Influence of Sex on the Microbiota of the Human Face
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Collection
2.3. DNA Processing
2.4. Microbiome and Statistical Analyses
3. Results
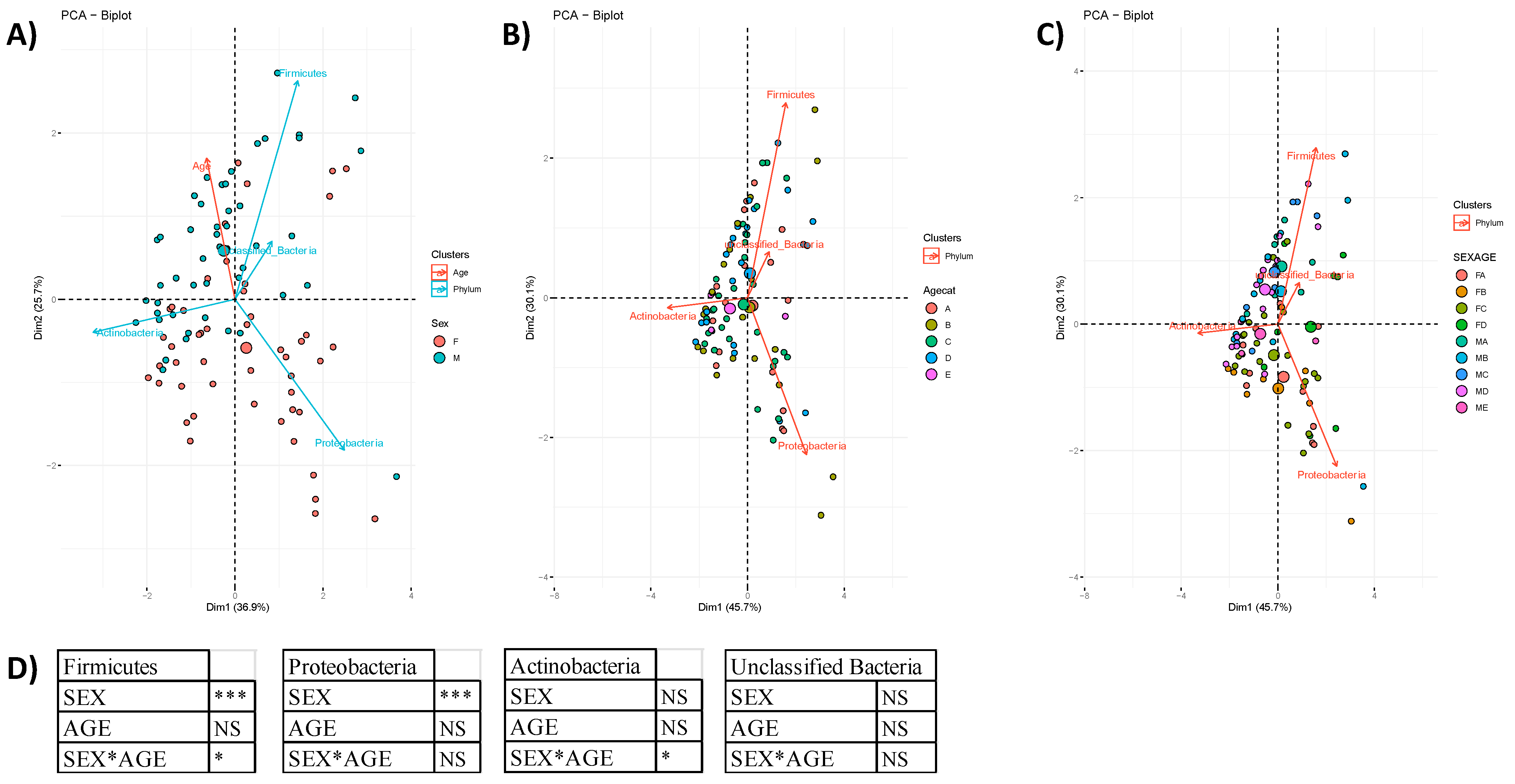
3.1. Community Profiling
3.1.1. Alpha-Diversity
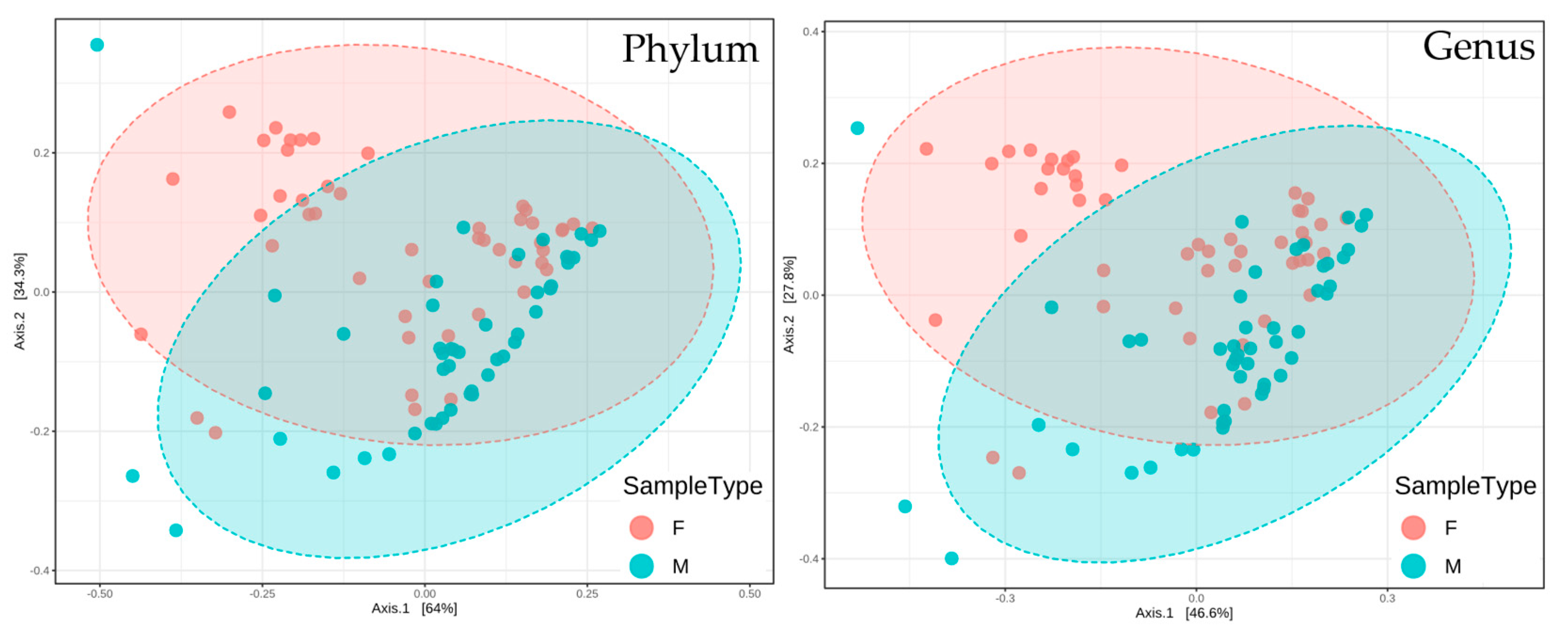
3.1.2. Beta-Diversity
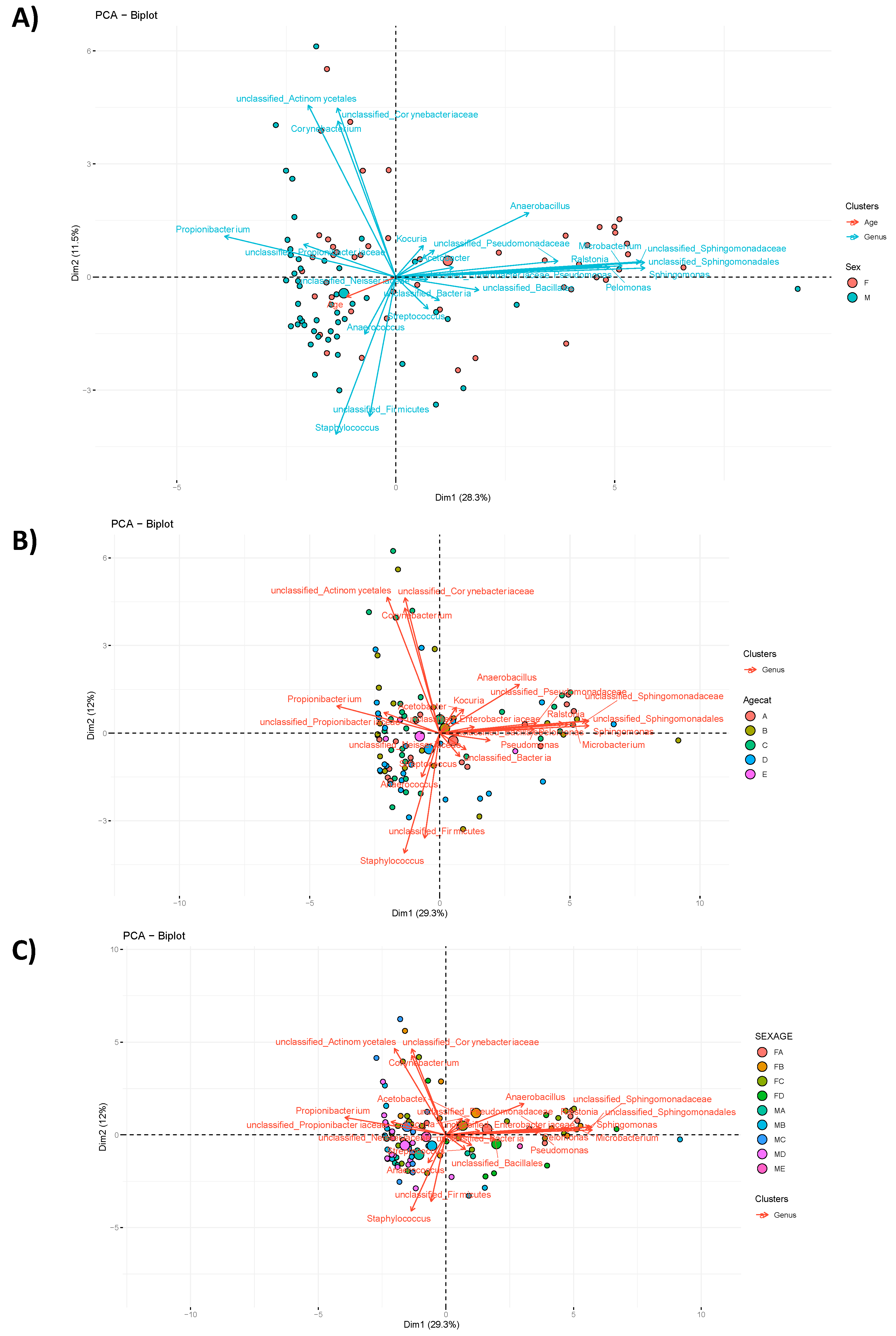
3.2. Community Composition
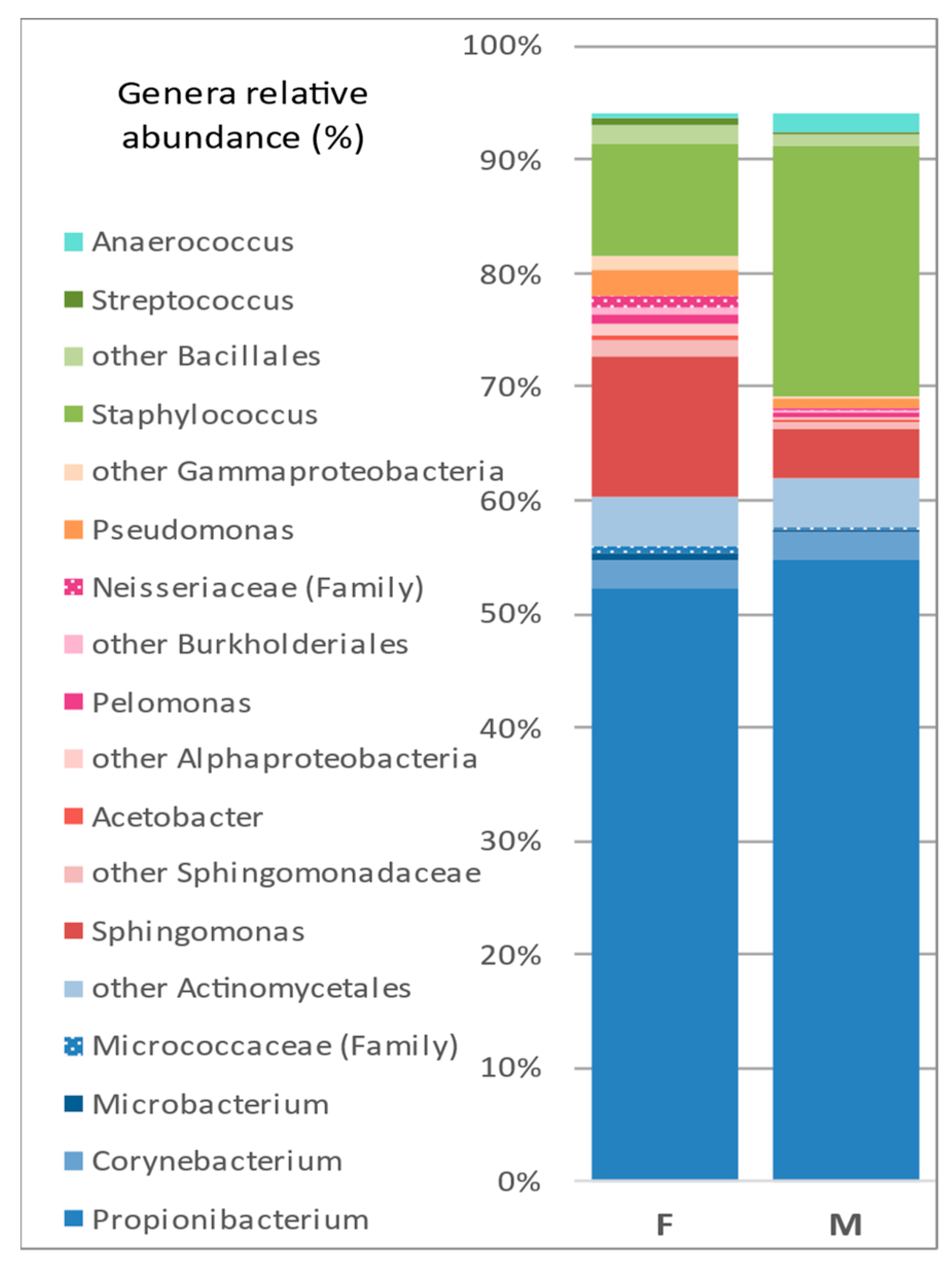
3.2.1. Taxa Abundance
3.2.2. LefSe Analysis of Signature Taxa
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fredricks, D.N. Microbial Ecology of Human Skin in Health and Disease. J. Investig. Dermatol. Symp. Proc. 2001, 6, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.R.; James, W.D. Microbial Ecology of the Skin. Annu. Rev. Microbiol. 1988, 42, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Iebba, V.; Totino, V.; Gagliardi, A.; Santangelo, F.; Cacciotti, F.; Trancassini, M.; Mancini, C.; Cicerone, C.; Corazziari, E.; Pantanella, F.; et al. Eubiosis and dysbiosis: The two sides of the microbiota. New Microbiol. 2016, 39, 1–12. [Google Scholar] [PubMed]
- Hamady, M.; Knight, R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009, 19, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The Human Microbiome Project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Noecker, C.; McNally, C.P.; Eng, A.; Borenstein, E. High-resolution characterization of the human microbiome. Transl. Res. J. Lab. Clin. Med. 2017, 179, 7–23. [Google Scholar] [CrossRef]
- Belkaid, Y.; Segre, J.A. Dialogue between skin microbiota and immunity. Science 2014, 346, 954–959. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The Skin Microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; Deal, C.; et al. The NIH Human Microbiome Project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The healthy human microbiome. Genome Med. 2016, 8, 51. [Google Scholar] [CrossRef]
- Parekh, P.J.; Balart, L.A.; Johnson, D.A. The Influence of the Gut Microbiome on Obesity, Metabolic Syndrome and Gastrointestinal Disease. Clin. Transl. Gastroenterol. 2015, 6, e91. [Google Scholar] [CrossRef] [PubMed]
- Lunjani, N.; Hlela, C.; O’Mahony, L. Microbiome and skin biology. Curr. Opin. Allergy Clin. Immunol. 2019, 19, 328–333. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Arweiler, N.B.; Netuschil, L. The Oral Microbiota. In Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2016; Volume 902, pp. 45–60. [Google Scholar]
