Nodulation and Growth Promotion of Chickpea by Mesorhizobium Isolates from Diverse Sources
Abstract
:1. Introduction
2. Material and Methods
2.1. Chickpea-Associated Mesorhizobium Strains
2.2. Nodulation and Plant Growth
2.3. Biochemical and Physiological Characterization
2.4. Data Analysis
3. Results
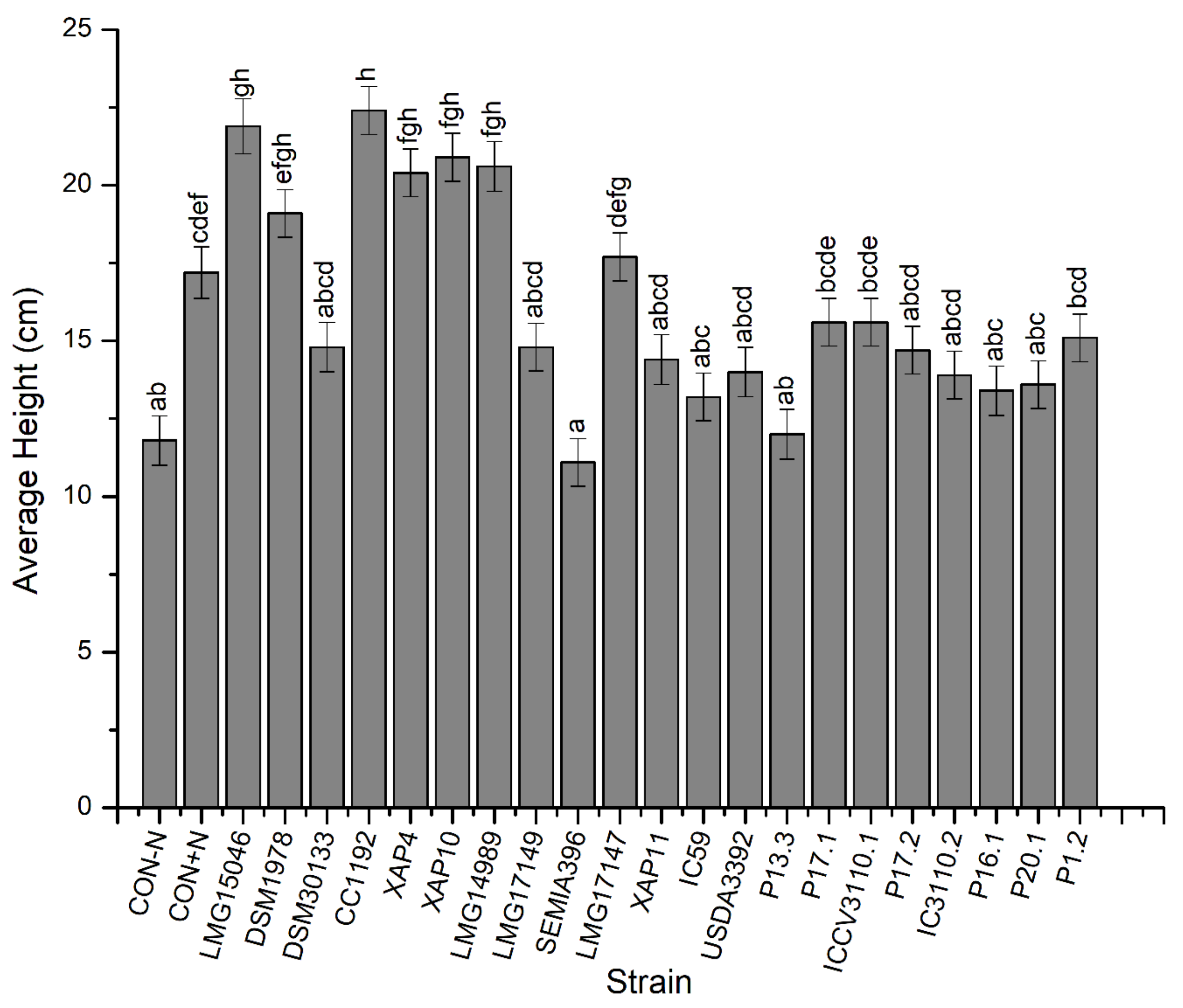
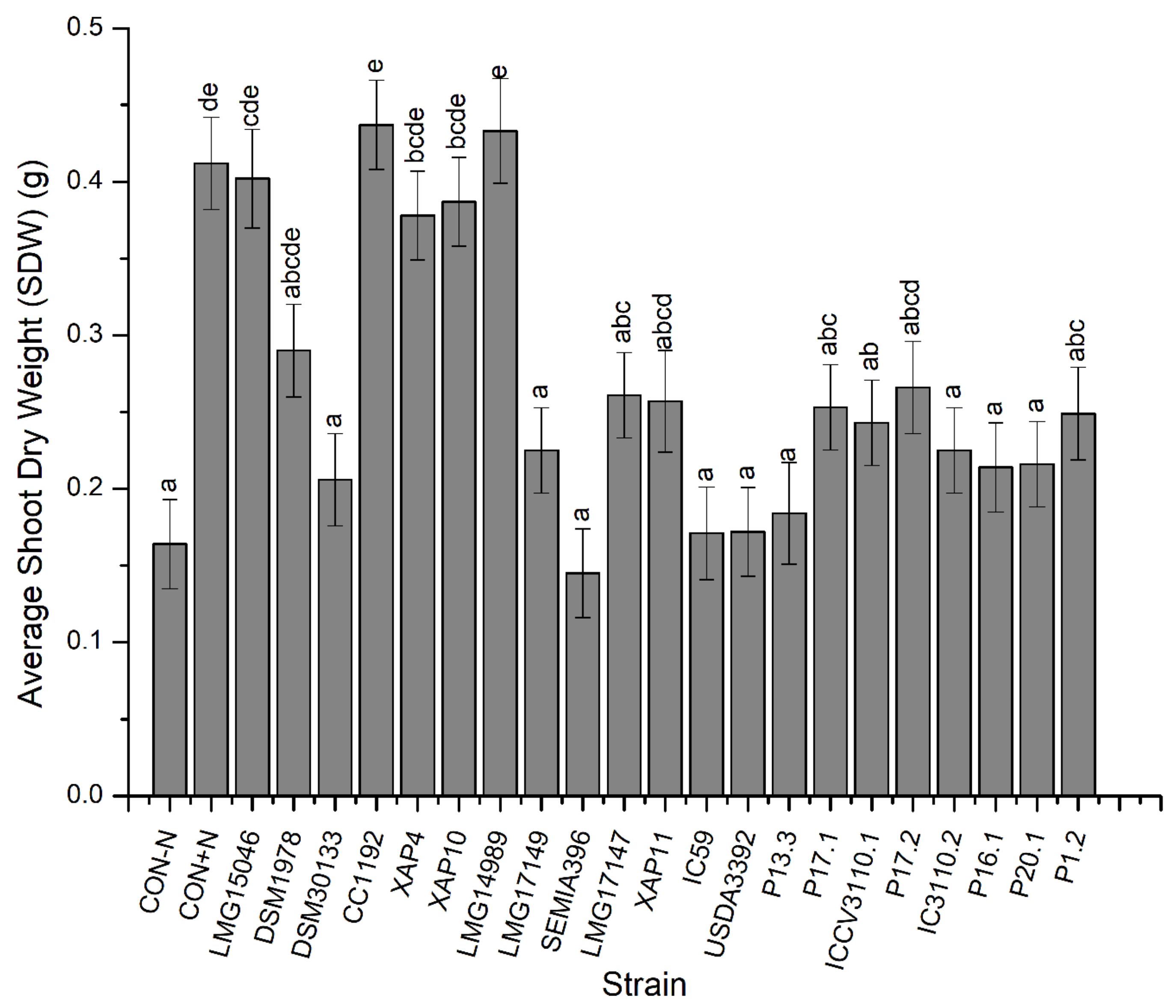
3.1. Nodulation and Plant Growth
3.2. Plant Growth Promotion Characteristics
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferguson, B.J.; Indrasumunar, A.; Hayashi, S.; Lin, M.-H.; Lin, Y.-H.; Reid, D.E.; Gresshoff, P.M. Molecular Analysis of Legume Nodule Development and Autoregulation. J. Integr. Plant Biol. 2010, 52, 61–76. [Google Scholar] [CrossRef]
- Miller, A.J.; Fan, X.; Orsel, M.; Smith, S.J.; Wells, D.M. Nitrate Transport and Signalling. J. Exp. Bot. 2007, 58, 2297–2306. [Google Scholar] [CrossRef]
- Murray, J.D.; Liu, C.-W.; Chen, Y.; Miller, A.J. Nitrogen Sensing in Legumes. J. Exp. Bot. 2016, 68, 1919–1926. [Google Scholar] [CrossRef] [Green Version]
- Rao, D.; Mohanty, S.; Chinmayee, A.; Atoliya, N. Rhizobial Taxonomy-Current Status. IUNFC Newsl. 2018, 3, 1–4. [Google Scholar]
- Zakhia, F.; de Lajudie, P. Taxonomy of Rhizobia. Agronomie 2001, 21, 569–576. [Google Scholar] [CrossRef]
- Zakhia, F.; Jeder, H.; Domergue, O.; Willems, A.; Cleyet-Marel, J.C.; Gillis, M.; Dreyfus, B.; De Lajudie, P. Characterisation of Wild Legume Nodulating Bacteria (LNB) in the Infra-Arid Zone of Tunisia. Syst. Appl. Microbiol. 2004, 27, 380–395. [Google Scholar] [CrossRef]
- Chandini; Randeep, K.; Ravendra, K.; Om, P. The Impact of Chemical Fertilizers on Our Environment and Ecosystem. Res. Trends Environ. Sci. 2019, 4, 69–86. [Google Scholar]
