A) | aac(6′)-Ib-cr | tet(A), tet(B) | - |
| BW-175 | 2014 | South Sudan | 38 | blaCTX-M-125, blaDHA-1, blaTEM-1B | aac(3)-IIa, aadA5 | - | dfrA17 | mph(A) | qnrB4 | - | - |
| BW-176 | 2014 | South Sudan | 38 | blaCTX-M-99-like (<99% sequence homology), blaTEM-1B | aac(3)-IIa, aadA5 | - | dfrA17 | mph(A) | - | - | - |
| BW-177 | 2014 | South Sudan | 38 | blaCTX-M-14, blaTEM-1B | aac(3)-IId, aadA5 | - | dfrA17, sul1 | mph(A) | - | - | - |
| BW-178 | 2014 | South Sudan | 636 | blaCTX-M-15, blaTEM-169, blaTEM-1B | aadA1, aph(3″)-Ib, aph(6)-Id | - | dfrA1, sul2 | mph(A) | - | tet(B) | - |
| BW-179 | 2014 | Uganda | 617 | blaCTX-M-15, blaOXA-1, blaTEM-1B | aac(3)-IIa, aac(6′)-Ib-cr, aadA5 | catA1 | dfrA17 | mph(A) | aac(6′)-Ib-cr | tet(B) | sitABCD |
| BW-180 | 2013 | South Sudan | 2624 | blaCTX-M-15, blaTEM-1B | aph(3″)-Ib, aph(6)-Id | - | sul2 | - | qnrS1 | - | - |
| BW-181 | 2014 | South Sudan | 616 | blaCTX-M-15, blaTEM-169 | aph(3″)-Ib, aph(6)-Id | - | dfrA14, sul2 | - | qnrS1 | tet(A) | - |
| BW-182 | 2014 | Nigeria | 38 | blaCTX-M-15, blaOXA-1, blaTEM-1B | aac(6′)-Ib-cr, aadA1, aph(3″)-Ib, aph(6)-Id | catA1 | dfrA1, sul2 | - | aac(6′)-Ib-cr | tet(A), tet(D) | - |
| BW-183 | 2014 | Democratic Republic of the Congo | 617 | blaCTX-M-15, blaOXA-1, blaTEM-1B | aac(3)-IIa, aac(6′)-Ib-cr, aadA5, aph(3″)-Ib, aph(6)-Id | catA1 | dfrA17, sul1, sul2 | mph(A) | aac(6′)-Ib-cr | tet(B) | sitABCD |
| BW-184 | 2014 | Mali | 38 | blaCTX-M-27 | aadA5, aph(3″)-Ib, aph(6)-Id | - | dfrA17, sul1, sul2 | mph(A) | - | - | - |
| BW-185 | 2014 | Mali | 101 | blaCTX-M-1 | aadA5 | - | dfrA17, sul2 | - | - | - | sitABCD |
| BW-186 | 2014 | South Sudan | 648 | blaCTX-M-15, blaOXA-1, blaTEM-1B | aac(3)-IIa, aac(6′)-Ib-cr, aadA5, aph(3″)-Ib, aph(6)-Id | catA1 | dfrA17, sul2 | mph(A) | aac(6′)-Ib-cr | tet(B) | sitABCD |
| BW-187 | 2015 | Mali | 38 | blaCTX-M-27 | aadA5, aph(3″)-Ib, aph(6)-Id | - | dfrA17, sul1, sul2 | mph(A) | - | - | - |
| BW-188 | 2015 | not further defined African destinations | 677 | blaCTX-M-15, blaTEM-1B | aadA1, aph(3″)-Ib, aph(6)-Id | - | dfrA1, sul1, sul2 | - | qnrS1 | - | - |
| BW-189 | 2014 | unknown or multiple deployment settings | 48 | blaCTX-M-15, blaOXA-1, blaTEM-1B | aac(3)-IIa, aac(6′)-Ib-cr, aadA12, aph(3″)-Ib, aph(6)-Id | - | dfrA14, dfrA5, sul2 | erm(B) | qnrB1, aac(6′)-Ib-cr | tet(A) | - |
| BW-192 | 2016 | Sudan & Uganda | 4305 | blaCTX-M-15, blaTEM-1B | aph(6)-Id | dfrA14, sul2 | - | mph(A) | - | tet(B) | - |
| BW-193 | 2016 | Nigeria | 88 | blaCTX-M-1, blaOXA-1 | aac(3)-IId, aadA5, ant(2″)-Ia, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id | - | dfrA17, dfrA36, sul1 | mph(A) | - | tet(B) | - |
| BW-194 | 2016 | Mali | 167 | blaCTX-M-15 | - | catA1 | - | erm(B), mph(A) | - | tet(B) | - |
| BW-195 | 2016 | Mali | 131 | blaCTX-M-15, blaTEM-1B | - | - | - | - | - | - | - |
| BW-198 | 2016 | Ethiopia | 405 | blaCTX-M-15, blaOXA-1 | aac(3)-IIa, aac(6′)-Ib-cr, aadA5 | catA1 | dfrA17, sul1 | mph(A) | aac(6′)-Ib-cr | tet(B) | |
| BW-199 | 2016 | Mali | 450 | blaCTX-M-15, blaTEM-1B | aph(3″)-Ib, aph(6)-Id | - | dfrA14, sul2 | - | qnrS1 | - | - |
| BW-201 | 2016 | India & Nepal | 1312 | blaCTX-M-15, blaTEM-1B | aac(3)-IId, aph(3″)-Ib, aph(6)-Id | catA1 | dfrA12, dfrA8, sul1, sul2 | mph(A) | - | tet(B) | sitABCD |
| BW-203 | 2016 | Sudan & Uganda | 394 | blaCTX-M-15 | aph(3″)-Ib, aph(6)-Id | - | dfrA1, dfrA7, sul2 | - | qnrS1 | - | - |
| BW-206 | 2016 | Mali | 38 | blaCTX-M-15 | aph(3″)-Ib, aph(6)-Id | - | dfrA1, sul2 | - | - | - | - |
| BW-207 | 2016 | Iraq | 131 | blaCTX-M-15, blaTEM-1B | aac(3)-IId, aadA5, | - | dfrA17, sul1 | - | - | sitABCD | |