- Boxberger, M.; Cenizo, V.; Cassir, N.; La Scola, B. Challenges in exploring and manipulating the human skin microbiome. Microbiome 2021, 9, 125. [Google Scholar] [CrossRef]
- Cundell, A.M. Microbial Ecology of the Human Skin. Microb. Ecol. 2018, 76, 113–120. [Google Scholar] [CrossRef]
- Grice, E.A. The intersection of microbiome and host at the skin interface: Genomic- and metagenomic-based insights. Genome Res. 2015, 25, 1514–1520. [Google Scholar] [CrossRef]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Deming, C.; Conlan, S.; Kong, H.H.; Segre, J.A.; Barnabas, B.; Blakesley, R.; Bouffard, G.; Brooks, S.; et al. Biogeography and individuality shape function in the human skin metagenome. Nature 2014, 514, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Moskovicz, V.; Gross, A.; Mizrahi, B. Extrinsic Factors Shaping the Skin Microbiome. Microorganisms 2020, 8, 1023. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Bowler, P.G.; Duerden, B.I.; Armstrong, D.G. Wound Microbiology and Associated Approaches to Wound Management. Clin. Microbiol. Rev. 2001, 14, 244–269. [Google Scholar] [CrossRef] [PubMed]
- Hosomi, K.; Ohno, H.; Murakami, H.; Natsume-Kitatani, Y.; Tanisawa, K.; Hirata, S.; Suzuki, H.; Nagatake, T.; Nishino, T.; Mizuguchi, K.; et al. Method for preparing DNA from feces in guanidine thiocyanate solution affects 16S rRNA-based profiling of human microbiota diversity. Sci. Rep. 2017, 7, 4339. [Google Scholar] [CrossRef]
- Ederveen, T.H.A.; Smits, J.P.H.; Boekhorst, J.; Schalkwijk, J.; van den Bogaard, E.H.; Zeeuwen, P.L.J.M. Skin microbiota in health and disease: From sequencing to biology. J. Dermatol. 2020, 47, 1110–1118. [Google Scholar] [CrossRef]
- Kong, H.H.; Andersson, B.; Clavel, T.; Common, J.E.; Jackson, S.A.; Olson, N.D.; Segre, J.A.; Traidl-Hoffmann, C. Performing Skin Microbiome Research: A Method to the Madness. J. Investig. Dermatol. 2017, 137, 561–568. [Google Scholar] [CrossRef]
- Loman, N.J.; Misra, R.V.; Dallman, T.J.; Constantinidou, C.; Gharbia, S.E.; Wain, J.; Pallen, M.J. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 2012, 30, 434–439. [Google Scholar] [CrossRef]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion torrent, pacific biosciences and illumina MiSeq sequencers. BMC Genom. 2012, 13, 341. [Google Scholar] [CrossRef]
- Castelino, M.; Eyre, S.; Moat, J.; Fox, G.; Martin, P.; Ho, P.; Upton, M.; Barton, A. Optimisation of methods for bacterial skin microbiome investigation: Primer selection and comparison of the 454 versus MiSeq platform. BMC Microbiol. 2017, 17, 23. [Google Scholar] [CrossRef]