- Giller, K.E.; Cadisch, G. Future Benefits from Biological Nitrogen Fixation: An Ecological Approach to Agriculture. Plant Soil 1995, 174, 255–277. [Google Scholar] [CrossRef]
- Herridge, D.F. Inoculation Technology for Legumes. In Nitrogen-fixing Leguminous Symbioses; Springer: Dordrecht, The Netherlands, 2008; pp. 77–115. [Google Scholar]
- Rao, D.L.N.; Balachandar, D. Nitrogen Inputs from Biological Nitrogen Fixation in Indian Agriculture. In The Indian Nitrogen Assessment. Sources of Reactive Nitrogen, Environmental and Climate Effects, Management Options, and Policies; Abrol, Y.P., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 117–132. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Menge, D.N.L.; Reed, S.C.; Cleveland, C.C. Biological Nitrogen Fixation: Rates, Patterns and Ecological Controls in Terrestrial Ecosystems. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20130119. [Google Scholar] [CrossRef] [Green Version]
- Rascio, N.; La Rocca, N. Biological Nitrogen Fixation. Ref. Modul. Earth Syst. Environ. Sci. 2013. [Google Scholar] [CrossRef]
- Burén, S.; Rubio, L.M. State of the Art in Eukaryotic Nitrogenase Engineering. FEMS Microbiol. Lett. 2018, 365, fnx274. [Google Scholar] [CrossRef]
- Udvardi, M.; Kahn, M. Evolution of the (Brady)Rhizobium-Legume Symbiosis: Why Do Bacteroids Fix Nitrogen? Symbiosis (Philadelphia, PA) 1993, 14, 87–101. [Google Scholar]
- Moulin, L.; Munive, A.; Dreyfus, B.; Boivin-Masson, C. Nodulation of Legumes by Members of the β-Subclass of Proteobacteria. Nature 2001, 411, 948–950. [Google Scholar] [CrossRef]
- Weir, B.S. The Current Taxonomy of Rhizobia|NZ Rhizobia. Available online: https://www.rhizobia.co.nz/taxonomy/rhizobia (accessed on 1 June 2021).
