Abstract
Total and diazotrophic bacteria were assessed in the rhizosphere soils of native and encroaching legumes growing in the Succulent Karoo Biome (SKB), South Africa. These were Calobota sericea, Lessertia diffusa, Vachellia karroo, and Wiborgia monoptera, of Fabaceae family near Springbok (Northern Cape Province) and neighboring refugia of the Fynbos biome for C. sericea for comparison purposes. Metabarcoding approach using 16S rRNA gene revealed Actinobacteria (26.7%), Proteobacteria (23.6%), Planctomycetes, and Acidobacteria (10%), while the nifH gene revealed Proteobacteria (70.3%) and Cyanobacteria (29.5%) of the total sequences recovered as the dominant phyla. Some of the diazotrophs measured were assigned to families; Phyllobacteriaceae (39%) and Nostocaceae (24.4%) (all legumes), Rhodospirillaceae (7.9%), Bradyrhizobiaceae (4.6%) and Methylobacteriaceae (3%) (C. sericea, V. karroo, W. monoptera), Rhizobiaceae (4.2%; C. sericea, L. diffusa, V. Karroo), Microchaetaceae (4%; W. monoptera, V. karroo), Scytonemataceae (3.1%; L. diffusa, W. monoptera), and Pseudomonadaceae (2.7%; V. karroo) of the total sequences recovered. These families have the potential to fix the atmospheric nitrogen. While some diazotrophs were specific or shared across several legumes, a member of Mesorhizobium species was common in all rhizosphere soils considered. V. karroo had statistically significantly higher Alpha and distinct Beta-diversity values, than other legumes, supporting its influence on soil microbes. Overall, this work showed diverse bacteria that support plant life in harsh environments such as the SKB, and shows how they are influenced by legumes.
1. Introduction
The Succulent Karoo Biome (SKB) in South Africa is one of the world’s most species rich biomes, with over 5000 plant taxa of which 40% are endemic to the region [1,2,3]. The main vegetation cover includes leaf-succulent, stem-succulent and non-succulent plant species [2,4]. The SKB is semi-arid, experiences winter rainfall, and the high levels of plant diversity have been ascribed to the region’s oscillating wet and dry climatic conditions, moderate climatic history and soils that are sandy, acidic and nutrient limited [4,5].
The need for conservation of this ecosystem has directed some research on the influence of rainfall and droughts on vegetation cover [3,6]. However, the fact that the success and survival of plant species in this relatively harsh environment are due to their interactions with microbial communities in the soil, is often underestimated [7,8].
Members of the Fabaceae family form a significant part of the non-succulent plant species in the SKB, particularly due to their ability to survive in extreme environments [9,10,11,12]. These plants generally possess the ability to fix atmospheric nitrogen in association with symbiotic and free-living soil bacteria [13]. For example, diverse species of Ensifer were reported to be the nodulating symbionts of Vachellia jacquemontii, thus promoting the plant’s growth in alkaline soils in the Indian Thar Desert [14]. Recent studies that focused on the root-nodule bacteria in the Fynbos biome, neighboring the SKB, revealed that diverse rhizobia (Azorhizobium, Bradyrhizobium, Ensifer, Mesorhizobium Rhizobium, and Paraburkholderia) are associated with a wide range of host legumes [15]. However, there is limited information about plant-microbe interactions in the SKB particularly using the next generation sequencing approaches. These include, the post-fire occurrences response of microbial communities using the polymerase chain reaction denaturing gradient gel electrophoresis technique [16] and the association of fungal communities with Aizoaceae plants in the Namaqua National Park in the SKB using the metabarcoding approach of the ITS1 region specific primers [8]. Based on sanger sequencing approach, there are some rhizobial interactions with Vachellia karroo and Lessertia spp. reported [17,18].
Plant-microbe interactions may also account for the establishment and subsequent invasiveness of non-native plants in new areas [19,20]. Such success is attributed to the ability of the invader plant to reconstitute the rhizosphere communities, either through the formation of new associations with the resident microorganisms or by co-introduction with its own interacting partners [19,21]. For instance, many invasive Acacia species have been co-introduced with their preferential diazotrophic Bradyrhizobium symbionts [22,23]. The consequences of such invasions include loss of native plant diversity and changes in the structure and composition of microbes in the soil [20,24]. This could also be the case for the noted encroachment of Vachellia karroo in the SKB from other biomes in South Africa [17,25], which is a topic that has not been addressed previously.
This study explored the diversity and abundance of bacteria associated with different indigenous legume species (i.e., Calobota sericea, Wiborgia monoptera, Lessertia diffusa) and an encroaching V. karroo species near Springbok (Northern Cape Province, SA). Microbes sustain ecosystems through nutrient cycling processes. Therefore, understanding plant-microbe interactions is important in the management of the SKB given its significance in the agricultural sector since fodder shrubs such as C. sericea support livestock [26]. The sampling sites included those from the understudied arid soils of the SKB and the neighboring refugia of the Fynbos biome, for comparative purposes. Of these legumes, C. sericea is commonly distributed in the Northern Cape, Western Cape, and Eastern Cape Provinces of South Africa [26]. L. diffusa occurs in the western part of the Northern Cape, in a small part of the Western Cape and North West Provinces [26] while W. monoptera occurs across the Greater Cape Floristic Region of South Africa [27,28]. Generally, C. sericea is more commonly distributed than L. diffusa and W. monoptera species. Although V. karroo is native to southern Africa, it is considered to be encroaching on the SKB [25].
We utilized a metabarcoding approach based on the 16S ribosomal RNA (rRNA) gene and the nifH gene encoding one of the nitrogenase subunits to unveil the total and diazotrophic bacteria in legumes’ rhizosphere soils considered.
2. Materials and Methods
2.1. Study Sites
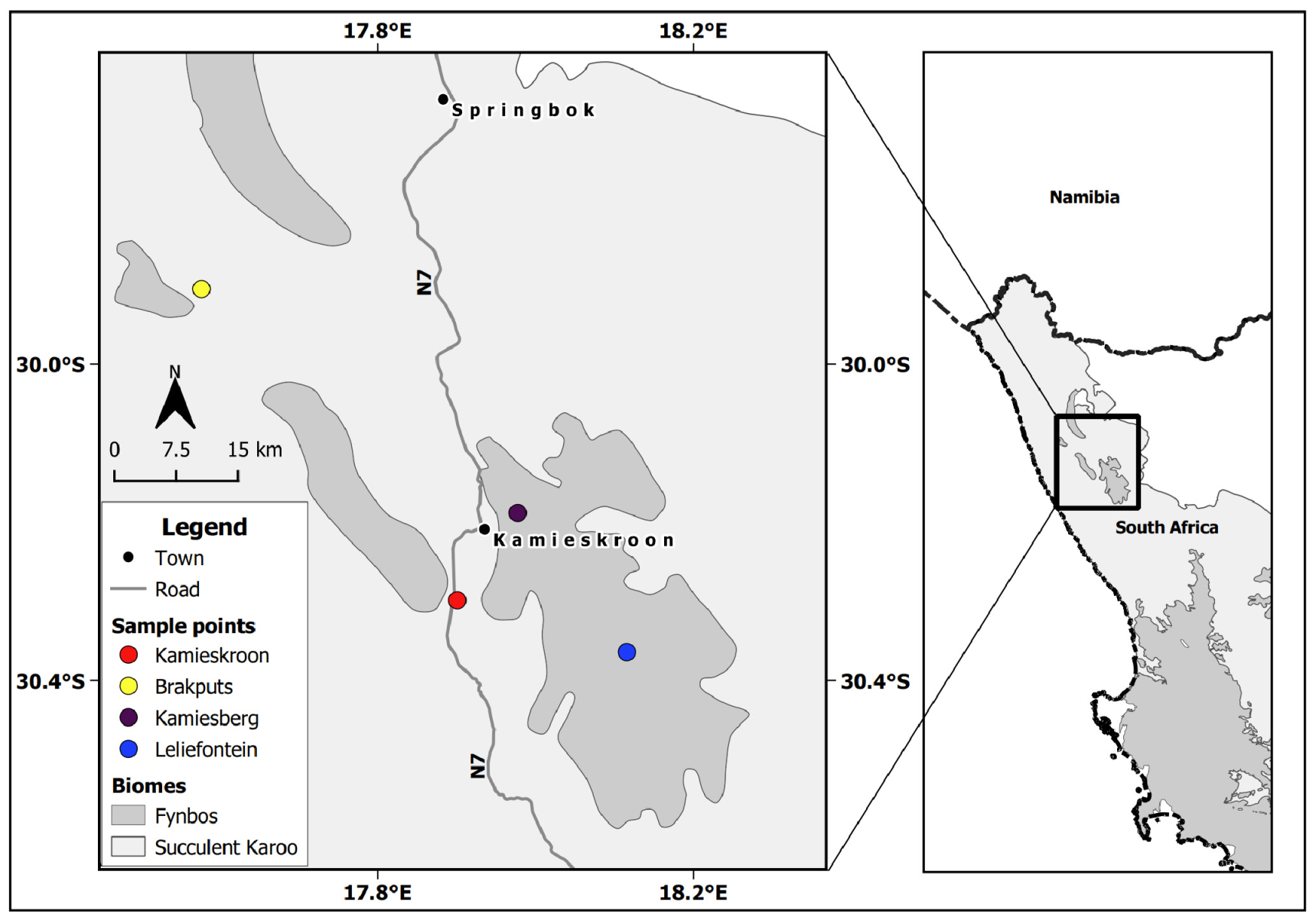
The SKB of South Africa is situated in the western part of the Northern Cape Province. It lies below 800 m but may reach up to 1500 m above the sea level (a.s.l) on the eastern side. It is characterized by semi-arid to arid conditions, receiving 20 to 290 mm of rainfall per year during the winter season. The temperature may rise beyond 40 °C during the summer periods. SKB soils are generally acidic, weakly developed and mostly found on rocks with limited nutrients [2,5,29]. Based on the occurrence of legumes, two sites within this biome were identified, which were Kamieskroon (30°17′15.74″ S, 17°51′43.81″ E), and Brakputs (29°54′19.43″ S, 17°34′37.41″ E). For comparative purposes, two nearby sites in the Kamiesberg Center, i.e., Leliefontein (30°21′50.09″ S, 18° 6′56.42″ E) and Kamiesberg (30°11′17.52″ S, 17°58′38.67″ E) were also selected (Figure 1). The Kamiesberg Center is an outlying area of the Fynbos, mostly above 1200 m a.s.l., with climatic conditions and soil properties that are generally similar to those of the SKB [30,31].
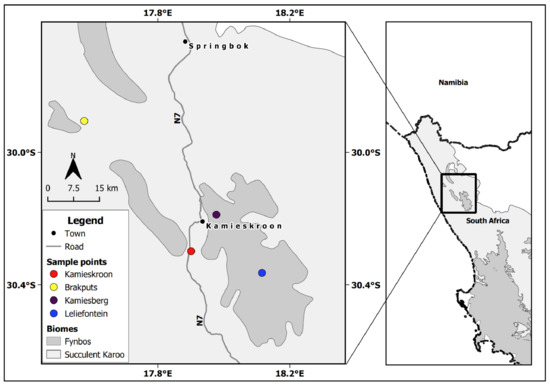
Figure 1.