- Liu, Z.; Lozupone, C.; Hamady, M.; Bushman, F.D.; Knight, R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007, 35, e120. [Google Scholar] [CrossRef]
- Kong, H.H.; Segre, J.A. Skin Microbiome: Looking Back to Move Forward. J. Investig. Dermatol. 2012, 132, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liang, H.; Zhou, M.; Song, L.; He, C. Skin bacterial structure of young females in China: The relationship between skin bacterial structure and facial skin types. Exp. Dermatol. 2021, 30, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Samaras, S.; Hoptroff, M. The Microbiome of Healthy Skin. In Skin Microbiome Handbook; Wiley: Hoboken, NJ, USA, 2020; pp. 1–32. [Google Scholar] [CrossRef]
- Baldwin, H.E.; Bhatia, N.D.; Friedman, A.; Martin, R.; Seité, S. The Role of Cutaneous Microbiota Harmony in Maintaining a Functional Skin Barrier. Ski. J. Cutan. Med. 2017, 1, s139. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Park, M.; Kong, H.H.; Segre, J.A. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Kim, J.J.; Myeong, N.R.; Kim, T.; Kim, D.A.; An, S.; Kim, H.; Park, T.; Jang, S.I.; Yeon, J.H.; et al. Segregation of age-related skin microbiome characteristics by functionality. Sci. Rep. 2019, 9, 16748. [Google Scholar] [CrossRef]
- Ying, S.; Zeng, D.-N.; Chi, L.; Tan, Y.; Galzote, C.; Cardona, C.; Lax, S.; Gilbert, J.; Quan, Z.-X. The Influence of Age and Gender on Skin-Associated Microbial Communities in Urban and Rural Human Populations. PLoS ONE 2015, 10, e0141842. [Google Scholar] [CrossRef]
- Kim, J.-H.; Son, S.-M.; Park, H.; Kim, B.K.; Choi, I.S.; Kim, H.; Huh, C.S. Taxonomic profiling of skin microbiome and correlation with clinical skin parameters in healthy Koreans. Sci. Rep. 2021, 11, 16269. [Google Scholar] [CrossRef]
- Somboonna, N.; Wilantho, A.; Srisuttiyakorn, C.; Assawamakin, A.; Tongsima, S. Bacterial communities on facial skin of teenage and elderly Thai females. Arch. Microbiol. 2017, 199, 1035–1042. [Google Scholar] [CrossRef]
- Zhai, W.; Huang, Y.; Zhang, X.; Fei, W.; Chang, Y.; Cheng, S.; Zhou, Y.; Gao, J.; Tang, X.; Zhang, X.; et al. Profile of the skin microbiota in a healthy Chinese population. J. Dermatol. 2018, 45, 1289–1300. [Google Scholar] [CrossRef]
- Gupta, V.K.; Paul, S.; Dutta, C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front. Microbiol. 2017, 8, 1162. [Google Scholar] [CrossRef] [PubMed]