- Berrada, H. Taxonomy of the Rhizobia: Current Perspectives. Br. Microbiol. Res. J. 2014, 4, 616–639. [Google Scholar] [CrossRef] [Green Version]
- Perret, X.; Staehelin, C.; Broughton, W.J. Molecular Basis of Symbiotic Promiscuity. Microbiol. Mol. Biol. Rev. 2000, 64, 180. [Google Scholar] [CrossRef] [Green Version]
- Keet, J.H.; Ellis, A.G.; Hui, C.; Le Roux, J.J. Legume-Rhizobium Symbiotic Promiscuity and Effectiveness Do Not Affect Plant Invasiveness. Ann. Bot. 2017, 119, 1319–1331. [Google Scholar] [CrossRef]
- Lima, A.S.; Nóbrega, R.S.A.; Barberi, A.; Da Silva, K.; Ferreira, D.F.; Moreira, F.M.D.S. Nitrogen-Fixing Bacteria Communities Occurring in Soils under Different Uses in the Western Amazon Region as Indicated by Nodulation of Siratro (Macroptilium Atropurpureum). Plant Soil 2009, 319, 127–145. [Google Scholar] [CrossRef]
- Ndungu, S.M.; Messmer, M.M.; Ziegler, D.; Gamper, H.A.; Mészáros, É.; Thuita, M.; Vanlauwe, B.; Frossard, E.; Thonar, C. Cowpea (Vigna Unguiculata L. Walp) Hosts Several Widespread Bradyrhizobial Root Nodule Symbionts across Contrasting Agro-Ecological Production Areas in Kenya. Agric. Ecosyst. Environ. 2018, 261, 161–171. [Google Scholar] [CrossRef]
- Silva, F.V.; Simões-Araújo, J.L.; Silva Júnior, J.P.; Xavier, G.R.; Rumjanek, N.G. Genetic Diversity of Rhizobia Isolates from Amazon Soils Using Cowpea (Vigna Unguiculata) as Trap Plant. Braz. J. Microbiol. 2012, 43, 682–691. [Google Scholar] [CrossRef] [Green Version]
- Laranjo, M.; Alexandre, A.; Rivas, R.; Velázquez, E.; Young, J.P.W.; Oliveira, S. Chickpea Rhizobia Symbiosis Genes Are Highly Conserved across Multiple Mesorhizobium Species. FEMS Microbiol. Ecol. 2008, 66, 391–400. [Google Scholar] [CrossRef]
- Rivas, R.; Laranjo, M.; Mateos, P.F.; Oliveira, S.; Martínez-Molina, E.; Velázquez, E. Strains of Mesorhizobium Amorphae and Mesorhizobium Tianshanense, Carrying Symbiotic Genes of Common Chickpea Endosymbiotic Species, Constitute a Novel Biovar (Ciceri) Capable of Nodulating Cicer arietinum. Lett. Appl. Microbiol. 2007, 44, 412–418. [Google Scholar] [CrossRef]
- Zhang, J.J.; Liu, T.Y.; Chen, W.F.; Wang, E.T.; Sui, X.H.; Zhang, X.X.; Li, Y.; Li, Y.; Chen, W.X. Mesorhizobium Muleiense Sp. Nov., Nodulating with Cicer arietinum L. Int. J. Syst. Evol. Microbiol. 2012, 62, 2737–2742. [Google Scholar] [CrossRef]
- Dekkiche, S.; Benguedouar, A.; Sbabou, L.; Taha, K.; Filali-Maltouf, A.; Béna, G. Chickpea (Cicer arietinum) Is Nodulated by Unexpected Wide Diversity of Mesorhizobium Species in Eastern Algeria. Arch. Agron. Soil Sci. 2018, 64, 285–297. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Sathya, A.; Vijayabharathi, R.; Varshney, R.K.; Gowda, C.L.L.; Krishnamurthy, L. Plant Growth Promoting Rhizobia: Challenges and Opportunities. 3 Biotech 2015, 5, 355–377. [Google Scholar] [CrossRef] [Green Version]
- Berg, G. Plant–Microbe Interactions Promoting Plant Growth and Health: Perspectives for Controlled Use of Microorganisms in Agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef]
- Gupta, S.; Pandey, S. ACC Deaminase Producing Bacteria with Multifarious Plant Growth Promoting Traits Alleviates Salinity Stress in French Bean (Phaseolus Vulgaris) Plants. Front. Microbiol. 2019, 10, 1–17. [Google Scholar] [CrossRef]
- Menéndez, E.; Pérez-Yépez, J.; Hernández, M.; Rodríguez-Pérez, A.; Velázquez, E.; León-Barrios, M. Plant Growth Promotion Abilities of Phylogenetically Diverse Mesorhizobium Strains: Effect in the Root Colonization and Development of Tomato Seedlings. Microorganisms 2020, 8, 412. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, S.K.; Mohammed, M.; Ibny, F.Y.I.; Dakora, F.D. Rhizobia as a Source of Plant Growth-Promoting Molecules: Potential Applications and Possible Operational Mechanisms. Front. Sustain. Food Syst. 2021, 4, 1–14. [Google Scholar] [CrossRef]
- Toukabri, W.; Ferchichi, N.; Hlel, D.; Jadlaoui, M.; Kheriji, O.; Mhamdi, R.; Trabelsi, D. Response of Intercropped Barley and Fenugreek to Mono- and Co-Inoculation with Sinorhizobium Meliloti F42 and Variovorax Paradoxus F310 under Contrasting Agroclimatic Regions. Arch. Microbiol. 2021, 203, 1657–1670. [Google Scholar] [CrossRef]
- Saleem, M.; Arshad, M.; Hussain, S.; Bhatti, A.S. Perspective of Plant Growth Promoting Rhizobacteria (PGPR) Containing ACC Deaminase in Stress Agriculture. J. Ind. Microbiol. Biotechnol. 2007, 34, 635–648. [Google Scholar] [CrossRef]
- Brígido, C.; Glick, B.R.; Oliveira, S. Survey of Plant Growth-Promoting Mechanisms in Native Portuguese Chickpea Mesorhizobium Isolates. Microb. Ecol. 2017, 73, 900–915. [Google Scholar] [CrossRef]