Sampling sites in the Succulent Karoo Biome and the neighbouring patches of the Fynbos biome in the western part of the Northern Cape Province in South Africa.
2.2. Soil Sampling and Physico-Chemical Properties
In total, 24 composite soil samples were obtained from the four sites under the four different legumes. Five soil sub-samples were randomly scooped at a depth of 0–10 cm using a soil auger in the root zone area under each legume. The five sub-samples were bulked to form one composite sample per plant. Such samples were collected for C. sericea from Brakputs (n = 3), Kamiesberg (n = 4), Kamieskroon (n = 2) and Leliefontein (n = 1), while those for V. karroo were collected from Brakputs (n = 5), and Kamieskroon (n = 1). For W. monoptera from Brakputs (n = 3) and L. diffusa from Brakputs (n = 5). The composite soil samples were stored at 4 °C. Prior to analysis, each composite soil sample was split into two proportions. The first proportion was sieved (2 mm mesh) and used for soil properties analysis of pH using the KCl method, total carbon (TC), and total nitrogen (TN) using the dry combustion method, ammonium (NH4+), nitrates (NO3−) measured calorimetrically and soil texture determined using the Hydrometer method at Bemlab, Cape Town, South Africa. The second proportion was kept at −70 °C for subsequent analyses.
2.3. Soil Metabarcoding Analysis
A culture-independent (metabarcoding) approach based on next generation sequencing of specific marker genes was used to investigate the diversity and abundance of total and diazotrophic bacteria in the collected soils [32]. Total soil genomic DNA was isolated from 0.5 g of each composite soil sample using the FastDNATM SPIN Kit for Soil (MP Biomedicals, Solon, OH, USA). The quality and quantity of extracted DNA were evaluated using 1.0% (w/v) agarose (Whitehead Scientific, Cape Town, South Africa) gel electrophoresis and a NanoDrop™ 2000/2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). For each soil DNA sample, two consistent NanoDrop readings, whose variation was less than 5%, were considered for subsequent determination of the DNA concentration.
The two metabarcoding markers used were the universal house-keeping 16S rRNA gene and the nifH gene specific to diazotrophs [33]. The extracted DNA was split into two proportions. The first proportion was sent to the Biomedical Core Research Facilities at the University of Michigan, Ann Arbor, USA for sequencing of the V4 hypervariable region of the 16S rRNA gene. For library preparation, forward and reverse 515F/806R primers contained Illumina-sequencing P5 and P7 dual-indexing adapters with a unique barcode for every sample for multiplexing and demultiplexing [34]. PCR conditions were as follows: 94 °C for 3 min, followed by 33 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s with a final elongation step at 72 °C for 5 min.
The second DNA proportion was sent to MR DNA (www.mrdnalab.com, accessed on 2 October 2021, Shallowater, TX, USA) for sequencing of the nifH gene using PolF/PolR primers [35]. For these reactions, the forward primer contained a barcode. The amplifications were carried out using the HotStarTaq Plus Master Mix Kit (Qiagen, Germantown, MD, USA) with cycling conditions as follows: 94 °C for 3 min, followed by 28 cycles of 94 °C for 30 s, 53 °C for 40 s, and 72 °C for 1 min and a final elongation step at 72 °C for 5 min.
For both genes, amplicons from different samples were pooled together in equal concentrations and purified using calibrated Agencourt Ampure XP beads (Agencourt Bioscience Corporation, Beverly, MA, USA). Paired-end sequencing was performed using the Illumina MiSeq platform to produce paired reads that were 250 nucleotides in length. All raw sequence data for the 16S rRNA and nifH genes have been deposited with links to BioProject accession numbers (PRJNA767582, PRJNA767584), respectively.
The 16S rRNA sequence data were processed and analysed using Mothur version 1.35.1 [36]. After joining the paired forward and reverse sequence reads, the sequence quality was checked by specifying a maximum length of 275 base pair (bp) and a minimum of 250 bp, as well as removal of all sequences with ambiguities and with more than 8 bp homopolymers. The sequences were then sorted into their respective samples by allowing of maximum of 2 bp mismatches with the primer and no mismatches in the multiplexing barcodes. The quality-filtered and sorted sequences were then aligned to the SILVA ribosomal RNA reference database [37] provided through Mothur and filtered (filter.seqs option in mothur) to ensure that all the sequences aligned to a similar region of the V4 region of the 16S rRNA gene. These sequences were then processed using a pseudo-single linkage algorithm with the pre.cluster command in Mothur using a 2 bp similarity cut-off [36,38]. In addition, chimeras were detected and removed [39] using the chimera.uchime command in Mothur. The remaining sequences were then associated with particular taxonomic lineages using the Greengenes database [40] with a cut-off confidence level of 80%. After removing sequences of unknown taxonomic lineages, the remaining data were classified into operational taxonomic units (OTUs) using average neighbour algorithm with a similarity cut-off of 97%. OTUs were assigned by obtaining a representative sequence from each OTU (get.oturep command in Mothur) and classifying it using the Greengenes database at a confidence level of 80% [40].
The nifH sequence data were processed using the analysis pipeline of MR DNA (www.mrdnalab.com, accessed on 2 October 2021, Shallowater, TX, USA) according to qiime pipeline using the divisive amplicon denoising algorithm (dada2) [41]. This entailed the joining of paired end sequences and quality filtering. For the latter, sequences shorter than 200 bp, containing more than two ambiguous characters, that had homopolymer runs longer than 6 bp, or that contained more than one mismatch to the sample-specific barcode or to the primers sequences were removed. Following removal of the barcode and primer sequences, the raw nifH reads were denoised to remove sequencing errors, separated into OTUs defined by clustering at 97% sequence similarity [42] using a curated and publicly available database of nifH sequences (https://wwwzehr.pmc.ucsc.edu/, accessed on 15 September 2021) [43], after which chimeras were removed [44,45,46]. Final OTUs were taxonomically classified using BLASTn against this database.
2.4. Statistical Analyses of Metabarcoding Data and Soil Physico-Chemical Properties
Prior to statistical analyses, singletons were removed using the filter.shared command in Mothur (version 1.35.1) [36]. Alpha and beta diversity analyses were conducted to compare samples based on OTU assignment. Alpha diversity indices (richness, non-parametric Shannon diversity and Shannon evenness) [47] were calculated using the summary.single command in Mothur [36]. Beta diversity OTU-based (Bray-Curtis and Jaccard) matrices to compare the structure (Bray-Curtis) and membership (Jaccard) of the samples were calculated using the dist.shared command in Mothur [36]. The least number of sequences across all the samples after removal of singletons were set as the sampling depths (i.e., n = 1465 for 16S rRNA and n = 2691 for nifH) and subsampled 1000 times (iters = 1000) to standardize both the alpha and beta diversity calculations. Good coverage estimate for the alpha diversity matrices was used to determine the % coverage within samples after sub-sampling.
Prior to analysis of variance (ANOVA) or permutations analysis of variance (PERMANOVA), a legume rhizosphere single soil sample from Leliefontein was eliminated. Therefore, four legume species (C. sericea, L. diffusa, V. karroo, and W. monoptera from three sampling sites (Brakputs, Kamiesberg, and Kamieskroon) totalling to 23 samples were considered. Alpha diversity and soil properties data were subjected to normality tests using histograms and Q-Q plots to check the homogeneity of variance in R (v.4.0.0) software (https://www.rstudio.com, accessed on 5 September 2021). One-way ANOVA was conducted to determine whether the Alpha diversity indices and soil properties were significantly different under legumes and sites where the samples were collected. The Akaike information criterion (AIC) was used to select the best-fit model to explain variation in the dependent variables. Table of p values for all possible pairwise comparisons of least square means (lsmeans), and the letters (letter display) were conducted using the Tukey’s honestly significant difference (Tukey’s HSD) post-hoc test in R (v.4.0.0). Pearson correlation and regression analysis with the soil properties were also conducted using R (v.4.0.0) to test if these data were correlated and by how much the properties explained the changes in the alpha diversity indices. PERMANOVA was conducted to test if legume species and sites influenced the Beta diversity matrices.
For graphical visualization of the structure of communities as shaped by the legumes in the respective sampling sites, non-metric multidimensional scaling (NMDS) and principal co-ordinate (PCoA) were performed in R (v.4.0.0) on OTU-based matrices (Bray-Curtis and Jaccard) [48]. Canonical analysis of principal coordinates (CAP) was also conducted in R (v.4.0.0) to emphasize the influence of soil properties on the structure of bacteria [49]. Heatmaps were generated using relative abundance data of OTUs using the R (v.4.0.0) package heatmap.
3. Results
3.1. Physico-Chemical Properties of the Studied Rhizosphere Soils from the Succulent Karoo Biome
The results of the soil properties analysis are presented in Table SI. Overall, soil properties were not significantly different among the sampling sites (Brakputs, Kamiesberg and Kamieskroon) (p > 0.05). Legume species, however, revealed significant differences for pH and TC (p < 0.05). Overall, pH values ranged from 4.00 (W. monoptera) to 6.47 (V. karroo). The pH values associated with V. karroo were significantly higher than those from W. monoptera and C. sericea (p < 0.05). The pH therefore ranged from extremely acidic for W. monoptera, to strongly acidic for C. sericea, L. diffusa and slightly acidic for V. karroo. TC values ranged from 0.61 to 1.93 (%) for L. diffusa and V. karroo, respectively. TC values under V. karroo were significantly higher than those from C. sericea and L. diffusa (p < 0.05). TC values were generally low for all the species, except V. karroo where it was high. TN values ranged from 0.05 to 0.16 (%) for rhizosphere soils from L. diffusa and V. karroo, respectively. Generally, TN values ranged from low to very low for all the legume rhizosphere soils analyzed. The NH4+ and NO3− content ranged from 9.46 to 17.1 mg-N kg−1 and 0.57 to 10.9 mg-N kg−1, respectively. There were no significant differences observed for TN, NH4+ and NO3− values between the different legumes, although V. karroo consistently maintained the highest values (p > 0.05).