- Hillebrand, G.G.; Dimitriu, P.; Malik, K.; Park, Y.; Qu, D.; Mohn, W.W.; Kong, R. Temporal Variation of the Facial Skin Microbiome: A 2-Year Longitudinal Study in Healthy Adults. Plast. Reconstr. Surg. 2020, 147, 50S–61S. [Google Scholar] [CrossRef] [PubMed]
- Torre, E.; Sola, D.; Caramaschi, A.; Mignone, F.; Bona, E.; Fallarini, S. A Pilot Study on Clinical Scores, Immune Cell Modulation, and Microbiota Composition in Allergic Patients with Rhinitis and Asthma Treated with a Probiotic Preparation. Int. Arch. Allergy Immunol. 2022, 183, 186–200. [Google Scholar] [CrossRef] [PubMed]
- Bona, E.; Massa, N.; Toumatia, O.; Novello, G.; Cesaro, P.; Todeschini, V.; Boatti, L.; Mignone, F.; Titouah, H.; Zitouni, A.; et al. Climatic Zone and Soil Properties Determine the Biodiversity of the Soil Bacterial Communities Associated to Native Plants from Desert Areas of North-Central Algeria. Microorganisms 2021, 9, 1359. [Google Scholar] [CrossRef] [PubMed]
- Willis, A.D. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef]
- R Foundation for Statistical Computing. R Core R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Cosseau, C.; Romano-Bertrand, S.; Duplan, H.; Lucas, O.; Ingrassia, I.; Pigasse, C.; Roques, C.; Jumas-Bilak, E. Proteobacteria from the human skin microbiota: Species-level diversity and hypotheses. One Health 2016, 2, 33–41. [Google Scholar] [CrossRef]
- Grogan, M.D.; Bartow-McKenney, C.; Flowers, L.; Knight, S.A.B.; Uberoi, A.; Grice, E.A. Research Techniques Made Simple: Profiling the Skin Microbiota. J. Investig. Dermatol. 2019, 139, 747–752.e1. [Google Scholar] [CrossRef]
- Dunbar, J.; Barns, S.M.; Ticknor, L.O.; Kuske, C.R. Empirical and Theoretical Bacterial Diversity in Four Arizona Soils. Appl. Environ. Microbiol. 2002, 68, 3035–3045. [Google Scholar] [CrossRef]
- Skowron, K.; Bauza-Kaszewska, J.; Kraszewska, Z.; Wiktorczyk-Kapischke, N.; Grudlewska-Buda, K.; Kwiecińska-Piróg, J.; Wałecka-Zacharska, E.; Radtke, L.; Gospodarek-Komkowska, E. Human Skin Microbiome: Impact of Intrinsic and Extrinsic Factors on Skin Microbiota. Microorganisms 2021, 9, 543. [Google Scholar] [CrossRef]
- Fierer, N.; Hamady, M.; Lauber, C.L.; Knight, R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 17994–17999. [Google Scholar] [CrossRef]
- Giacomoni, P.U.; Mammone, T.; Teri, M. Gender-linked differences in human skin. J. Dermatol. Sci. 2009, 55, 144–149. [Google Scholar] [CrossRef]