- Zoundji, M.C.C.; Houngnandan, P.; Boko, F.; Toukourou, F. Characterization of Indigenous Rhizobia Strains Associated to Soybean [Glycine max (L.) Merrill] in Benin. Int. J. Plant Soil Sci. 2020, 32, 35–46. [Google Scholar] [CrossRef]
- Sridevi, M.; Mallaiah, K.V. Phosphate Solubilization by Rhizobium Strains. Indian J. Microbiol. 2009, 49, 98–102. [Google Scholar] [CrossRef] [Green Version]
- Ahemad, M.; Khan, M.S. Effects of Pesticides on Plant Growth Promoting Traits of Mesorhizobium Strain MRC4. J. Saudi Soc. Agric. Sci. 2012, 11, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Shahid, M.; Khan, M.S.; Syed, A.; Marraiki, N.; Elgorban, A.M. Mesorhizobium Ciceri as Biological Tool for Improving Physiological, Biochemical and Antioxidant State of Cicer aritienum (L.) under Fungicide Stress. Sci. Rep. 2021, 11, 1–18. [Google Scholar] [CrossRef]
- Nascimento, F.; Brígido, C.; Alho, L.; Glick, B.R.; Oliveira, S. Enhanced Chickpea Growth-Promotion Ability of a Mesorhizobium Strain Expressing an Exogenous ACC Deaminase Gene. Plant Soil 2012, 353, 221–230. [Google Scholar] [CrossRef]
- Uchiumi, T.; Ohwada, T.; Itakura, M.; Mitsui, H.; Nukui, N.; Dawadi, P.; Kaneko, T.; Tabata, S.; Yokoyama, T.; Tejima, K.; et al. Expression Islands Clustered on the Symbiosis Island of the Mesorhizobium Loti Genome. J. Bacteriol. 2004, 186, 2439–2448. [Google Scholar] [CrossRef] [Green Version]
- Valeria, C.P.; Echeverria, M.; Sánchez, C.; Ugalde, R.A.; Menéndez, A.B.; Lepek, V.C. Engineered ACC Deaminase-Expressing Free-Living Cells of Mesorhizobium Loti Show Increased Nodulation Effi Ciency and Competitiveness on Lotus Spp. Gen. Appl. Microbiol. 2010, 56, 331–338. [Google Scholar]
- Nukui, N.; Minamisawa, K.; Ayabe, S.-I.; Aoki, T. Expression of the 1-Aminocyclopropane-1-Carboxylic Acid Deaminase Gene Requires Symbiotic Nitrogen-Fixing Regulator Gene NifA2 in Mesorhizobium Loti MAFF303099. Appl. Environ. Microbiol. 2006, 72, 4964–4969. [Google Scholar] [CrossRef] [Green Version]
- Purushothaman, R.; Upadhyaya, H.D.; Gaur, P.M.; Gowda, C.L.L.; Krishnamurthy, L. Kabuli and Desi Chickpeas Differ in Their Requirement for Reproductive Duration. Field Crop. Res. 2014, 163, 24–31. [Google Scholar] [CrossRef] [Green Version]
- Saxena, M.C. Recent Advances in Chickpea Agronomy. In Proceedings of the International Workshop on Chickpea Improvement, Hyderabad, India, 28 February–2 March 1979; ICRISAT: Hyderabad, India, 1979. [Google Scholar]
- Upadhyaya, H.D.; Dwivedi, S.L.; Baum, M.; Varshney, R.K.; Udupa, S.M.; Gowda, C.L.; Hoisington, D.; Singh, S. Genetic Structure, Diversity, and Allelic Richness in Composite Collection and Reference Set in Chickpea (Cicer arietinum L.). BMC Plant Biol. 2008, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mpai, T.; Maseko, S.T. Possible Benefits and Challenges Associated with Production of Chickpea in Inland South Africa. Acta Agric. Scand. Sect. B Soil Plant Sci. 2018, 68, 479–488. [Google Scholar] [CrossRef]
- Jukanti, A.K.; Gaur, P.M.; Gowda, C.L.L.; Chibbar, R.N. Nutritional Quality and Health Benefits of Chickpea (Cicer arietinum L.): A Review. Br. J. Nutr. 2012, 108, S11–S26. [Google Scholar] [CrossRef]
- Fikre, A.; Desmae, H.; Ahmed, S. Tapping the Economic Potential of Chickpea in Sub-Saharan Africa. Agronomy 2020, 10, 2015–2018. [Google Scholar] [CrossRef]
- Naseer, I.; Ahmad, M.; Nadeem, S.M.; Ahmad, I.; Najm-ul-Seher; Zahir, Z.A. Rhizobial Inoculants for Sustainable Agriculture: Prospects and Applications. In In Biofertilizers for Sustainable Agriculture and Environment; Springer: Cham, Switzerland, 2019; pp. 245–283. [Google Scholar] [CrossRef]
- van der Maesen, L.J.G. Origin, History and Taxonomy of Chickpea. In The Chickpea; C.A.B. International: Wallingford, UK, 1987; pp. 11–34. [Google Scholar]
- De Candolle, A. Origin of Cultivated Plants; Hather: New York, NY, USA, 1885. [Google Scholar]
- Howieson, J.G.; Dilworth, M.J. Working with Rhizobia; Australia Government: Canberra, Australia, 2016. [Google Scholar]
- Zhao, C.T.; Wang, E.T.; Zhang, Y.M.; Chen, W.F.; Sui, X.H.; Chen, W.X.; Liu, H.C.; Zhang, X.X. Mesorhizobium Silamurunense Sp. Nov., Isolated from Root Nodules of Astragalus Species. Int. J. Syst. Evol. Microbiol. 2012, 62, 2180–2186. [Google Scholar] [CrossRef] [Green Version]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank: Update. Nucleic Acids Res. 2004, 32, D23–D26. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Cadahia, E.; Leyva, A.; Ruiz-Argiieso, T. Indigenous Plasmids and Cultural Characteristics of Rhizobia Nodulating Chickpeas (Cicer arietinum L.). Arch Microbiol 1986, 146, 239–244. [Google Scholar] [CrossRef]