3.2. The Dominant Bacterial Communities in the Rhizosphere Soils Considered
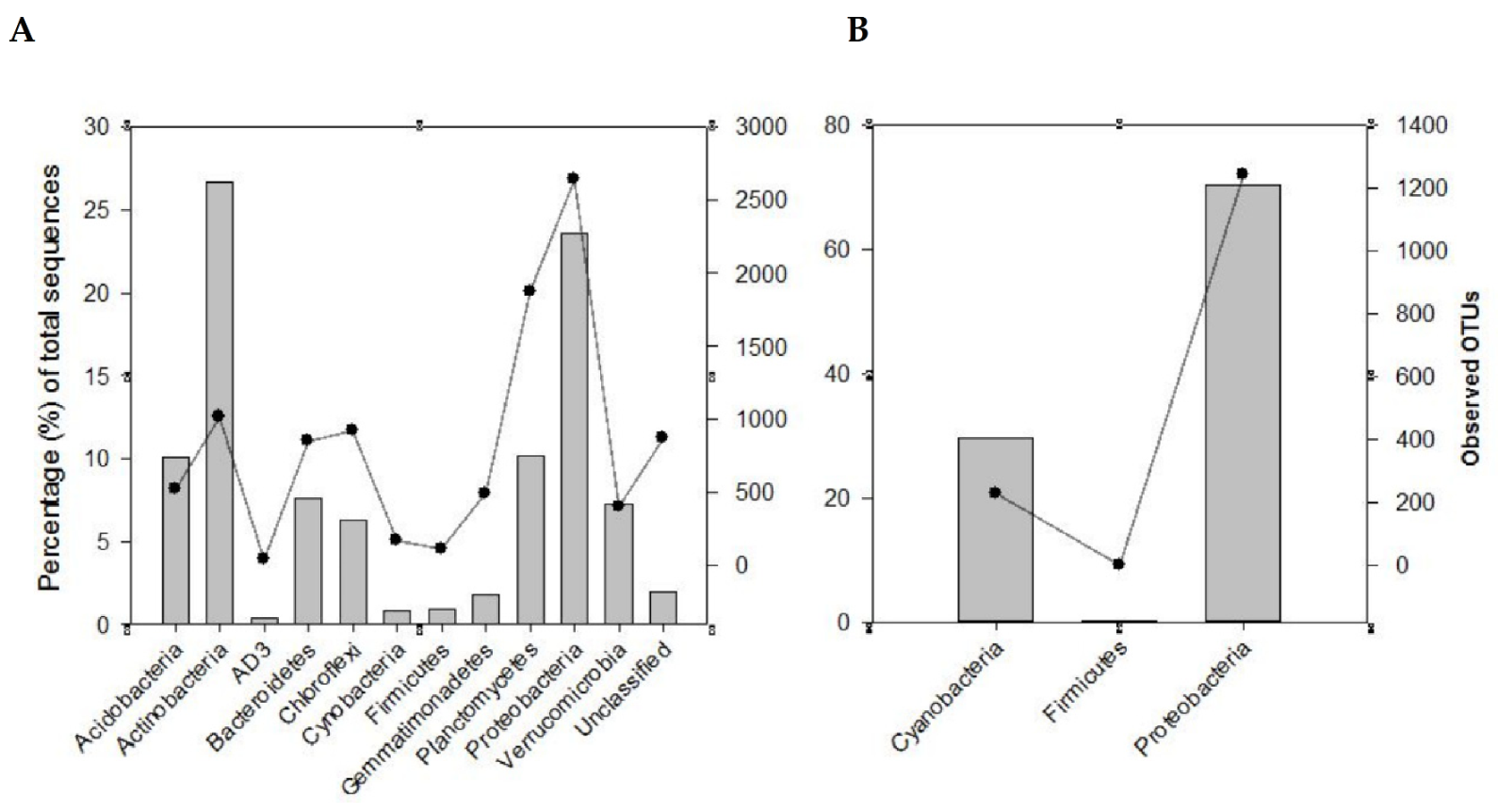
Using the 16S rRNA metabarcoding, a total of 362,185 sequences were classified into 10,643 OTUs (at 97% similarity threshold) from the 24 rhizosphere soil samples examined. Although good coverage (range 66–91%, average 79%) estimates suggested that dominant taxa would be identified, analysis of a larger volume of sequences is recommended for a comprehensive sampling of the diversity of these communities. The identified OTUs were assigned to biologically and taxonomically diverse species from a range of bacterial phyla (Table S2; Figure 2A). Among these were Actinobacteria (26.7%), Proteobacteria (23.6%), Planctomycetes and Acidobacteria (10%), Bacteroidetes (7.6%), Verrucomicrobia (7.3%), Chloroflexi (6.3%), Gemmatimonadetes (1.8%), Firmicutes (0.9%), and Cyanobacteria (0.8%). These corresponded to 1018, 2641, 1878, 519, 852, 404, 924, 598, 118, and 161 OTUs, respectively (Figure 2A and Figure 3A). These phyla represented approximately 96% and 85% of the total sequences and OTUs observed, respectively. Thirty-eight OTUs representing approximately 18% of the total sequences were shared across all the soil samples from the two biomes. Most of these OTUs belonged to Actinobacteria (17 OTUs) and Proteobacteria (11 OTUs). Forty OTUs representing approximately 13.6% of the total recovered sequences were shared among the SKB samples. These OTUs were also dominated by Actinobacteria and Proteobacteria.
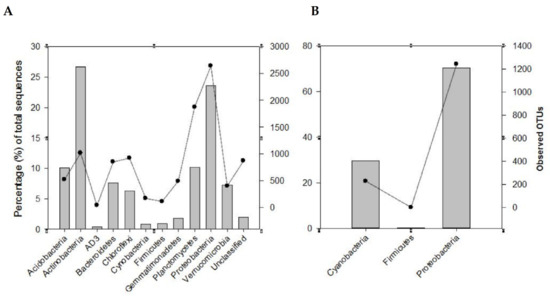
Figure 2.
Recovery of the major phyla, using metabarcoding with the 16S rRNA (A) and nifH (B) genes, from the rhizosphere soils of the different legume species from various sites in the study area. The bars represent the percentage total sequences while the lines with a filled circle symbol represent the observed OTUs.
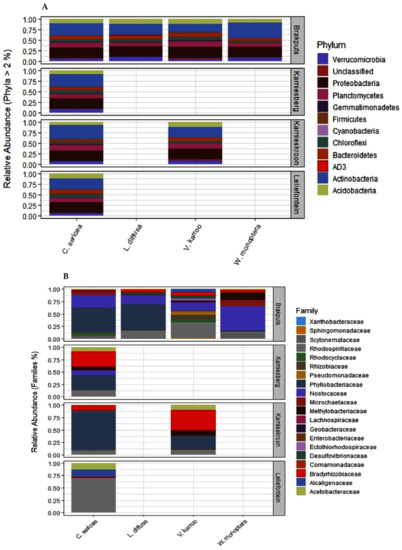
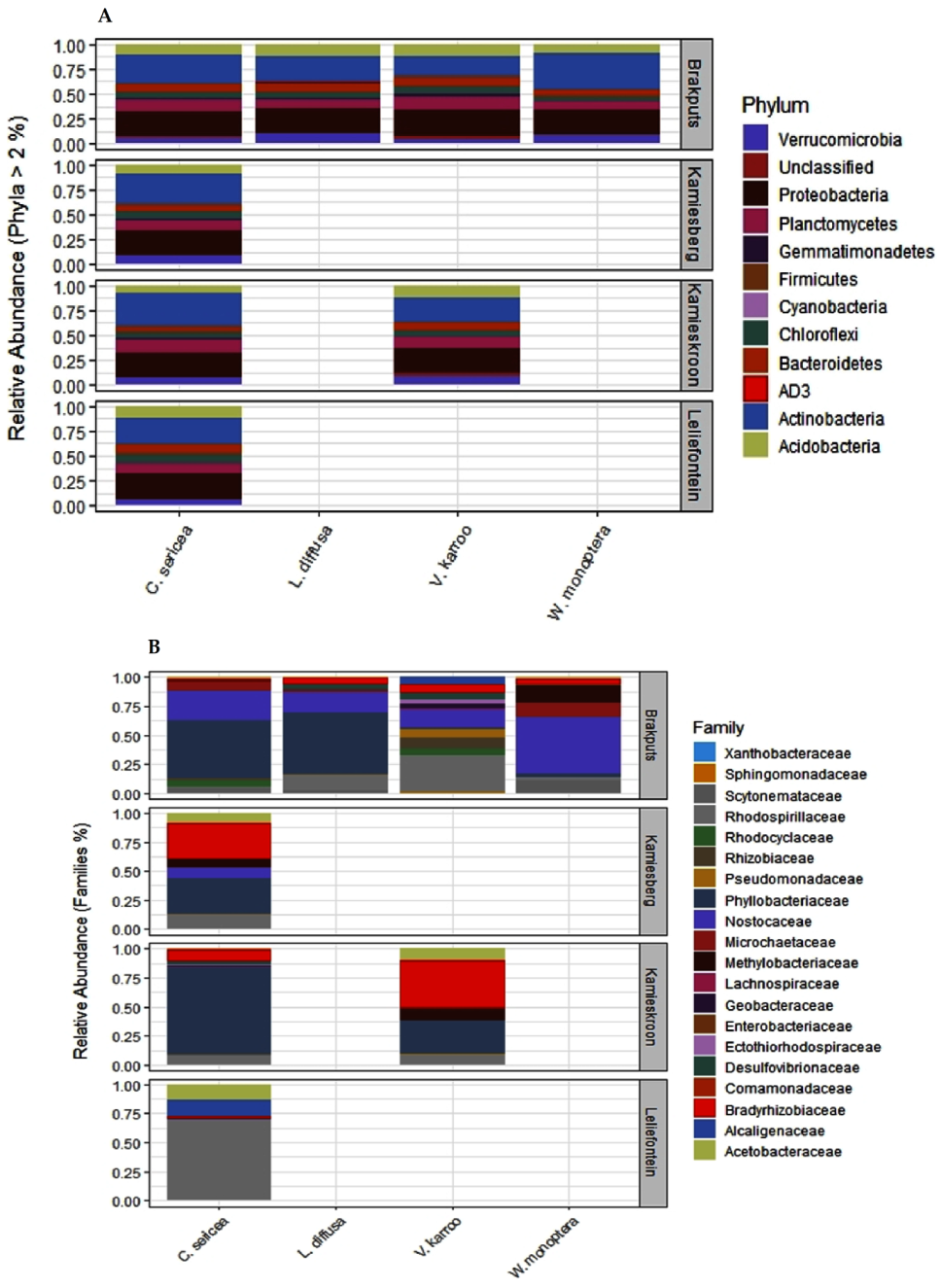
Figure 3.
Distribution of relative abundances of microbial communities in the rhizosphere soils of different legume species in the study sites. (A) Phylum level relative abundances of total bacteria (16S rRNA), (B) relative abundances of dominant nitrogen-fixing families (nifH gene).
Metabarcoding using the nifH gene allowed for the classification of 1,094,932 sequences into 1471 OTUs using a similarity threshold of 97%. Good coverage (range 96.4–99.8%, average 99%) indicated that sub-sampling at 2691 sequences covered the majority of the diversity of communities for all the samples. Proteobacteria and Cyanobacteria (respectively represented by 70.3% and 29.5% of the total sequences) were the most dominant and diverse phyla (Figure 2B). They contained 1241 (84.4%) and 229 (15.6%), respectively, of the total OTUs observed. Overall, these two phyla represented approximately 99.8% and 99.9% respectively, of the nifH total sequences and OTUs observed. A small proportion of the sequences (0.20%, 1 OTU) originated from the phylum Firmicutes.