- Wallen-Russell, C.; Wallen-Russell, S. A New Benchmark to Determine What Healthy Western Skin Looks Like in Terms of Biodiversity Using Standardised Methodology. Cosmetics 2020, 7, 79. [Google Scholar] [CrossRef]
- Hwang, B.K.; Lee, S.; Myoung, J.; Hwang, S.J.; Lim, J.M.; Jeong, E.T.; Park, S.G.; Youn, S.H. Effect of the skincare product on facial skin microbial structure and biophysical parameters: A pilot study. MicrobiologyOpen 2021, 10, e1236. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Ciardiello, T.; Franzoni, M.; Pasini, F.; Giuliani, G.; Rinaldi, F. Effect of commonly used cosmetic preservatives on skin resident microflora dynamics. Sci. Rep. 2021, 11, 8695. [Google Scholar] [CrossRef] [PubMed]
- Dimitriu, P.A.; Iker, B.; Malik, K.; Leung, H.; Mohn, W.W.; Hillebrand, G.G. New Insights into the Intrinsic and Extrinsic Factors That Shape the Human Skin Microbiome. mBio 2019, 10, e00839-19. [Google Scholar] [CrossRef] [PubMed]
- Si, J.; Lee, S.; Park, J.M.; Sung, J.; Ko, G.P. Genetic associations and shared environmental effects on the skin microbiome of Korean twins. BMC Genom. 2015, 16, 992. [Google Scholar] [CrossRef]
- Oh, J.; Conlan, S.; Polley, E.C.; Segre, J.A.; Kong, H.H. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012, 4, 77. [Google Scholar] [CrossRef]
- Leung, M.H.Y.; Tong, X.; Wilkins, D.; Cheung, H.H.L.; Lee, P.K.H. Individual and household attributes influence the dynamics of the personal skin microbiota and its association network. Microbiome 2018, 6, 16–21. [Google Scholar] [CrossRef]
- Lee, H.; Jeong, J.; Oh, Y.; Lee, C.-J.; Mun, S.; Lee, D.-G.; Jo, H.W.; Heo, Y.M.; Baek, C.; Heo, C.Y.; et al. Comparative analysis of human facial skin microbiome between topical sites compared to entire face. Genes Genom. 2021, 43, 1483–1495. [Google Scholar] [CrossRef]
- Cobo, F.; Navarro-Marí, J.M. First description of Anaerococcus octavius as cause of bacteremia. Anaerobe 2020, 61, 102130. [Google Scholar] [CrossRef]
- Lagier, J.-C.; El Karkouri, K.; Nguyen, T.-T.; Armougom, F.; Raoult, D.; Fournier, P.-E. Non-contiguous finished genome sequence and description of Anaerococcus senegalensis sp. nov. Stand. Genom. Sci. 2012, 6, 116. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luebberding, S.; Krueger, N.; Kerscher, M. Skin physiology in men and women: In vivo evaluation of 300 people including TEWL, SC hydration, sebum content and skin surface pH. Int. J. Cosmet. Sci. 2013, 35, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Mounier, J.; Rea, M.C.; O’Connor, P.M.; Fitzgerald, G.F.; Cogan, T.M. Growth Characteristics of Brevibacterium, Corynebacterium, Microbacterium, and Staphylococcus spp. Isolated from Surface-Ripened Cheese. Appl. Environ. Microbiol. 2007, 73, 7732–7739. [Google Scholar] [CrossRef] [PubMed]