- Nour, S.M.; Fernandez, M.P.; Normand, P.; Cleyet-Marel, J.-C. Rhizobium Ciceri Sp. Nov., Consisting of Strains That Nodulate Chickpeas. Int. J. Syst. Bacteriol. 1994, 44, 511–522. [Google Scholar] [CrossRef] [Green Version]
- Jarvis, B.D.W.; Van Berkum, P.; Chen, W.X.; Nour, S.M.; Fernandez, M.P.; Cleyet-Marel, J.C.; Gillis, M. Transfer of Rhizobium Loti, Rhizobium Huakuii, Rhizobium Ciceri, Rhizobium Mediterraneum, and Rhizobium Tianshanense to Mesorhizobium Gen. Nov. Int. J. Syst. Bacteriol. 1997, 47, 895–898. [Google Scholar] [CrossRef] [Green Version]
- Hill, Y.; Colombi, E.; Bonello, E.; Haskett, T.; Ramsay, J.; O’Hara, G.; Terpolilli, J. Evolution of Diverse Effective N2-Fixing Microsymbionts of Cicer arietinum Following Horizontal Transfer of the Mesorhizobium Ciceri CC1192 Symbiosis Integrative and Conjugative Element. Appl. Environ. Microbiol. 2021, 87, 1–16. [Google Scholar] [CrossRef]
- Nour, S.M.; Cleyet-Marel, J.-C.; Normand, P.; Fernandez, M.P. Genomic Heterogeneity of Strains Nodulating Chickpeas (Cicer arietinum L.) and Description of Rhizobiurn Mediterraneurn Sp. Nov. Int. J. Syst. Bacteriol. 1995, 45, 640–648. [Google Scholar] [CrossRef]
- de Toledo, B.F.B.; Marcondes, J.; de Macedo Lemos, E.G. Caracterização de Rizóbios Indicados Para Produção de Inoculantes Por Meio de Sequenciamento Parcial Do 16S RRNA. Pesqui. Agropecu. Bras. 2009, 44, 384–391. [Google Scholar] [CrossRef] [Green Version]
- Kamble, P.M.; Singh, A.; Kashyap, L.R. Characterization of Intrinsic Variability of Mesorhizobium Ciceri Isolates of Cultivated Fields. Indian J. Exp. Biol. 2006, 44, 671–674. [Google Scholar]
- Kumar, R.O.P.; Rao, J.V.D.K.; Sudarshan, M.R.; Usha, K. Rhizobium Germplasm Resources at ICRISAT Center; International Crops Research Institute for the Semi-arid Tropics: Patancheru, India, 1991; ISBN 9780333227794. [Google Scholar]
- Moreno, M.T.; Cubero, J.I. Variation in Cicer arietinum L. Euphytica 1978, 27, 465–485. [Google Scholar] [CrossRef]
- Sauer, D.B.; Burroughs, R. Disinfection of Seeds Surfaces with Sodium Hypochlorite. Phytopathology 1986, 76, 745–749. [Google Scholar] [CrossRef]
- Tzfira, T. Plant Physiology. Third Edition. By Lincoln Taiz and, Eduardo Zeiger. Sunderland (Massachusetts): Sinauer Associates. ISBN: 0–87893–823–0. 2002. Q. Rev. Biol. 2003, 78, 231–232. [Google Scholar] [CrossRef]
- Birhan, A.; Tadele, T.; Yilkal, B.; Seble, W.Y. Phenotypic Characterization and Symbiotic Effectiveness Test of Chickpea (Cicer arietinum L.) Rhizobia Isolated from Dejen and Aneded Districts, East Gojjam Zone, Amahara Region, Ethiopia. Afr. J. Biotechnol. 2018, 17, 730–738. [Google Scholar] [CrossRef] [Green Version]
- Gordon, S.A.; Weber, R.P. Colorimetric Estimation of Indoleacetic Acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef] [Green Version]
- Zafar, M.; Ahmed, N.; Mustafa, G.; Zahir, Z.A.; Simms, E.L. Molecular and Biochemical Characterization of Rhizobia from Chickpea (Cicer arietinum). Pakistan J. Agric. Sci. 2017, 54, 373–381. [Google Scholar] [CrossRef] [Green Version]
- Swinehart, D.F. The Beer-Lambert Law. J. Chem. Educ. 1962, 39, 333–335. [Google Scholar] [CrossRef]
- Pikovskaya, R. Mobilization of Phosphorus in Soil in Connection with the Vital Activity of Some Microbial Species—ScienceOpen. Available online: https://www.scienceopen.com/document?vid=44a8aa7f-fad0-4038-a97b-a766d1a3b6fb (accessed on 25 August 2021).
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. 2020. Available online: https://www.r-project.org/ (accessed on 16 July 2022).
- Schwyn, B.; Neilands, J.B. Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Penrose, D.M.; Glick, B.R. Methods for Isolating and Characterizing ACC Deaminase-Containing Plant Growth-Promoting Rhizobacteria. Physiol. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef]
- Adissie, S.; Adgo, E.; Feyisa, T. Effect of Rhizobial Inoculants and Micronutrients on Yield and Yield Components of Faba Bean (Vicia Faba L.) on Vertisol of Wereillu District, South Wollo, Ethiopia. Cogent Food Agric. 2020, 6, 1747854. [Google Scholar] [CrossRef]
- Gunnabo, A.H.; van Heerwaarden, J.; Geurts, R.; Wolde-meskel, E.; Degefu, T.; Giller, K.E. Symbiotic Interactions between Chickpea (Cicer arietinum L.) Genotypes and Mesorhizobium Strains. Symbiosis 2020, 82, 235–248. [Google Scholar] [CrossRef]