Based on the nifH data, several diazotrophic families were measured (Figure 3B; Table S3). These consisted of OTUs from the families Nostoceae, Microchaetaceae and Scytonemataceae (24.4%, 4.0%, and 3.1%), respectively, (Cyanobacteria) of the total sequences (Figure 3B; Table S3). Others OTUs belonged to the families Phyllobacteriaceae (39.0%), Rhodospirillaceae (7.9%), Bradyrhizobiaceae (4.6%), Rhizobiaceae (4.2%) Rhodocyclaceae (3.7%), Methylobacteriaceae (3.0%), Pseudomonadaceae (2.7%), Geobacteraceae (1.78%), Alcaligenaceae (1.2%), Acetobacteraceae (1.1%), Desulfovibrionaceae (0.62%), Comamonadaceae (0.18%), Ectothiorhodospiraceae (0.16%), Sphingomonadaceae (0.12%), and Enterobacteriaceae (0.06%) (Proteobacteria) of the total sequences. There was also one OTU belonging to the family Lachnospiraceae (0.2%) (Firmicutes) of the total sequences (Table S3). The high dominance of Phyllobacteriaceae was also supported by the recovery of one OTU in all 24 soil samples examined. This OTU represented a member of the genus Mesorhizobium and formed 27.3% of the total nifH sequences. Two OTUs, members of Proteobacteria representing approximately 27.3% of the total sequences were shared among the SKB samples.
3.3. OTU-Based Diversity of Diazotrophs as Influenced by Legume Species
Legumes showed common and distinct influences on the diazotrophs at family level in the arid soils of the Succulent Karoo or Fynbos biomes based on nifH metabarcoding analysis (Figure 3B; Table S3). For example, the rhizosphere soils of C. sericea and V. karroo shared most members of the identified families. These included OTUs from Bradyrhizobiaceae, Acetobacteraceae, Phyllobacteriaceae, Rhodospirillaceae, Comamonadaceae, Rhizobiaceae, Rhodocyclaceae, Nostocaceae, Methylobacteriaceae, and Sphingomonadaceae. On the other hand, L. diffusa and W. monoptera showed few families whose OTUs were mainly shared either with C. sericea, V. karroo or both. These were OTUs from Bradyrhizobiaceae, Nostocaceae, Microchaetaceae, Rhodospirillaceae, Methylobacteriaceae, Acetobacteraceae, and Scytonemataceae in the rhizosphere soils of W. monoptera and OTUs from Phyllobacteriaceae, Rhizobiaceae, Nostocaceae, and Scytonemataceae in the rhizosphere soils of L. diffusa (Table S3).
In terms of the association of diazotrophs with specific legumes, the rhizosphere soils of V. karroo specifically showed a high diversity and abundance of OTUs representing Alcaligenaceae, Desulfovibrionaceae, Geobacteraceae, Lachnospiraceae, Ectothiorhodospiraceae and Pseudomonadaceae among others (Table S3). Enterobacteriaceae was specifically found in the rhizosphere of W. monoptera (Table S3).
3.4. Alpha Diversity
The richness (observed taxa) and structure-based alpha diversity metrics (Shannon Index and Shannoneven Index) were strongly influenced by legumes species (p < 0.01) but not the sampling sites or the interactions between legume species and sites (p > 0.05) for both 16S rRNA and nifH genes (Table S4). Therefore, only the results of the influence of legume species on alpha diversity indices are presented.
Based on the 16S rRNA, V. karroo revealed the highest species richness (687 ± 31.6), Shannon index (structure) (6.09 ± 0.08) and Shannoneven index (evenness) (0.932 ± 0.006) (p < 0.05) (Table 1). The lowest alpha diversity indices were revealed for W. monoptera, species richness (482 ± 44.8), Shannon index (5.45 ± 0.11) and Shannoneven index (0.883 ± 0.008) (p < 0.05) (Table 1). Richness was significantly positively correlated with the soil pH, TN and TC (r = 0.70, p < 0.001; r = 0.54, p < 0.01; r = 0.52, p < 0.05) respectively (Table 2). A similar trend was observed for Shannon and Shannoneven indices correlations with soil properties as follows; Shannon index (r = 0.77, p < 0.001 (soil pH), r = 0.53, p < 0.01 (TN) and r = 0.50, p < 0.05) for TC. Shannoneven index (r = 0.79, p < 0.001; r = 0.49, p < 0.05; r = 0.42, p < 0.05) for pH, TN and TC respectively (Table 2). Regression analysis revealed significant effects of soil pH (r2 = 0.46; p < 0.001), TN (r2 = 0.21; p < 0.05) and TC (r2 = 0.20; p ≤ 0.05) for richness. Soil pH (r2 = 0.56; p < 0.001), TN (r2 = 0.22; p < 0.01) and TC (r2 = 0.18; p ≤ 0.05) for Shannon index. Soil pH (r2 = 0.59; p < 0.001), TN (r2 = 0.21; p < 0.05) and TC (r2 = 0.15; p < 0.05) for Shannoneven index (Table 2).

Table 1.
Alpha diversity indices determined using the 16S rRNA and nifH barcodes for different legume species.

Table 2.
Pearson linear correlation (r) and regression (r2) analysis of the Alpha diversity indices with soil physico-chemical properties.
Based on the nifH, V. karroo revealed significantly higher richness (115.0 ± 12.1) than C. sericea (65.5 ± 10.0) and L. diffusa (45.2 ± 13.6) (p < 0.05) (Table 1). V. karroo also had the highest structure (2.91 ± 0.25) while W. monoptera revealed the highest evenness (0.67 ± 0.08) (p < 0.05) (Table 1). Interestingly, pH did not reveal significant correlations with any of the alpha diversity indices (Table 2). Species richness was significantly positively correlated with TN (r = 0.62; p ≤ 0.01), TC (r = 0.66; p ≤ 0.001), NH4+ (r = 0.42; p ≤ 0.05) and NO3− (r = 0.46; p < 0.05) (Table 2). Shannon index was significantly positively correlated with TN and TC (r = 0.55; p < 0.01, r = 0.59; p < 0.01) respectively, while Shannoneven index was only significantly positively correlated with TC (r = 0.45; p < 0.05) (Table 2). Regression analysis revealed significant r2 values for TN (r2 = 0.38; p < 0.01), TC (r2 = 0.43; p < 0.001), NH4+ (r2 = 0.18; p ≤ 0.05) and NO3− (r2 = 0.21; p ≤ 0.05) for richness. Shannon index revealed (r2 = 0.30; p < 0.01) TN and (r2 = 0.35; p < 0.01) TC. R2 = 0.20; p < 0.05 (TC) was revealed for Shannoneven Index (Table 2).
3.5. Beta-Diversity
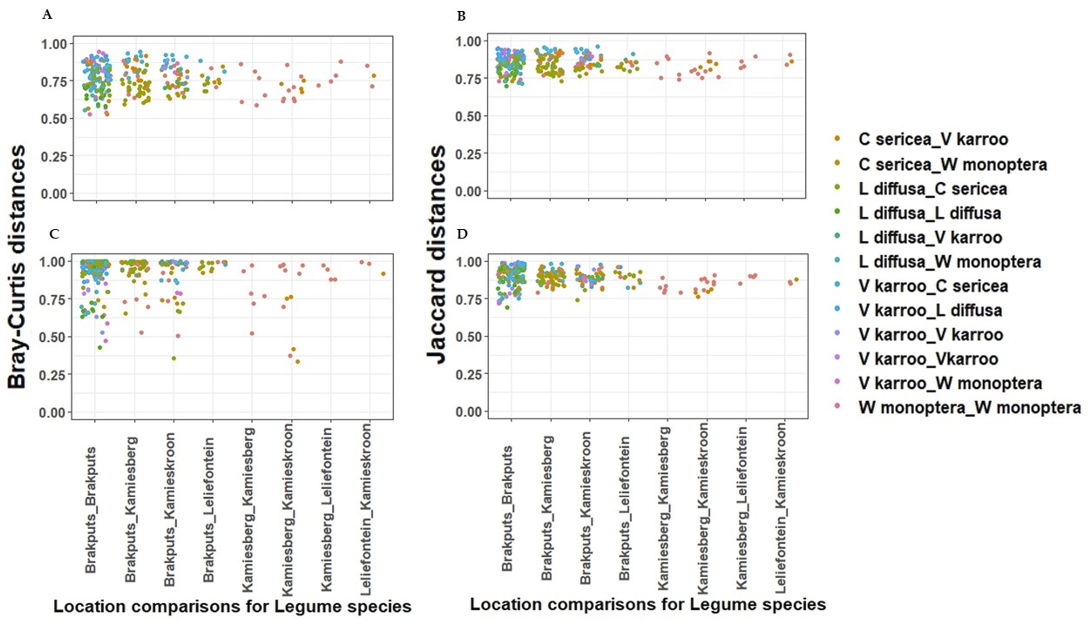
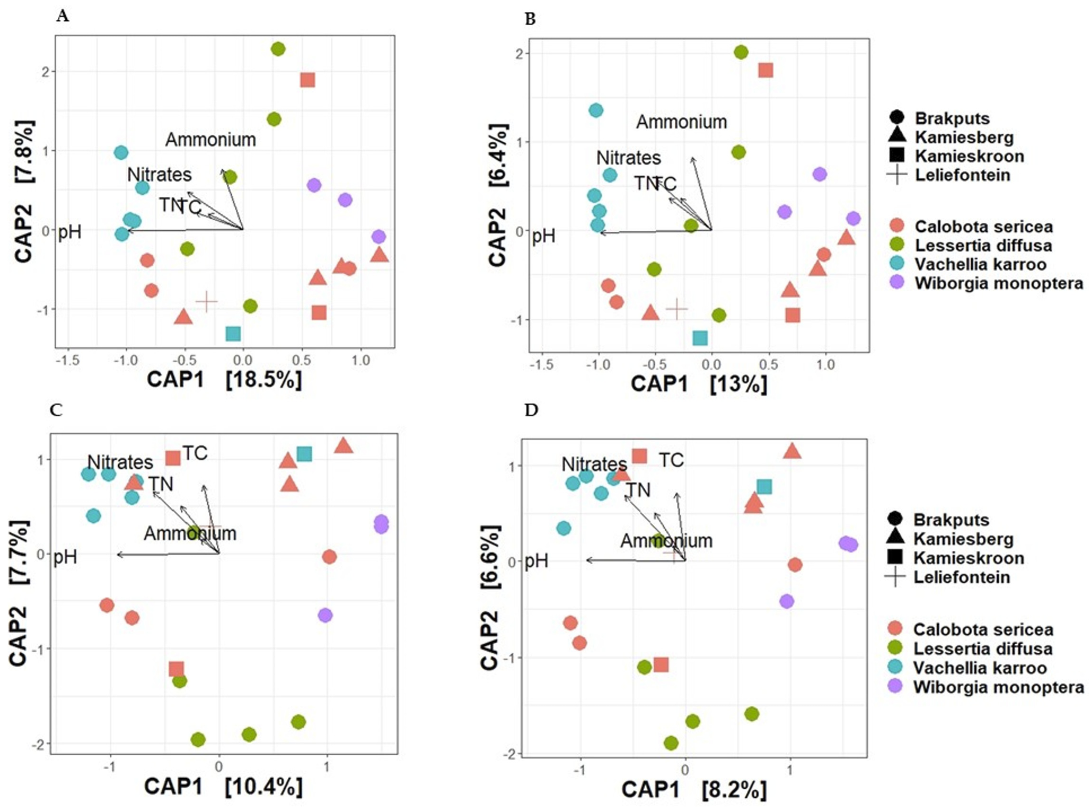
Based on 16S rRNA gene, the average dissimilarity in community structure between legumes and sites was 76 ± 9% (Bray-Curtis), and 85 ± 5% (Jaccard) (Figure 4A,B). These high indices suggested a low level of similarity between the samples examined. Prior to PERMANOVA analysis, ordination analysis (NMDS and PCoA) based on Bray-Curtis and Jaccard matrices revealed legumes specific clustering of bacteria particularly for V. karroo (results not shown). PERMANOVA analysis based on both matrices revealed that variability among samples was best explained by the legumes (p < 0.001) (Table S4). CAP analysis showed pH, TN, and TC as important factors influencing the 16S rRNA Beta-diversity (Figure 5A,B). For example, communities clustered together in the direction of high pH, TN and TC particularly those associated with V. karroo.