- Corretto, E.; Antonielli, L.; Sessitsch, A.; Höfer, C.; Puschenreiter, M.; Widhalm, S.; Swarnalakshmi, K.; Brader, G. Comparative Genomics of Microbacterium Species to Reveal Diversity, Potential for Secondary Metabolites and Heavy Metal Resistance. Front. Microbiol. 2020, 11, 1869. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; Murray, P.R.; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef] [PubMed]
- White, D.C.; Sutton, S.D.; Ringelberg, D.B. The genus Sphingomonas: Physiology and ecology. Curr. Opin. Biotechnol. 1996, 7, 301–306. [Google Scholar] [CrossRef]
- Leys, N.M.E.J.; Ryngaert, A.; Bastiaens, L.; Verstraete, W.; Top, E.M.; Springael, D. Occurrence and Phylogenetic Diversity of Sphingomonas Strains in Soils Contaminated with Polycyclic Aromatic Hydrocarbons. Appl. Environ. Microbiol. 2004, 70, 1944–1955. [Google Scholar] [CrossRef]
- Bay, L.; Barnes, C.J.; Fritz, B.G.; Thorsen, J.; Restrup, M.E.M.; Rasmussen, L.; Sørensen, J.K.; Hesselvig, A.B.; Odgaard, A.; Hansen, A.J.; et al. Universal Dermal Microbiome in Human Skin. mBio 2020, 11, e02945-19. [Google Scholar] [CrossRef]
- Laughlin, R.S.; Musch, M.W.; Hollbrook, C.J.; Rocha, F.M.; Chang, E.B.; Alverdy, J.C. The Key Role of Pseudomonas aeruginosa PA-I Lectin on Experimental Gut-Derived Sepsis. Ann. Surg. 2000, 232, 133–142. [Google Scholar] [CrossRef]
- Fujii, T.; Shinozaki, J.; Kajiura, T.; Iwasaki, K.; Fudou, R. A newly discovered Anaerococcus strain responsible for axillary odor and a new axillary odor inhibitor, pentagalloyl glucose. FEMS Microbiol. Ecol. 2014, 89, 198–207. [Google Scholar] [CrossRef][Green Version]

| Sex | Age | Sex*Age | |
|---|---|---|---|
| Acetobacter | ** | NS | * |
| Anaerobacillus | * | NS | NS |
| Anaerococcus | ** | NS | NS |
| Corynebacterium | NS | NS | NS |
| Microbacterium | *** | NS | NS |
| Pelomonas | ** | NS | NS |
| Propionibacterium | NS | NS | ** |
| Pseudomonas | *** | ** | ** |
| Ralstonia | ** | NS | NS |
| Sphingomonas | *** | NS | NS |
| Staphylococcus | *** | NS | * |
| Streptococcus | NS | NS | NS |
| unclassified_Actinomycetales | NS | NS | NS |
| unclassified_Bacillales | ** | NS | NS |