- Ben Romdhane, S.; Tajini, F.; Trabelsi, M.; Aouani, M.E.; Mhamdi, R. Competition for Nodule Formation between Introduced Strains of Mesorhizobium Ciceri and the Native Populations of Rhizobia Nodulating Chickpea (Cicer arietinum) in Tunisia. World J. Microbiol. Biotechnol. 2007, 23, 1195–1201. [Google Scholar] [CrossRef]
- Afzal, I.; Rehman, H.U.; Naveed, M.; Basra, S.M.A. Recent Advances in Seed Enhancements. In New Challenges in Seed Biology—Basic and Translational Research Driving Seed Technology; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef] [Green Version]
- Gaur, P.M.; Kumar, J.; Gowda, C.L.; Pande, S.; Siddique, K.H.M.; Khan, T.; Warkentin, T.; Chaturvedi, S.K.; Than, A.M.; Abdi, K.D. Breeding Chickpea for Early Phenology: Perspectives, Progress and Prospects. In Proceedings of the Fourth International Food Legumes Research Conference, New Delhi, India, 18–22 October 2008; pp. 1–11. [Google Scholar]
- Thudi, M.; Chitikineni, A.; Liu, X.; He, W.; Roorkiwal, M.; Yang, W.; Jian, J.; Doddamani, D.; Gaur, P.M.; Rathore, A.; et al. Recent Breeding Programs Enhanced Genetic Diversity in Both Desi and Kabuli Varieties of Chickpea (Cicer arietinum L.). Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Gul, R.; Khan, H.; Khan, N.U.; Khan, F.U. Characterization of Chickpea Germplasm for Nodulation and Effect of Rhizobium Inoculation on Nodules Number and Seed Yield. J. Anim. Plant Sci. 2014, 24, 1421–1429. [Google Scholar]
- Muniz, A.; Nunes, R.H.; Silva, T.A.; Brose, E.; Dala, C. Seleção de Estirpes de Rizóbio Para Grão-de-Bico; Congreso Latinoamericano De La Ciencia Del Suelo: Mar del Plata, Argentina, 2012. [Google Scholar]
- Crow, V.L.; Jarvis, B.D.W.; Greenwood, R.M. Deoxyribonucleic Acid Homologies among Acid-Producing Strains of Rhizobium. Int. J. Syst. Bacteriol. 1981, 31, 152–172. [Google Scholar] [CrossRef] [Green Version]
- Ramsay, J.P.; Sullivan, J.T.; Stuart, G.S.; Lamont, I.L.; Ronson, C.W. Excision and Transfer of the Mesorhizobium Loti R7A Symbiosis Island Requires an Integrase IntS, a Novel Recombination Directionality Factor RdfS, and a Putative Relaxase RlxS. Mol. Microbiol. 2006, 62, 723–734. [Google Scholar] [CrossRef]
- Sullivan, J.T.; Ronson, C.W. Evolution of Rhizobia by Acquisition of a 500-Kb Symbiosis Island That Integrates into a Phe-TRNA Gene. Proc. Natl. Acad. Sci. USA 1998, 95, 5145–5149. [Google Scholar] [CrossRef] [Green Version]
- Greenlon, A.; Chang, P.L.; Damtew, Z.M.; Muleta, A.; Carrasquilla-Garcia, N.; Kim, D.; Nguyen, H.P.; Suryawanshi, V.; Krieg, C.P.; Yadav, S.K.; et al. Global-Level Population Genomics Reveals Differential Effects of Geography and Phylogeny on Horizontal Gene Transfer in Soil Bacteria. Proc. Natl. Acad. Sci. USA 2019, 116, 15200–15209. [Google Scholar] [CrossRef] [Green Version]
- Haskett, T.L.; Terpolilli, J.J.; Bekuma, A.; O’Hara, G.W.; Sullivan, J.T.; Wang, P.; Ronson, C.W.; Ramsay, J.P. Assembly and Transfer of Tripartite Integrative and Conjugative Genetic Elements. Proc. Natl. Acad. Sci. USA 2016, 113, 12268–12273. [Google Scholar] [CrossRef]
- Nandasena, K.G.; O’Hara, G.W.; Tiwari, R.P.; Sezmiş, E.; Howieson, J.G. In Situ Lateral Transfer of Symbiosis Islands Results in Rapid Evolution of Diverse Competitive Strains of Mesorhizobia Suboptimal in Symbiotic Nitrogen Fixation on the Pasture Legume Biserrula pelecinus L. Environ. Microbiol. 2007, 9, 2496–2511. [Google Scholar] [CrossRef]
- Wardell, G.E.; Hynes, M.F.; Young, P.J.; Harrison, E. Why Are Rhizobial Symbiosis Genes Mobile? Philos. Trans. R. Soc. B Biol. Sci. 2022, 377. [Google Scholar] [CrossRef]
- Porter, S.S.; Faber-Hammond, J.; Montoya, A.P.; Friesen, M.L.; Sackos, C. Dynamic Genomic Architecture of Mutualistic Cooperation in a Wild Population of Mesorhizobium. ISME J. 2019, 13, 301–315. [Google Scholar] [CrossRef] [Green Version]
- Elias, N.V.; Herridge, D.F. Naturalised Populations of Mesorhizobia in Chickpea (Cicer arietinum L.) Cropping Soils: Effects on Nodule and Productivity of Commercial Chickpea. Plant Soil 2015, 387, 233–249. [Google Scholar] [CrossRef]
- Benezech, C.; Doudement, M.; Gourion, B. Legumes Tolerance to Rhizobia Is Not Always Observed and Not Always Deserved. Cell. Microbiol. 2020, 22, 1–9. [Google Scholar] [CrossRef]
- Nziguheba, G.; Zingore, S.; Kihara, J.; Merckx, R.; Njoroge, S.; Otinga, A.; Vandamme, E.; Vanlauwe, B. Phosphorus in Smallholder Farming Systems of Sub-Saharan Africa: Implications for Agricultural Intensification. Nutr. Cycl. Agroecosyst. 2016, 104, 321–340. [Google Scholar] [CrossRef]