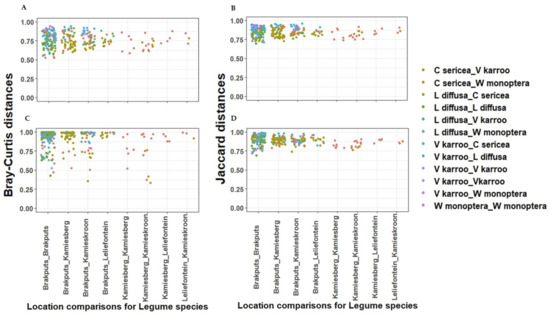
Figure 4.
Dissimilarities between sites and legume species using pair wise structure-based Bray-Curtis and Jaccard distances respectively. (A,B) = 16S rRNA gene; (C,D) = nifH gene.
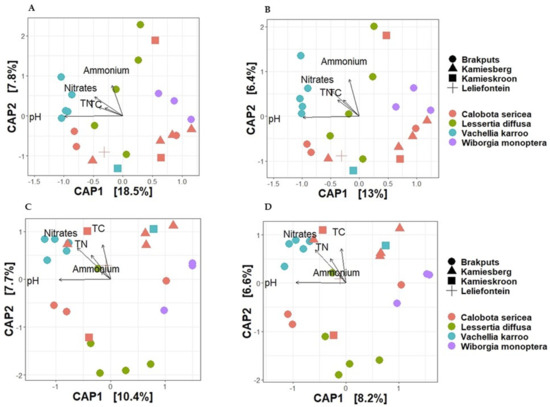
Figure 5.
Canonical analysis of principal coordinates (CAP) of Beta-diversity based on Bray-Curtis and Jaccard distances for visual presentations of patterns of composition of bacterial communities with respect to legume species and their relationship with soil chemical properties. (A) = 16S rRNA Bray-Curtis, (B) = 16S rRNA Jaccard, (C) = nifH Bray-Curtis and (D) = nifH Jaccard. TC, total carbon; TN, total nitrogen.
The average dissimilarity for the nifH gene was 91 ± 13% (Bray-Curtis) and 89 ± 6% (Jaccard) (Figure 4C,D). These indicated a low level of similarity between the samples. Similar to the 16S rRNA, nifH gene-based ordination analysis on diazotrophic communities revealed legumes specific clustering particularly for V. karroo (Figure 5C,D). PERMANOVA analysis based on both matrices revealed that legumes and sites influenced structure and membership matrices (p < 0.01) (Table S4). For example, CAP analysis showed clear clustering patterns among legumes (e.g., L. diffusa, W. monoptera and V. karroo). In addition, nitrates and TN were found as the environmental properties that mainly influenced the nifH Beta diversity, as communities clustered in the direction of high nitrates and TN particularly those associated with V. karroo and some C. sericea species (Figure 5C,D).
4. Discussion
4.1. Bacteria Composition in Rhizosphere Soils Used Is Associated with Environmental Conditions
Based on the metabarcoding of the 16S rRNA, several of the taxa identified (e.g., Proteobacteria, Actinobacteria, Bacteroidetes, and Cyanobacteria) have previously been shown to dominate rhizosphere and desert soils [7,50]. Most of them are reported to have inherent properties making them well suited to the SKB soils, such as adaptation to high temperatures and soil acidity as is the case for the SKB. For instance, many species of Cyanobacteria, Planctomycetes and Verrucomicrobia are known to thrive in higher temperatures and are tolerant to low soil pH [51,52,53].
The consistent dominance of the Proteobacteria phylum using both 16S rRNA and nifH genes in this study was in agreement with several other studies which reported their abundance in different soil ecosystems such as agricultural, forests, grasslands, saline as well as semi-arid soils [54]. This is attributed to their role in sustaining these environments through various biogeochemical processes. Cyanobacteria members are aquatic, but have also consistently been reported in soil crusts and rhizosphere soils in arid and semi-arid environments, particularly due to their role in atmospheric nitrogen fixation [51,54]. This was consistent with the nifH findings in this study. Their low measurements according to the universal 16S rRNA gene could be due to its low-resolution power compared to functional and group specific primers such as the nifH gene [55]. Firmicutes on the other hand, were less diverse and abundant in the considered rhizosphere soils using the both metabarcoding approaches. This was not surprising as members of this phylum are mainly associated with contaminated environments such as sludges and soil impacted by acid mine drainage [54,56].
Environmental factors strongly influenced the total and diazotrophic bacteria in the soils examined. This was particularly evident for soil pH and nutrients. Significantly positive correlations of >70% for pH and >40% for TN and TC, were observed with the richness and structure of total bacteria. These factors accounted for much of the variation within communities (at least 45% for pH, 20% for TN and at least 15% for TC). These factors further influenced the diazotrophs, as significant positive correlations of >40% were observed for species richness for all the soil properties except pH. TC and TN influenced the structure of the diazotrophs as >55% positive correlations were observed for the species structure, while TC showed 45% positive correlations with species evenness. These factors accounted for at least 18% of variations within the diazotrophs. The influence of environmental factors on microbes was also revealed for beta-diversity analyses. Similar effects of pH, TN, TC, and NO3− on soil bacteria were found in the bulk or rhizosphere soils of Acacia dealbata in the grassland biome in South Africa [57], and in an agricultural soil in Kenya [58]. Soil pH has also been reported as a main driver of changes in the structure and composition of total soil bacteria [58,59].
It is likely that the different groups of bacteria identified in this study form part of specialized communities, some of which interact with plants to facilitate or enhance their ability to colonize and become established in the harsh environmental conditions of the SKB. Ways in which this facilitation can occur is through nitrogen, carbon and phosphorus compounds cycling, thereby improving the plants’ access to nutrients [60,61]. Many species of the Gemmatimonadetes, Cyanobacteria, Proteobacteria, Chloroflexi, Firmicutes, and Acidobacteria, for example, are known to facilitate carbon assimilation via (bacterio) chlorophyll-based photosynthesis [62,63], while certain Proteobacteria and Firmicutes are capable of phosphorus solubilization, thus making this element available to plants [64,65]. However, in the nitrogen-poor soils of the SKB, cycling of this element is perhaps equally important, and various members of the Proteobacteria [66,67], Planctomycetes [53], Bacteroidetes [68], Verrucomicrobia [69], Firmicutes [64], and Cyanobacteria [51,52] are known to be capable of fixing atmospheric nitrogen to provide ammonium and/or nitrates for plant growth.
The bacteria in the rhizosphere soils of the legumes investigated are involved in the promotion of the plants’ growth and might even protect them against pathogens. Many Acidobacteria, Firmicutes and Proteobacteria are well known to produce indole-3-acetic acids (IAAs), siderophores and the enzyme aminocyclopropane carboxylate (ACC) deaminase [64,70]. IAA is a plant growth hormone [13,64], and siderophores chelate iron and avail it to plants for growth, while making it unavailable to other organisms, particularly pathogens [64,71]. Under stressful conditions (e.g., drought and heat), the ACC deaminase promotes root growth by lowering ethylene levels in plants [68,71,72,73]. Taken together, the diverse bacteria measured thus underscore the crucial role they play in supporting plants in the SKB.
4.2. Legume Species Influence Diazotrophs in the Succulent Karoo Biome
Metabarcoding with the nifH allowed a more in-depth analysis of the diazotrophic taxa occurring in the rhizosphere soils examined. Our findings revealed diverse OTUs which were separated into at least 19 families of diazotrophs. The distribution, diversity and abundance of these diazotrophs were however, modulated by legumes considered. For example, the rhizosphere soils of L. diffusa were mainly dominated by OTUs belonging to Phyllobacteriaceae, Rhizobiaceae, Nostocaceae, and Scytonemataceae. These families contain species known to fix atmospheric nitrogen. For example, Phyllobacteriaceae contained the rhizobial genus Mesorhizobium and Rhizobiaceae contained the genera Sinorhizobium/Ensifer among others. These findings support previous reports which demonstrated the symbiotic relationships of Mesorhizobium and Sinorhizobium/Ensifer with Lessertia species [17,66]. Nostocaceae and Scytonemataceae on the other hand, are known to contain species with free-living nitrogen fixation abilities [51]. The rhizosphere soils of W. monoptera mainly contained species belonging to Bradyrhizobiaceae, Methylobacteriaceae, Nostocaceae, Rhodospirillaceae, Acetobacteraceae, Microchaetaceae, Scytonemataceae, and Enterobacteriaceae families. Bradyrhizobiaceae and Methylobacteriaceae members have symbiotic rhizobial capabilities with these legumes, which is consistent with previous reports regarding the distribution of Bradyrhizobium and its association with a wide range of legumes [17,28,66]. Methylobacteriaceae members have also been reported to possess symbiotic associations with diverse legumes in the papilionoid tribe Crotalarieae [74,75]. Members from the other families contain free-nitrogen fixation capacities. For example, members from Rhodospirillaceae [76,77] and Microchaetaceae [51] as well as Enterobacteriaceae that occurred specifically in rhizosphere soils of W. monoptera [78]. Moreover, some diazotrophic members belonging to Acetobacteraceae promote plant growth potentially through phosphorus solubilization and antagonism against plant pathogens [65]. The rhizosphere soils of L. diffusa and W. monoptera therefore, contained diazotrophic communities with the ability to mainly fix atmospheric nitrogen and solubilize phosphorus for W. monoptera species. According to the alpha-diversity analyses, generally low species richness, structure and evenness was measured under the rhizospheres of W. monoptera and L. diffusa. Consistently, the communities under these two legumes clustered separately, being distinct from those under C. sericea and V. karroo. Both legumes are not widely distributed in the SKB, which explains the low diversity, abundance and specialization of their associated diazotrophs.