| unclassified_Bacteria | NS | NS | NS |
| unclassified_Corynebacteriaceae | NS | NS | NS |
| unclassified_Firmicutes | ** | NS | * |
| unclassified_Neisseriaceae | * | NS | NS |
| unclassified_Propionibacteriaceae | NS | NS | NS |
| unclassified_Sphingomonadaceae | *** | NS | NS |
| unclassified_Sphingomonadales | *** | NS | NS |
| p-Value | MW/KW 1 | Median | Spread | ||||
|---|---|---|---|---|---|---|---|
| F | M | F | M | ||||
| Shannon | Phylum | 7.67E-03 | 1514 | 1.009 | 0.839 | 0.573 | 0.442 |
| Genus | 2.28E-04 | 1647 | 1.888 | 1.349 | 1.008 | 0.731 | |
| Simpson | Phylum | 2.29E-02 | 1462 | 0.560 | 0.491 | 0.501 | 0.528 |
| Genus | 3.63E-03 | 1546 | 0.709 | 0.597 | 0.553 | 0.595 | |
| Number of Reads | Phyla Relative Abundance | p-Values | FDR | LDA Score | ||
|---|---|---|---|---|---|---|
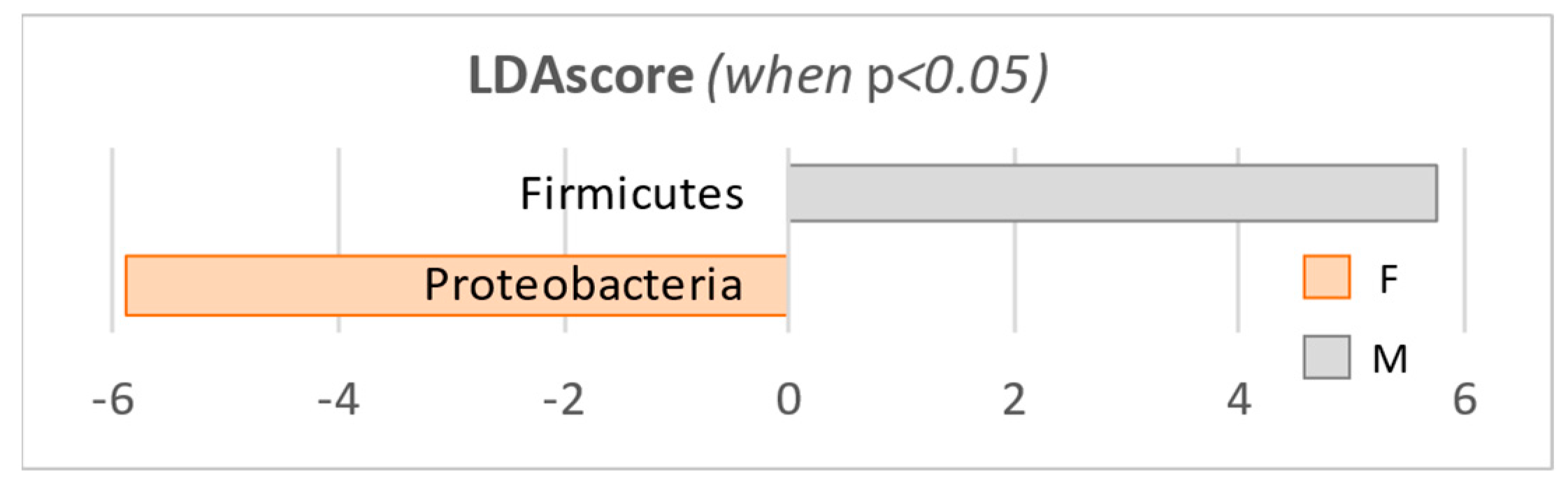
| Proteobacteria | W | 2,152,800 | 21.9% | 7.40E-09 | 2.96E-08 | −5.89 |
| M | 616,800 | 7.4% | ||||
| Firmicutes | W | 1,514,800 | 13.2% | 4.62E-05 | 9.24E-05 | 5.75 |
| M | 2,638,200 | 25.7% | ||||
| Actinobacteria | W | 5,878,800 | 60.3% | 2.47E-01 | 3.29E-01 | 5.26 |
| M | 6,246,500 | 62.0% |
| Phylum | Class | Order | Family | Genus | F | M |
|---|---|---|---|---|---|---|
| Actinobacteria | Actinobacteria | Actinomycetales | Propionibacteriaceae | Propionibacterium | 52.18% | 54.65% |
| Corynebacteriaceae | Corynebacterium | 2.60% | 2.61% | |||
| Microbacteriaceae | Microbacterium | 0.54% | 0.16% | |||
| Micrococcaceae (Family) | 0.61% | 0.23% | ||||
| other Actinomycetales | 4.30% | 4.25% | ||||
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | 12.48% | 4.41% |
| other Sphingomonadaceae | 1.38% | 0.50% | ||||
| Rhodospirillales | Acetobacteraceae | Acetobacter | 0.45% | 0.14% | ||
| other Alphaproteobacteria | 0.97% | 0.33% | ||||
| Betaproteobacteria | Burkholderiales | Comamonadaceae | Pelomonas | 0.88% | 0.35% | |
| other Burkholderiales | 0.62% | 0.23% | ||||
| Neisseriales | Neisseriaceae (Family) | 1.11% | 0.33% | |||
| other Betaproteobacteria | 0.25% | 0.06% | ||||
| Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | 2.20% | 0.71% | |
| other Gammaproteobacteria | 1.24% | 0.23% | ||||
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | 9.88% | 22.12% |
| other Bacillales | 1.58% | 1.00% | ||||
| Lactobacillales | Streptococcaceae | Streptococcus | 0.77% | 0.21% | ||
| other Lactobacillales | 0.19% | 0.03% | ||||
| Clostridia | Clostridiales | Clostridia Incertae Sedis XI | Anaerococcus | 0.40% | 1.59% | |