- Rao, I.M.; Sommer, R. Phosphorus Fertilization and Management in Soils of Sub- Saharan Africa. In Soil Phosphorus (Advances in Soil Science); La, R., Stewart, B.A., Eds.; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Yadav, A.; Singh, R.P.; Singh, A.L.; Singh, M. Identification of Genes Involved in Phosphate Solubilization and Drought Stress Tolerance in Chickpea Symbiont Mesorhizobium Ciceri Ca181. Arch. Microbiol. 2021, 203, 1167–1174. [Google Scholar] [CrossRef]
- Muleta, A.; Tesfaye, K.; Haile Selassie, T.H.; Cook, D.R.; Assefa, F. Phosphate Solubilization and Multiple Plant Growth Promoting Properties of Mesorhizobium Species Nodulating Chickpea from Acidic Soils of Ethiopia. Arch. Microbiol. 2021, 203, 2129–2137. [Google Scholar] [CrossRef]
- Kryvoruchko, I.S. Zn-Use Efficiency for Optimization of Symbiotic Nitrogen Fixation in Chickpea (Cicer arietinum L.). Turk. J. Botany 2017, 41, 423–441. [Google Scholar] [CrossRef]
- Howieson, J.G.; Malden, J.; Yates, R.J.; O’Hara, G.W. Techniques for the Selection and Development of Elite Inoculant Strains of Rhizobium Leguminosarum in Southern Australia. Symbiosis 2000, 28, 33–48. [Google Scholar]
- Ben Romdhane, S.; Aouani, M.E.; Trabelsi, M.; De Lajudie, P.; Mhamdi, R. Selection of High Nitrogen-Fixing Rhizobia Nodulating Chickpea (Cicer arietinum) for Semi-Arid Tunisia. J. Agron. Crop Sci. 2008, 194, 413–420. [Google Scholar] [CrossRef]
- L’Taief, B.; Sifi, B.; Gtari, M.; Zaman-Allah, M.; Lachaâl, M. Phenotypic and Molecular Characterization of Chickpea Rhizobia Isolated from Different Areas of Tunisia. Can. J. Microbiol. 2007, 53, 427–434. [Google Scholar] [CrossRef]
- Haile, W.; Beyene, S. Response of chickpea (Cicer arietinum L.) to Nitrogen and Phosphorus Fertilizers in Halaba and Taba, Southern Ethiopia. Ethiop. J. Nat. Resour. 2013, 13, 115–128. [Google Scholar]
- Wolde-meskel, E.; van Heerwaarden, J.; Abdulkadir, B.; Kassa, S.; Aliyi, I.; Degefu, T.; Wakweya, K.; Kanampiu, F.; Giller, K.E. Additive Yield Response of Chickpea (Cicer arietinum L.) to Rhizobium Inoculation and Phosphorus Fertilizer across Smallholder Farms in Ethiopia. Agric. Ecosyst. Environ. 2018, 261, 144–152. [Google Scholar] [CrossRef]



| Strain 1 | Original Host 2 | Region of Original Isolation | References |
|---|---|---|---|
| Mesorhizobium ciceri LMG14989T | Cicer arietinum L. | Spain | [56,57,58] |
| Mesorhizobium ciceri CC1192 | Cicer arietinum L. | Israel | [59] |
| Mesorhizobium mediterranean USDA3392 | Cicer arietinum L. | Brazil | [58,60] |
| Mesorhizobium sp. SEMIA396 | Cicer arietinum L. | North America | [61] |
| Mesorhizobium sp. DSM30133 | Cicer arietinum L. | Belgium | [56] |
| Mesorhizobium sp. DSM1978 | Cicer arietinum L. | India | [62] |
| Mesorhizobium sp. XAP4 | Cicer arietinum L. | USA (Wisconsin) | Unknown |
| Mesorhizobium sp. XAP10 | Cicer arietinum L. | Australia | Unknown |
| Mesorhizobium sp. XAP11 | Cicer arietinum L. | India | Unknown |
| Mesorhizobium sp. LMG15046 | Cicer arietinum L. | India | [56] |
| Mesorhizobium sp. LMG17147 | Cicer arietinum L. | India | [56] |
| Mesorhizobium sp. LMG17149 | Cicer arietinum L. | Russia | [56] |
| Mesorhizobium sp. IC59 * | Cicer arietinum L. | India | [63] |
| Mesorhizobium sp. ICCV3110.1 | Cicer arietinum L. (ICCV3110) | Mbombela, Mpumalanga, South Africa | This study |
| Mesorhizobium sp. P16.1 | Cicer arietinum L. (ICCV3110) | Mbombela, Mpumalanga, South Africa | This study |
| Mesorhizobium sp. P13.3 | Cicer arietinum L. (ICCV92944) | Mbombela, Mpumalanga, South Africa | This study |
| Mesorhizobium sp. P17.1 | Cicer arietinum L. (ICCV4105) | Mbombela, Mpumalanga, South Africa | This study |
| Mesorhizobium sp. P17.2 | Cicer arietinum L. (ICCV4105) | Mbombela, Mpumalanga, South Africa | This study |
| Mesorhizobium sp. P20.1 | Cicer arietinum L. (ICCV4110) | Mbombela, Mpumalanga, South Africa | This study |
| Mesorhizobium sp. P1.2 | Cicer arietinum L. (ICCV4110) | Mbombela, Mpumalanga, South Africa | This study |
| Mesorhizobium sp. IC3110.2 | Cicer arietinum L. (ICCV3110) | Mbombela, Mpumalanga, South Africa | This study |
| Measured Parameters | Sources of Variations | ||
|---|---|---|---|
| Strain | Genotype | Genotype × Strain | |
| Height | p < 0.001 | p < 0.001 | p > 0.05 |
| SDW | p < 0.001 | p < 0.001 | p > 0.05 |
| NFW | p < 0.001 | p < 0.001 | p > 0.05 |
| Light Variety | Dark Variety | ICCV3110 | ICCV3111 | ICCV3203 | ICCV4105 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | SDW | RSE | Score | SDW | RSE | Score | SDW | RSE | Score | SDW | RSE | Score | SDW | RSE | Score | SDW | RSE | Score |