The rhizosphere of C. sericea was dominated by OTUs from Bradyrhizobiaceae, Rhodocyclaceae, Rhizobiaceae, Nostocaceae, Phyllobacteriaceae, Rhodospirillaceae, Acetobacteraceae, Sphingomonadaceae, Methylobacteriaceae, and Comamonadaceae families. Species from Bradyrhizobiaceae, Rhizobiaceae, Methylobacteriaceae and Phyllobacteriaceae have symbiotic characteristics with legumes in the papilionoid tribe Crotalarieae [66,79,80,81]. Members of the Rhodocyclaceae, Nostocaceae, Rhodospirillaceae, Acetobacteraceae, and Comamonadaceae are free-living nitrogen fixers [82,83,84,85]. In addition, some members from Acetobacteraceae are known to promote plant growth through phosphorus solubilization and antagonism against pathogens [65]. Some members from Comamonadaceae promote plant growth through carbon cycling [86], while members from Sphingomonadaceae are known to promote plant growth through the production of IAAs [87]. These communities did not have specific clustering pattern with C. sericea according to the ordination analysis. Therefore, the diazotrophs associated with C. sericea are not only diverse and abundant, but also possess different plant growth promoting properties, ranging from atmospheric nitrogen fixation, phosphorus solubilization, production of IAAs and antagonism against pathogens. This could be the reason this legume is commonly distributed in the SKB and also in the neighboring Fynbos biome.
V. karroo species had higher diversity and abundance of diazotrophs than all the other legumes. Significantly higher species richness and diversity under V. karroo were observed using 16S rRNA-based metabarcoding. These were supported by ordination analysis, where beta-diversity indices showed clustering of members associated with V. karroo away from those associated with the other legumes. Additionally, higher pH, TC, TN, NH4+, and NO3− were measured in the V. karroo rhizosphere soils than for other legumes. Increased levels of nutrients and pH associated with V. karroo are believed to support its ability to attract diverse and abundant bacteria [57,59]. This gives it a competitive advantage in modifying the nutrient status of its rhizosphere soils more strongly than other legumes in the same habitat. In Acacia sensu lato, such advantages have been suggested to be as a result of nitrogen fixation, organic matter production via root exudates, and leave fall under their canopy [57]. These mechanisms drive the invasiveness of certain legumes [14,19,20] and may also play an important role in the encroaching behavior of V. karroo in the SKB.
The bacterial distribution patterns observed in the rhizosphere communities associated with V. karroo suggest that it modified its rhizosphere environment, favouring or enhancing the establishment of particular communities [19,20,24,88]. For example, OTUs from Geobacteraceae, Alcaligenaceae, Lachnospiraceae, Desulfovibrionaceae, Ectothiorhodospiraceae, and Pseudomonadaceae taxa were specifically associated with the rhizosphere soils of V. karroo. These diazotrophs are free-nitrogen fixers [61,89,90,91,92,93,94,95]. Moreover, some members from Alcaligenaceae promote plant growth through production and control of IAAs levels [87,96]. OTUs such as those from Pseudomonadaceae promote plant growth through phosphorus solubilization, production of IAAs and siderophores [97,98,99]. Although this study is the first to report on the metabarcoding-based rhizosphere bacteria associated with V. karroo, different invasive trees such as Berberis thunbergii DC. (Japanese barberry) have been shown to attract different taxa including Pseudomonadaceae in their rhizosphere environment [88]. Acacia dealbata, an Australian invasive legume in South Africa, mainly attracted members from Bradyrhizobiaceae and Pseudomonadeceae among others in its rhizosphere [19,57]. Therefore, interactions between V. karroo and particular soil bacteria lead to significant changes in the composition of rhizosphere communities. Other free nitrogen-fixers which were associated with V. Karroo, but also found in the rhizosphere of the other legumes included members from Rhodocyclaceae, Nostocaceae, Rhodospirillaceae, Acetobacteraceae, Microchaetaceae, Sphingomonadaceae, and Comamonadaceae families [51,65,77,84,86,87]. In addition, some members from Acetobacteraceae, Sphingomonadaceae, and Comamonadaceae also promote plants growth through phosphorus solubilization, the production of IAAs, siderophore production for protection against pathogens, and carbon cycling [85,86,100].
With regards to possible rhizobial symbionts, V. karroo showed the ability to associate with diverse rhizobia. For example, several OTUs that were affiliated with Phyllobacteriaceae (Mesorhizobium), Bradyrhizobium, and a few with Rhizobiaceae (Rhizobium and Ensifer) formed part of its rhizosphere. These findings are consistent with other studies [14,17,66,101]. Undoubtedly, the ability of V. karroo to establish symbiotic interactions with a wide range of rhizobia also provides it with a competitive advantage over other legumes to colonize and encroach on new areas [25]. The high diversity of its potential rhizobial symbionts, combined with unique bacteria in V. karroo’s rhizosphere soils in the SKB, support its involvement in reconstructing its microbiome, a strategy used by most invasive members of the mimosoid clade of legumes to invade and colonize new environments [19]. The rhizosphere of V. karroo therefore contained the most diverse and abundant diazotrophs that possess a wide range of plant growth promotion properties such as atmospheric nitrogen fixation, phosphorus solubilization, the production of IAAs and siderophores, carbon cycling as well as antagonism effects against pathogens.
Most studies that investigated the influence of plant species and environmental conditions on soil microbial communities have mainly focused on non-legumes versus legumes [55]. For example, legume species Stylosanthes guianensis (Aubl.) Sw., Trifolium pratense L., and Medicago sativa L. enriched soil microbial communities compared to grass species (Paspalum natatum, Festuca arundinacea L., Lolium perenne L.) in China [102]. In Nigeria, a legume species Pterocarpus erinaceus improved the soil physico-chemical properties and selected for ten bacterial species compared to a non-legume Anoigessus leiocarpa, which was associated with low values of soil properties and only six bacterial species [103]. This study further revealed that legume species, which fall under the same family Fabaceae, select for specific microbial communities and also share some microbial communities across their rhizospheres.
5. Conclusions
This study revealed diverse diazotrophs associated with legumes in the SKB in South Africa. Environmental factors such as soil pH, soil nutrients, and legume species influenced the microbial communities. A member of Mesorhizobium species was common in all rhizosphere soils considered. Other diazotrophs such as Bradyrhizobiaceae, Nostocaceae, among others were shared across several legume rhizosphere soils, while others such as Enterobacteriaceae and Pseudomonadaceae were specific to specific legume rhizosphere soils. Legumes such as L. diffusa and W. monoptera that are not widely distributed in the SKB haboured less diverse and abundant diazotrophs which mainly possess atmospheric nitrogen fixation properties compared to the widely distributed C. sericea and the encroaching V. karroo species. These legumes both contained diverse diazotrophic communities, associated with diverse plant growth promotion properties such as atmospheric nitrogen fixation, phosphorus solubilization, production of IAAs and siderophores, carbon cycling and plant protection against pathogens.
Since the metabarcoding approach of 16S rRNA and nifH genes only allowed us to identify communities that are potentially involved in important processes such as nitrogen fixation, metatranscriptomics analyses are recommended to study gene expression involved in crucial biological processes. Future research will also focus on root-bacteria nodulation and nitrogen-fixation with legumes and respective rhizosphere soils used in this study. If effective, such strains will be characterized and recommended for development as potential inoculum in the agricultural industry.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10020216/s1, Table S1: Statistical analysis of soil chemical properties of the study sites showing main effects of different legume species and sites, Table S2: Diverse bacterial phyla in the Succulent Karoo biome in South Africa, their inherent properties and possible impacts on plant communities, Table S3: Families containing diazotrophic bacteria (nifH gene) in the rhizosphere soils examined and their possible roles in plant growth promotion of legume species in the Succulent Karoo biome in South Africa, Table S4: Analysis of variance (ANOVA) and Permutations analysis of variance (PERMANOVA) to test the effects of factors ‘legume species’, ‘sites’ and their interactions on the respective alpha and beta diversity matrices, using the 16S rRNA and nifH gene barcodes.
Author Contributions
Conceptualization, E.K.M., E.T.S. and S.N.V.; Formal analysis, E.K.M.; Funding acquisition, E.T.S.; Investigation, E.K.M.; Methodology, E.K.M.; Project administration, E.T.S.; Supervision, E.T.S. and S.N.V.; Validation, E.K.M.; Visualization, E.K.M.; Writing—original draft, E.K.M.; Writing—review & editing, E.T.S. and S.N.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
This research was funded by DSI-NRF Centre of Excellence in Plant Health and Biotechnology (CPHB) through the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa. We are grateful to the farmers, and Kevin Murray and Martella du Preez for assistance with legume plants identifications and sampling, as well as Mariaan le Roux for confirming our samples identifications. The first author also thanks Sarah Potgieter, Solize Vosloo and Zander Human of the Department of Biochemistry, Genetics and Microbiology for their support in sequence data processing and analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cowling, R.; Rundel, P.; Desmet, P.; Esler, K. Extraordinary high regional-scale plant diversity in southern African arid lands: Subcontinental and global comparisons. Divers. Distrib. 1998, 4, 27–36. [Google Scholar]