| BW-208 | 2016 | Mali | 648 | blaCTX-M-15, blaOXA-1, blaTEM-1B | aac(6′)-Ib-cr, aadA5, aph(3″)-Ib, aph(6)-Id | catB3 | dfrA17, sul2 | - | aac(6′)-Ib-cr | tet(B) | - |
| BW-209 | 2016 | Iraq | 48 | blaCTX-M-15, blaTEM-1B | aac(3)-IId, aph(3″)-Ib, aph(6)-Id | catA1 | dfrA12, sul1, sul2 | mph(A) | qnrS1 | tet(B) | sitABCD |
| Antimicrobially Active Drug or Drug Combination | Minimum Inhibitory Concentration in Isolates without Detected Specific Resistance Genes, Mean Value (± Standard Deviation) | Minimum Inhibitory Concentration in Isolates with Detected Specific Resistance Genes, Mean Value (± Standard Deviation) | Significance P * |
|---|---|---|---|
| Gentamicin | 1 (±0) | 6 (±7.0) | n.e. # |
| Trimethoprim/sulfamethoxazole | 80 (±106.1) | 264.2 (±118.1) | 0.002 |
| Ciprofloxacin | 1.0 (±1.5) | 2.6 (±1.7) | 0.0006 |
| Moxifloxacin | 2.0 (±3.1) | 5.4 (±3.3) | 0.0002 |
| Anonymized Strain Identity | Year of Isolation | Deployment Site | Cytotoxic Necrotizing Factor (cnf-1) Gene 1 | Heat-Stable Enterotoxin (east1) Gene 2 | Hemolysin A (hlyA) Gene 3 | Secreted Autotransporter Toxin (sat) Gene 4 | Hemoglobin-Binding Protease (tsh) Gene 5 |
|---|---|---|---|---|---|---|---|
| BW-151 | 2007 | Uzbekistan | X | ||||
| BW-156 | 2007 | Democratic Republic of the Congo | X | X | |||
| BW-165 | 2010 | Sudan | X | ||||
| BW-166 | 2010 | Thailand | X | ||||
| BW-167 | 2011 | Afghanistan | X | ||||
| BW-168 | 2011 | Sudan | X | ||||
| BW-169 | 2011 | Sudan | X | X | |||
| BW-181 | 2014 | South Sudan | X | ||||
| BW-184 | 2014 | Mali | X | ||||
| BW-187 | 2015 | Mali | X | ||||
| BW-188 | 2015 | Not further defined African destinations | X | ||||
| BW-192 | 2016 | Sudan and Uganda | X | ||||
| BW-195 | 2016 | Mali | X | ||||
| BW-199 | 2016 | Mali | X | ||||
| BW-203 | 2016 | Sudan and Uganda | X | ||||
| BW-207 | 2016 | Iraq | X | X |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pankok, F.; Fuchs, F.; Loderstädt, U.; Kaase, M.; Balczun, C.; Scheithauer, S.; Frickmann, H.; Hagen, R.M. Molecular Epidemiology of Escherichia coli with Resistance against Third-Generation Cephalosporines Isolated from Deployed German Soldiers—A Retrospective Assessment after Deployments to the African Sahel Region and Other Sites between 2007 and 2016. Microorganisms 2022, 10, 2448. https://doi.org/10.3390/microorganisms10122448
Pankok F, Fuchs F, Loderstädt U, Kaase M, Balczun C, Scheithauer S, Frickmann H, Hagen RM. Molecular Epidemiology of Escherichia coli with Resistance against Third-Generation Cephalosporines Isolated from Deployed German Soldiers—A Retrospective Assessment after Deployments to the African Sahel Region and Other Sites between 2007 and 2016. Microorganisms. 2022; 10(12):2448. https://doi.org/10.3390/microorganisms10122448
Chicago/Turabian StylePankok, Frederik, Frieder Fuchs, Ulrike Loderstädt, Martin Kaase, Carsten Balczun, Simone Scheithauer, Hagen Frickmann, and Ralf Matthias Hagen. 2022. "Molecular Epidemiology of Escherichia coli with Resistance against Third-Generation Cephalosporines Isolated from Deployed German Soldiers—A Retrospective Assessment after Deployments to the African Sahel Region and Other Sites between 2007 and 2016" Microorganisms 10, no. 12: 2448. https://doi.org/10.3390/microorganisms10122448
APA StylePankok, F., Fuchs, F., Loderstädt, U., Kaase, M., Balczun, C., Scheithauer, S., Frickmann, H., & Hagen, R. M. (2022). Molecular Epidemiology of Escherichia coli with Resistance against Third-Generation Cephalosporines Isolated from Deployed German Soldiers—A Retrospective Assessment after Deployments to the African Sahel Region and Other Sites between 2007 and 2016. Microorganisms, 10(12), 2448. https://doi.org/10.3390/microorganisms10122448