| CON− N | 0.28 | 66.3 | IE | 0.18 | 70.4 | E | 0.12 | 27.6 | IE | 0.17 | 27.5 | IE | 0.07 | 19.2 | IE | 0.16 | 42.3 | LE |
| CON+ N | 0.43 | 100.0 | HE | 0.25 | 100.0 | HE | 0.44 | 100.0 | HE | 0.63 | 100.0 | HE | 0.35 | 100.0 | HE | 0.39 | 100.0 | HE |
| P16.1 | 0.40 | 94.1 | HE | 0.34 | 132.4 | HE | 0.13 | 28.7 | IE | 0.13 | 21.3 | IE | 0.18 | 50.9 | E | 0.11 | 28.5 | IE |
| P13.3 | 0.34 | 80.3 | HE | 0.22 | 87.0 | HE | 0.15 | 34.0 | IE | 0.17 | 26.9 | IE | 0.10 | 29.3 | IE | 0.13 | 33.3 | IE |
| P17.1 | 0.38 | 88.5 | HE | 0.37 | 145.1 | HE | 0.20 | 46.0 | LE | 0.27 | 43.7 | LE | 0.16 | 44.7 | LE | 0.14 | 37.6 | LE |
| P17.2 | 0.41 | 96.0 | HE | 0.15 | 58.5 | E | 0.37 | 85.5 | HE | 0.25 | 39.5 | LE | 0.26 | 75.3 | E | 0.16 | 41.9 | LE |
| P20.1 | 0.27 | 62.5 | E | 0.19 | 76.3 | E | 0.21 | 47.6 | LE | 0.17 | 26.9 | IE | 0.22 | 61.9 | E | 0.25 | 63.9 | E |
| P1.2 | 0.33 | 76.6 | E | 0.30 | 118.6 | HE | 0.13 | 29.2 | IE | 0.29 | 46.7 | LE | 0.18 | 50.9 | E | 0.27 | 69.1 | E |
| IC3110.2 | 0.30 | 70.3 | E | 0.21 | 81.8 | HE | 0.29 | 66.2 | E | 0.24 | 37.6 | LE | 0.13 | 36.0 | LE | 0.19 | 50.1 | E |
| ICCV3110.1 | 0.34 | 78.9 | E | 0.28 | 109.5 | HE | 0.17 | 37.9 | LE | 0.35 | 55.2 | E | 0.16 | 46.5 | LE | 0.17 | 45.3 | LE |
| IC59 | 0.29 | 67.4 | E | 0.14 | 56.5 | E | 0.18 | 41.4 | LE | 0.13 | 20.0 | IE | 0.13 | 38.4 | LE | 0.16 | 41.9 | LE |
| LMG17147 | 0.40 | 94.1 | HE | 0.22 | 86.2 | HE | 0.24 | 54.7 | E | 0.25 | 40.5 | LE | 0.19 | 54.7 | E | 0.26 | 68.7 | E |
| XAP11 | 0.37 | 87.1 | HE | 0.32 | 125.3 | HE | 0.16 | 36.8 | LE | 0.30 | 48.0 | LE | 0.16 | 46.1 | LE | 0.24 | 62.6 | E |
| SEMIA396 | 0.23 | 53.4 | E | 0.22 | 85.0 | HE | 0.07 | 14.9 | IE | 0.18 | 28.8 | IE | 0.08 | 24.0 | IE | 0.10 | 25.5 | IE |
| LMG17149 | 0.35 | 80.8 | HE | 0.31 | 121.7 | HE | 0.20 | 46.7 | LE | 0.14 | 22.4 | IE | 0.18 | 52.8 | E | 0.17 | 44.5 | LE |
| DSM1978 | 0.43 | 101.2 | HE | 0.29 | 115.4 | HE | 0.16 | 35.6 | LE | 0.45 | 72.3 | E | 0.20 | 57.6 | E | 0.20 | 53.1 | E |
| DSM30133 | 0.27 | 62.1 | E | 0.20 | 78.3 | E | 0.23 | 51.7 | E | 0.27 | 42.4 | LE | 0.15 | 42.7 | LE | 0.14 | 35.9 | LE |
| USDA3392 | 0.31 | 73.1 | E | 0.17 | 68.4 | E | 0.08 | 18.4 | IE | 0.19 | 29.9 | IE | 0.12 | 34.3 | IE | 0.15 | 39.7 | LE |
| LMG15046 | 0.48 | 112.9 | HE | 0.40 | 156.1 | HE | 0.38 | 88.0 | HE | 0.56 | 88.8 | HE | 0.31 | 89.2 | HE | 0.29 | 75.2 | E |
| XAP4 | 0.72 | 167.9 | HE | 0.32 | 126.5 | HE | 0.28 | 63.9 | E | 0.40 | 64.5 | E | 0.28 | 79.6 | E | 0.28 | 72.2 | E |
| CC1192 | 0.69 | 161.6 | HE | 0.41 | 163.2 | HE | 0.30 | 69.4 | E | 0.46 | 73.9 | E | 0.32 | 91.1 | HE | 0.44 | 114.5 | HE |
| LMG14989 | 0.66 | 154.6 | HE | 0.27 | 107.1 | HE | 0.42 | 96.6 | HE | 0.49 | 77.9 | E | 0.24 | 69.1 | E | 0.51 | 132.2 | HE |
| XAP10 | 0.61 | 142.2 | HE | 0.30 | 118.6 | HE | 0.44 | 101.6 | HE | 0.31 | 49.1 | LE | 0.21 | 60.5 | E | 0.45 | 117.9 | HE |
| Strains | Indole Acetic Acid (IAA) | Phosphate Solubilization Index (PSI) | ||
|---|---|---|---|---|
| Concentration (µg/mL) | Score | PSI | Score | |
| P16.1 | 108 | VHP | ns | NPS |
| P13.3 | 105 | VHP | ns | NPS |
| P17.1 | 92 | VHP | ns | NPS |
| P17.2 | 97 | VHP | ns | NPS |
| P20.1 | 104 | VHP | ns | NPS |
| P1.2 | 96 | VHP | ns | NPS |
| IC3110.2 | 103 | VHP | ns | NPS |
| ICCV3110.1 | 105 | VHP | ns | NPS |
| IC59 | 163 | VHP | 1.3 | LPS |
| LMG17147 | 38 | HP | 2.4 | HPS |
| XAP11 | 27 | MP | 2.3 | HPS |
| SEMIA396 | 99 | VHP | 2.7 | HPS |
| LMG17149 | 10 | LP | 3.3 | HPS |
| DSM1978 | 48 | VHP | 1.7 | LPS |
| DSM30133 | 46 | VHP | 2.4 | HPS |
| USDA3392 | 31 | HP | 2.4 | HPS |
| LMG15046 | 73 | VHP | 2.2 | HPS |
| XAP4 | 105 | VHP | 3.5 | HPS |
| CC1192 | 47 | VHP | 2.6 | HPS |
| LMG14989 | 78 | VHP | 1.4 | LPS |
| XAP10 | 94 | VHP | 2.3 | HPS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wanjofu, E.I.; Venter, S.N.; Beukes, C.W.; Steenkamp, E.T.; Gwata, E.T.; Muema, E.K. Nodulation and Growth Promotion of Chickpea by Mesorhizobium Isolates from Diverse Sources. Microorganisms 2022, 10, 2467. https://doi.org/10.3390/microorganisms10122467
Wanjofu EI, Venter SN, Beukes CW, Steenkamp ET, Gwata ET, Muema EK. Nodulation and Growth Promotion of Chickpea by Mesorhizobium Isolates from Diverse Sources. Microorganisms. 2022; 10(12):2467. https://doi.org/10.3390/microorganisms10122467
Chicago/Turabian StyleWanjofu, Edwin I., Stephanus N. Venter, Chrizelle W. Beukes, Emma T. Steenkamp, Eastonce T. Gwata, and Esther K. Muema. 2022. "Nodulation and Growth Promotion of Chickpea by Mesorhizobium Isolates from Diverse Sources" Microorganisms 10, no. 12: 2467. https://doi.org/10.3390/microorganisms10122467