- Mucina, L.; Jürgens, N.; Le Roux, A.; Rutherford, M.C.; Schmiedel, U.; Esler, K.J.; Powrie, L.W.; Desmet, P.G.; Milton, S.J.; Boucher, C. Succulent Karoo Biome. In The Vegetation of South Africa, Lesotho and Swaziland; Strelitzia: Pretoria, South Africa, 2006; Volume 19, pp. 221–299. [Google Scholar]
- Carrick, P.J. Shrub Community Dynamics in a South African Semi-Desert. Ph.D. Thesis, University of Cambridge, Cambridge, UK, 2001. [Google Scholar]
- Hoffman, M.T.; Carrick, P.; Gillson, L.; West, A. Drought, climate change and vegetation response in the succulent karoo, South Africa. S. Afr. J. Sci. 2009, 105, 54–60. [Google Scholar] [CrossRef]
- Mucina, L.; Rutherford, M.C. Biomes and Bioregions of Southern Africa. In The Vegetation of South Africa, Lesotho and Swaziland; Strelitzia: Pretoria, South Africa, 2006; Volume 19, pp. 31–51. [Google Scholar]
- Milton, S.; Dean, W.; Marincowitz, C.; Kerley, G. Effects of the 1990/91 drought on rangeland in the Steytlerville Karoo. S. Afr. J. Sci. 1995, 91, 78–84. [Google Scholar]
- Valverde, A.; De Maayer, P.; Oberholster, T.; Henschel, J.; Louw, M.K.; Cowan, D. Specific microbial communities associate with the rhizosphere of Welwitschia mirabilis, a living fossil. PLoS ONE 2016, 11, e0153353. [Google Scholar]
- Pieterse, Z.; Aveling, T.A.; Jacobs, A.; Cowan, D.A. Diversity and seasonality of fungal communities in soil from the Succulent Karoo biodiversity hotspot, South Africa. J. Arid. Environ. 2020, 172, 104020. [Google Scholar] [CrossRef]
- Lewis, G.P.; Schrire, B. Tribe Indigofereae. In Legumes of the World; Royal Botanic Gardens Kew: Richmond, UK, 2005; pp. 361–365. [Google Scholar]
- Schrire, B. Tribe Millettieae. Legumes of the World; Royal Botanic Gardens Kew: Richmond, UK, 2005; pp. 367–387. [Google Scholar]
- Sprent, J.I.; Ardley, J.; James, E.K. Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol. 2017, 215, 40–56. [Google Scholar] [CrossRef]
- Azani, N.; Babineau, M.; Bailey, C.D.; Banks, H.; Barbosa, A.R.; Pinto, R.B.; Boatwright, J.S.; Borges, L.M.; Brown, G.K.; Bruneau, A. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny The Legume Phylogeny Working Group (LPWG). Taxon 2017, 66, 44–77. [Google Scholar] [CrossRef]
- Martínez-Hidalgo, P.; Hirsch, A.M. The nodule microbiome: N2-fixing rhizobia do not live alone. Phytobiomes 2017, 1, 70–82. [Google Scholar] [CrossRef]
- Sankhla, I.S.; Tak, N.; Meghwal, R.R.; Choudhary, S.; Tak, A.; Rathi, S.; Sprent, J.I.; James, E.K.; Gehlot, H.S. Molecular characterization of nitrogen fixing microsymbionts from root nodules of Vachellia (Acacia) jacquemontii, a native legume from the Thar Desert of India. Plant Soil 2017, 410, 21–40. [Google Scholar] [CrossRef]
- Lemaire, B.; Dlodlo, O.; Chimphango, S.; Stirton, C.; Schrire, B.; Boatwright, S.; Honnay, O.; Smets, E.; Sprent, J.; James, E.; et al. Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the Core Cape Subregion (South Africa). FEMS Microbiol. Ecol. 2015, 91, 2–17. [Google Scholar] [CrossRef]
- Lucas, L. Post-Fire Response of Botanical and Microbial Communities in the Succulent Karoo; University of the Western Care: Bellville, South Africa, 2018. [Google Scholar]
- Beukes, C.W.; Boshoff, F.S.; Phalane, F.L.; Hassen, A.I.; le Roux, M.M.; Stepkowski, T.; Venter, S.N.; Steenkamp, E.T. Both alpha-and beta-rhizobia occupy the root nodules of Vachellia karroo in South Africa. Front. Microbiol. 2019, 10, 1195. [Google Scholar] [CrossRef]
- Gerding, M.; O’Hara, G.W.; Bräu, L.; Nandasena, K.; Howieson, J.G. Diverse Mesorhizobium spp. with unique nodA nodulating the South African legume species of the genus Lessertia. Plant Soil 2012, 358, 385–401. [Google Scholar] [CrossRef]
- Kamutando, C.N.; Vikram, S.; Kamgan-Nkuekam, G.; Makhalanyane, T.P.; Greve, M.; Le Roux, J.J.; Richardson, D.M.; Cowan, D.A.; Valverde, A. The functional potential of the rhizospheric microbiome of an invasive tree species, Acacia dealbata. Microb. Ecol. 2019, 77, 191–200. [Google Scholar] [CrossRef]
- Lazzaro, L.; Giuliani, C.; Fabiani, A.; Agnelli, A.E.; Pastorelli, R.; Lagomarsino, A.; Benesperi, R.; Calamassi, R.; Foggi, B. Soil and plant changing after invasion: The case of Acacia dealbata in a Mediterranean ecosystem. Sci. Total Environ. 2014, 497, 491–498. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef]
- Rodríguez-Echeverría, S. Rhizobial hitchhikers from Down Under: Invasional meltdown in a plant-bacteria mutualism? J. Biogeogr. 2010, 37, 1611–1622. [Google Scholar] [CrossRef]
- Crisóstomo, J.A.; Rodríguez-Echeverría, S.; Freitas, H. Co-introduction of exotic rhizobia to the rhizosphere of the invasive legume Acacia saligna, an intercontinental study. Appl. Soil Ecol. 2013, 64, 118–126. [Google Scholar] [CrossRef]
- Le Roux, J.J.; Hui, C.; Keet, J.H.; Ellis, A.G. Co-introduction vs ecological fitting as pathways to the establishment of effective mutualisms during biological invasions. New Phytol. 2017, 215, 1354–1360. [Google Scholar] [CrossRef]
- Dingaan, M.; du Preez, P.J. Vachellia (Acacia) karroo Communities in South Africa: An Overview. In Pure and Applied Biogeography; IntechOpen: London, UK, 2017; pp. 109–141. [Google Scholar]
- Müller, F.L.; Raitt, L.M.; Chimphango, S.B.; Samuels, M.I.; Cupido, C.F.; Boatwright, J.S.; Knight, R.; Trytsman, M. Prioritisation of native legume species for further evaluation as potential forage crops in water-limited agricultural systems in South Africa. Environ. Monit. Assess. 2017, 189, 512. [Google Scholar] [CrossRef]
- Moiloa, N.; Chimphango, S.; Muasya, A. A phylogenetic study of the genus Wiborgia (Crotalarieae, Fabaceae). S. Afr. J. Bot. 2018, 115, 179–193. [Google Scholar] [CrossRef]
- Moiloa, N. Phylogenetic Relationships and the Effects of Edaphic Heterogeneity on the Distribution of Wiborgia (Fabaceae) in the Greater Cape Floristic Region. Ph.D. Thesis, University of Cape Town, Cape Town, South Africa, 2016. [Google Scholar]
- Esler, K.; Von Staden, L.; Midgley, G. Determinants of the Fynbos/Succulent Karoo biome boundary: Insights from a reciprocal transplant experiment. S. Afr. J. Bot. 2015, 101, 120–128. [Google Scholar] [CrossRef]
- Van Wyk, A.E.; Smith, G.F. Regions of Floristic Endemism in Southern Africa: A Review with Emphasis on Succulents; Umdaus Press: Pretoria, South Africa, 2001; pp. 1–160. [Google Scholar]
- Rebelo, A.G.; Boucher, C.; Helme, N.; Mucina, L.; Rutherford, M.C. Fynbos Biome. In The Vegetation of South Africa, Lesotho and Swaziland; Strelitzia: Pretoria, South Africa, 2006; Volume 19, pp. 53–219. [Google Scholar]
- Pavan-Kumar, A.; Gireesh-Babu, P.; Lakra, W. DNA metabarcoding: A new approach for rapid biodiversity assessment. J. Cell Sci. Mol. Biol. 2015, 2, 111. [Google Scholar]
- Flores, M.; Morales, L.; Avila, A.; González, V.; Bustos, P.; García, D.; Mora, Y.; Guo, X.; Collado-Vides, J.; Pinero, D. Diversification of DNA sequences in the symbiotic genome of Rhizobium etli. J. Bacteriol. 2005, 187, 7185–7192. [Google Scholar] [CrossRef]
- Pichler, M.; Coskun, Ö.K.; Ortega-Arbulú, A.S.; Conci, N.; Wörheide, G.; Vargas, S.; Orsi, W.D. A 16S rRNA gene sequencing and analysis protocol for the Illumina MiniSeq platform. Microbiol. Open 2018, 7, e00611. [Google Scholar] [CrossRef]
- Poly, F.; Ranjard, L.; Nazaret, S.; Gourbière, F.; Monrozier, L.J. Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl. Environ. Microbiol. 2001, 67, 2255–2262. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Welch, D.M.; Morrison, H.G.; Sogin, M.L. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 2010, 12, 1889–1898. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Gaby, J.C.; Buckley, D.H. A comprehensive aligned nifH gene database: A multipurpose tool for studies of nitrogen-fixing bacteria. Database 2014, 2014, bau001. [Google Scholar] [CrossRef] [PubMed]
- Gaby, J.C.; Rishishwar, L.; Valderrama-Aguirre, L.C.; Green, S.J.; Valderrama-Aguirre, A.; Jordan, I.K.; Kostka, J.E. Diazotroph community characterization via a high-throughput nifH amplicon sequencing and analysis pipeline. Appl. Environ. Microbiol. 2018, 84, e01512–e01517. [Google Scholar] [CrossRef]
- Swanson, K.S.; Dowd, S.E.; Suchodolski, J.S.; Middelbos, I.S.; Vester, B.M.; Barry, K.A.; Nelson, K.E.; Torralba, M.; Henrissat, B.; Coutinho, P.M. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 2011, 5, 639–649. [Google Scholar] [CrossRef]
- Dowd, S.E.; Callaway, T.R.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Hagevoort, R.G.; Edrington, T.S. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 2008, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Dowd, S.E.; Sun, Y.; Secor, P.R.; Rhoads, D.D.; Wolcott, B.M.; James, G.A.; Wolcott, R.D. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008, 8, 43. [Google Scholar] [CrossRef]
- Pinto, A.J.; Schroeder, J.; Lunn, M.; Sloan, W.; Raskin, L. Spatial-temporal survey and occupancy-abundance modeling to predict bacterial community dynamics in the drinking water microbiome. mBio 2014, 5, e01135-14. [Google Scholar] [CrossRef]
- Benito, X.; Fritz, S.C.; Steinitz-Kannan, M.; Vélez, M.I.; McGlue, M.M. Lake regionalization and diatom metacommunity structuring in tropical South America. Ecol. Evol. 2018, 8, 7865–7878. [Google Scholar] [CrossRef]
- Anderson, M.J.; Willis, T.J. Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology 2003, 84, 511–525. [Google Scholar] [CrossRef]
- Makhalanyane, T.P.; Valverde, A.; Gunnigle, E.; Frossard, A.; Ramond, J.-B.; Cowan, D.A. Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 2015, 39, 203–221. [Google Scholar] [CrossRef]
- Dojani, S.; Kauff, F.; Weber, B.; Büdel, B. Genotypic and phenotypic diversity of cyanobacteria in biological soil crusts of the Succulent Karoo and Nama Karoo of southern Africa. Microb. Ecol. 2014, 67, 286–301. [Google Scholar] [CrossRef]
- Op den Camp, H.J.; Islam, T.; Stott, M.B.; Harhangi, H.R.; Hynes, A.; Schouten, S.; Jetten, M.S.; Birkeland, N.K.; Pol, A.; Dunfield, P.F. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 2009, 1, 293–306. [Google Scholar] [CrossRef]