| other Clostridia | 0.08% | 0.30% | ||||
| uncl. Bacteria | uncl. Bacteria | 4.60% | 4.97% | |||
| p-Values | FDR | Median Read Number | LDA Score | ||
|---|---|---|---|---|---|
| F | M | ||||
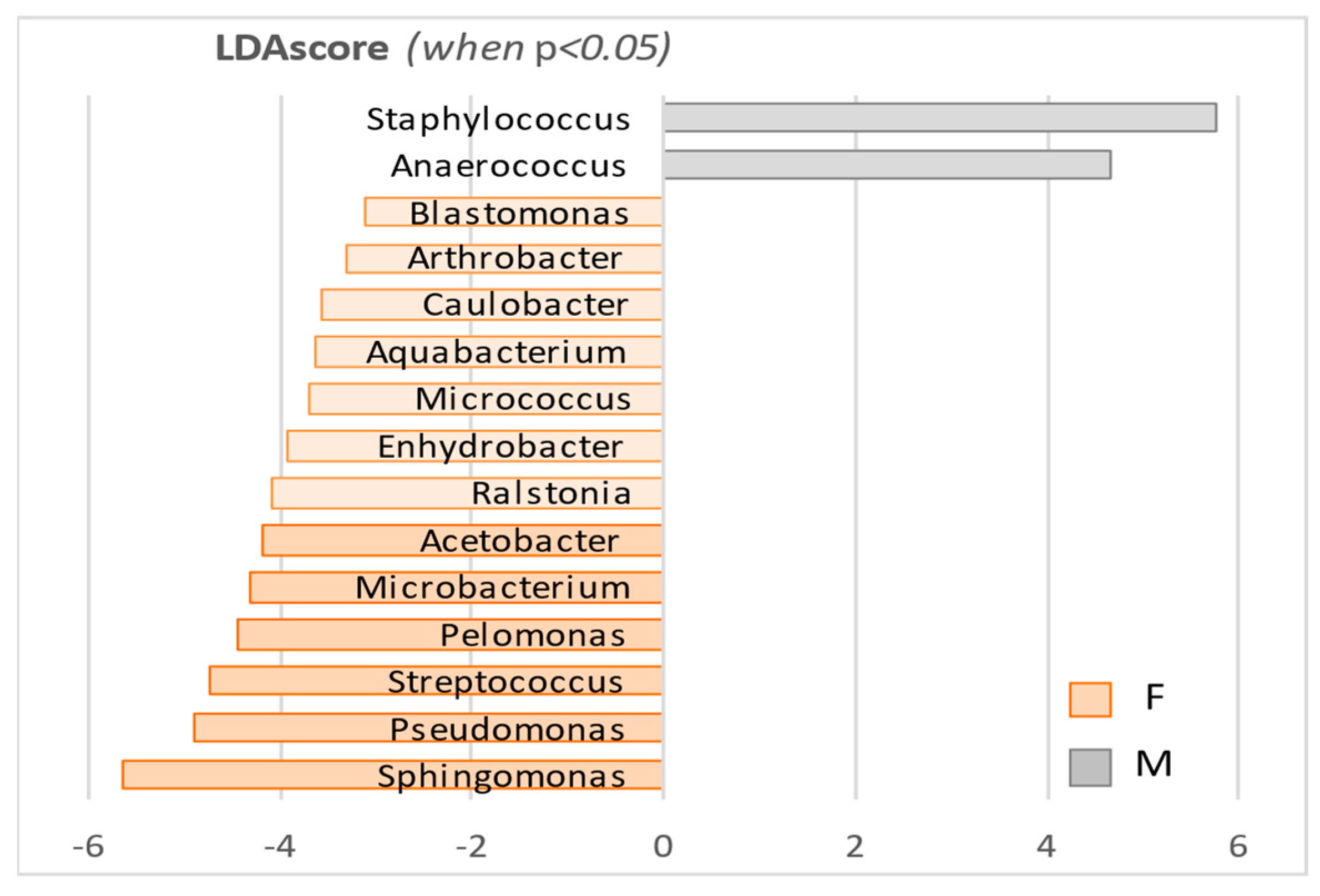
| Microbacterium A | 3.63E-09 | 5.72E-08 | 55,004 | 13,319 | −4.32 |
| Ralstonia β‡ | 4.67E-09 | 5.72E-08 | 36,469 | 11,060 | −4.10 |
| Arthrobacter A* | 7.53E-09 | 7.38E-08 | 5514.1 | 1367.8 | −3.32 |
| Sphingomonas α•° | 1.35E-08 | 9.34E-08 | 1,239,200 | 383,550 | −5.63 |
| Blastomonas α•° | 2.41E-08 | 1.07E-07 | 3575.1 | 947.4 | −3.12 |
| Pelomonas ⇆ | 4.35E-08 | 1.78E-07 | 92,260 | 34,183 | −4.46 |
| Staphylococcus FB | 7.02E-07 | 2.29E-06 | 1,088,700 | 2,297,500 | 5.78 |
| Caulobacter α | 1.87E-05 | 4.16E-05 | 9776.9 | 2312.5 | −3.57 |
| Micrococcus A* | 5.88E-05 | 1.25E-04 | 13,784 | 3772.4 | −3.70 |
| Pseudomonas γp | 8.32E-05 | 1.70E-04 | 207,110 | 43019 | −4.91 |
| Aquabacterium ⇆ | 1.04E-04 | 1.91E-04 | 14,808 | 6208.9 | −3.63 |
| Enhydrobacter γp | 1.05E-04 | 1.91E-04 | 20,008 | 2465.2 | −3.94 |
| Streptococcus FL | 2.03E-04 | 3.55E-04 | 132,430 | 23,639 | −4.74 |
| Acetobacter α | 2.91E-03 | 4.60E-03 | 48,747 | 17,091 | −4.20 |
| Anaerococcus FC | 3.54E-03 | 5.26E-03 | 52,957 | 146,280 | 4.67 |
| Propionibacterium A | 1.68E-01 | 2.17E-01 | 5,004,700 | 5,385,600 | 5.28 |
| Corynebacterium A | 5.73E-01 | 6.53E-01 | 301,120 | 349,850 | 4.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robert, C.; Cascella, F.; Mellai, M.; Barizzone, N.; Mignone, F.; Massa, N.; Nobile, V.; Bona, E. Influence of Sex on the Microbiota of the Human Face. Microorganisms 2022, 10, 2470. https://doi.org/10.3390/microorganisms10122470
Robert C, Cascella F, Mellai M, Barizzone N, Mignone F, Massa N, Nobile V, Bona E. Influence of Sex on the Microbiota of the Human Face. Microorganisms. 2022; 10(12):2470. https://doi.org/10.3390/microorganisms10122470
Chicago/Turabian StyleRobert, Clémence, Federica Cascella, Marta Mellai, Nadia Barizzone, Flavio Mignone, Nadia Massa, Vincenzo Nobile, and Elisa Bona. 2022. "Influence of Sex on the Microbiota of the Human Face" Microorganisms 10, no. 12: 2470. https://doi.org/10.3390/microorganisms10122470
APA StyleRobert, C., Cascella, F., Mellai, M., Barizzone, N., Mignone, F., Massa, N., Nobile, V., & Bona, E. (2022). Influence of Sex on the Microbiota of the Human Face. Microorganisms, 10(12), 2470. https://doi.org/10.3390/microorganisms10122470