- Kovaleva, O.; Merkel, A.Y.; Novikov, A.; Baslerov, R.; Toshchakov, S.; Bonch-Osmolovskaya, E. Tepidisphaera mucosa gen. nov., sp. nov., a moderately thermophilic member of the class Phycisphaerae in the phylum Planctomycetes, and proposal of a new family, Tepidisphaeraceae fam. nov., and a new order, Tepidisphaerales ord. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 549–555. [Google Scholar] [CrossRef]
- Mhete, M.; Eze, P.N.; Rahube, T.O.; Akinyemi, F.O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Sci. Afr. 2020, 7, e00246. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Dutta, A.; Sarkar, J.; Panigrahi, M.K.; Sar, P. Low-abundance members of the Firmicutes facilitate bioremediation of soil impacted by highly acidic mine drainage from the Malanjkhand copper project, India. Front. Microbiol. 2018, 9, 2882. [Google Scholar] [CrossRef] [PubMed]
- Kamutando, C.N.; Vikram, S.; Kamgan-Nkuekam, G.; Makhalanyane, T.P.; Greve, M.; Le Roux, J.J.; Richardson, D.M.; Cowan, D.; Valverde, A. Soil nutritional status and biogeography influence rhizosphere microbial communities associated with the invasive tree Acacia dealbata. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Muema, E.K.; Cadisch, G.; Röhl, C.; Vanlauwe, B.; Rasche, F. Response of ammonia-oxidizing bacteria and archaea to biochemical quality of organic inputs combined with mineral nitrogen fertilizer in an arable soil. Appl. Soil Ecol. 2015, 95, 128–139. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed]
- Mitter, E.K.; Tosi, M.; Obregón, D.; Dunfield, K.E.; Germida, J.J. Rethinking crop nutrition in times of modern microbiology: Innovative biofertilizer technologies. Front. Sustain. Food Syst. 2021, 5, 29. [Google Scholar] [CrossRef]
- Song, J.; Min, L.; Wu, J.; He, Q.; Chen, F.; Wang, Y. Response of the microbial community to phosphate-solubilizing bacterial inoculants on Ulmus chenmoui Cheng in Eastern China. PLoS ONE 2021, 16, e0247309. [Google Scholar] [CrossRef]
- Zeng, Y.; Koblížek, M. Phototrophic Gemmatimonadetes: A New “Purple” Branch on the Bacterial Tree of Life. In Modern Topics in the Phototrophic Prokaryotes; Springer: Basel, Switzerland, 2017; pp. 163–192. [Google Scholar]
- Xia, Y.; Kong, Y.; Thomsen, T.R.; Nielsen, P.H. Identification and ecophysiological characterization of epiphytic protein-hydrolyzing Saprospiraceae (“Candidatus Epiflobacter” spp.) in activated sludge. Appl. Environ. Microbiol. 2008, 74, 2229–2238. [Google Scholar] [CrossRef] [PubMed]
- Felestrino, É.B.; Santiago, I.F.; Freitas, L.d.S.; Rosa, L.H.; Ribeiro, S.P.; Moreira, L.M. Plant growth promoting bacteria associated with langsdorffia hypogaea-rhizosphere-host biological interface: A neglected model of bacterial prospection. Front. Microbiol. 2017, 8, 172. [Google Scholar] [CrossRef] [PubMed]
- Reis, V.M.; Teixeira, K.R.d.S. Nitrogen fixing bacteria in the family Acetobacteraceae and their role in agriculture. J. Basic Microbiol. 2015, 55, 931–949. [Google Scholar] [CrossRef] [PubMed]
- Andrews, M.; Andrews, M.E. Specificity in legume-rhizobia symbioses. Int. J. Mol. Sci. 2017, 18, 705. [Google Scholar] [CrossRef]
- Hegyi, A.; Nguyen, T.B.K.; Posta, K. Metagenomic Analysis of Bacterial Communities in Agricultural Soils from Vietnam with Special Attention to Phosphate Solubilizing Bacteria. Microorganisms 2021, 9, 1796. [Google Scholar] [CrossRef] [PubMed]
- Jorquera, M.A.; Shaharoona, B.; Nadeem, S.M.; de la Luz Mora, M.; Crowley, D.E. Plant growth-promoting rhizobacteria associated with ancient clones of creosote bush (Larrea tridentata). Microb. Ecol. 2012, 64, 1008–1017. [Google Scholar] [CrossRef]
- Khadem, A.F.; Pol, A.; Jetten, M.S.; den Camp, H.J.O. Nitrogen fixation by the verrucomicrobial methanotroph ‘Methylacidiphilum fumariolicum’SolV. Microbiology 2010, 156, 1052–1059. [Google Scholar] [CrossRef]
- Kielak, A.M.; Cipriano, M.A.; Kuramae, E.E. Acidobacteria strains from subdivision 1 act as plant growth-promoting bacteria. Arch. Microbiol. 2016, 198, 987–993. [Google Scholar] [CrossRef]
- Glick, B.R.; Todorovic, B.; Czarny, J.; Cheng, Z.; Duan, J.; McConkey, B. Promotion of plant growth by bacterial ACC deaminase. Crit. Rev. Plant Sci. 2007, 26, 227–242. [Google Scholar] [CrossRef]
- Saleem, M.; Arshad, M.; Hussain, S.; Bhatti, A.S. Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. J. Ind. Microbiol. Biotechnol. 2007, 34, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Mellado, J.; Onofre-Lemus, J.; Estrada-De Los Santos, P.; Martínez-Aguilar, L. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 2007, 73, 5308–5319. [Google Scholar] [CrossRef] [PubMed]
- Green, P.N.; Ardley, J.K. Review of the genus Methylobacterium and closely related organisms: A proposal that some Methylobacterium species be reclassified into a new genus, Methylorubrum gen. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 2727–2748. [Google Scholar] [CrossRef] [PubMed]
- Sy, A.; Giraud, E.; Samba, R.; Gillis, M.; Dreyfus, B. Nodulation of certain legumes of the genus Crotalaria by the new species Methylobacterium. Can. J. Microbiol. 2001, 47, 503–508. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Selao, T. Regulation of Nitrogen Fixation in Rhodospirillum Rubrum: Through Proteomics and Beyond. Ph.D. Thesis, Stockholm University, Stovkholm, Sweden, 2010. [Google Scholar]
- Bao, Z.; Sasaki, K.; Okubo, T.; Ikeda, S.; Anda, M.; Hanzawa, E.; Kakizaki, K.; Sato, T.; Mitsui, H.; Minamisawa, K. Impact of Azospirillum sp. B510 inoculation on rice-associated bacterial communities in a paddy field. Microbes Environ. 2013, 28, 487–490. [Google Scholar] [CrossRef]
- Schmitz, R.A.; Klopprogge, K.; Grabbe, R. Regulation of nitrogen fixation in Klebsiella pneumoniae and Azotobacter vinelandii: NifL, transducing two environmental signals to the nif transcriptional activator NifA. J. Mol. Microbiol. Biotechnol. 2002, 4, 235–242. [Google Scholar]
- Jourand, P.; Giraud, E.; Bena, G.; Sy, A.; Willems, A.; Gillis, M.; Dreyfus, B.; de Lajudie, P. Methylobacterium nodulans sp. nov., for a group of aerobic, facultatively methylotrophic, legume root-nodule-forming and nitrogen-fixing bacteria. Int. J. Syst. Evol. Microbiol. 2004, 54, 2269–2273. [Google Scholar] [CrossRef]
- Liu, X.Y.; Wang, E.T.; Li, Y.; Chen, W.X. Diverse bacteria isolated from root nodules of Trifolium, Crotalaria and Mimosa grown in the subtropical regions of China. Arch. Microbiol. 2007, 188, 1–14. [Google Scholar] [CrossRef]
- Phalane, F.L.; Steenkamp, E.; Law, I.; Botha, W. The Diversity of Root Nodule Bacteria Associated with Lebeckia Species in South Africa. Master’s Thesis, University of Pretoria, Pretoria, South Africa, 2008. [Google Scholar]
- Madigan, M.; Cox, S.S.; Stegeman, R.A. Nitrogen fixation and nitrogenase activities in members of the family Rhodospirillaceae. J. Bacteriol. 1984, 157, 73–78. [Google Scholar] [CrossRef]
- Alef, K.; Kleiner, D. Regulatory aspects of inorganic nitrogen metabolism in the Rhodospirillaceae. Arch. Microbiol. 1982, 133, 239–241. [Google Scholar] [CrossRef]
- Bae, H.-S.; Rash, B.A.; Rainey, F.A.; Nobre, M.F.; Tiago, I.; da Costa, M.S.; Moe, W.M. Description of Azospira restricta sp. nov., a nitrogen-fixing bacterium isolated from groundwater. Int. J. Syst. Evol. Microbiol. 2007, 57, 1521–1526. [Google Scholar] [CrossRef] [PubMed]
- Hanson, B.T.; Yagi, J.M.; Jeon, C.O.; Madsen, E.M. Role of nitrogen fixation in the autecology of Polaromonas naphthalenivorans in contaminated sediments. Environ. Microbiol. 2012, 14, 1544–1557. [Google Scholar] [CrossRef] [PubMed]
- Schmalenberger, A.; Hodge, S.; Bryant, A.; Hawkesford, M.J.; Singh, B.K.; Kertesz, M.A. The role of Variovorax and other Comamonadaceae in sulfur transformations by microbial wheat rhizosphere communities exposed to different sulfur fertilization regimes. Environ. Microbiol. 2008, 10, 1486–1500. [Google Scholar] [CrossRef] [PubMed]
- Tsavkelova, E.A.; Cherdyntseva, T.A.; Klimova, S.Y.; Shestakov, A.I.; Botina, S.G.; Netrusov, A.I. Orchid-associated bacteria produce indole-3-acetic acid, promote seed germination, and increase their microbial yield in response to exogenous auxin. Arch. Microbiol. 2007, 188, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Coats, V.C.; Pelletreau, K.N.; Rumpho, M.E. Amplicon pyrosequencing reveals the soil microbial diversity associated with invasive J apanese barberry (B erberis thunbergii DC.). Mol. Ecol. 2014, 23, 1318–1332. [Google Scholar] [CrossRef]
- Berthrong, S.T.; Yeager, C.M.; Gallegos-Graves, L.; Steven, B.; Eichorst, S.A.; Jackson, R.B.; Kuske, C.R. Nitrogen fertilization has a stronger effect on soil nitrogen-fixing bacterial communities than elevated atmospheric CO2. Appl. Environ. Microbiol. 2014, 80, 3103–3112. [Google Scholar] [CrossRef]
- Pedersen, J.N.; Bombar, D.; Paerl, R.W.; Riemann, L. Diazotrophs and N2-fixation associated with particles in coastal estuarine waters. Front. Microbiol. 2018, 9, 2759. [Google Scholar] [CrossRef]
- Sarria-Guzmán, Y.; Chávez-Romero, Y.; Gómez-Acata, S.; Montes-Molina, J.A.; Morales-Salazar, E.; Dendooven, L.; Navarro-Noya, Y.E. Bacterial communities associated with different Anthurium andraeanum L. plant tissues. Microbes Environ. 2016, 31, 321–328. [Google Scholar] [CrossRef]
- Bertics, V.J.; Löscher, C.; Salonen, I.; Dale, A.W.; Gier, J.; Schmitz, R.; Treude, T. Occurrence of benthic microbial nitrogen fixation coupled to sulfate reduction in the seasonally hypoxic Eckernförde Bay, Baltic Sea. Biogeosciences 2013, 10, 1243–1258. [Google Scholar] [CrossRef]
- Tourova, T.P.; Spiridonova, E.M.; Berg, I.A.; Slobodova, N.V.; Boulygina, E.S.; Sorokin, D.Y. Phylogeny and evolution of the family Ectothiorhodospiraceae based on comparison of 16S rRNA, cbbL and nifH gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2387–2398. [Google Scholar] [CrossRef]
- Holmes, D.E.; Nevin, K.P.; Lovley, D.R. In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl. Environ. Microbiol. 2004, 70, 7251–7259. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Q.; Ling, J.; Van Nostrand, J.D.; Shi, Z.; Zhou, J.; Dong, J. Diversity and structure of diazotrophic communities in mangrove rhizosphere, revealed by high-throughput sequencing. Front. Microbiol. 2017, 8, 2032. [Google Scholar] [CrossRef]
- Xiang, Q.; Luo, L.; Liang, Y.; Chen, Q.; Zhang, X.; Gu, Y. The diversity, growth promoting abilities and anti-microbial activities of bacteria isolated from the fruiting body of Agaricus bisporus. Pol. J. Microbiol. 2017, 66, 201–207. [Google Scholar] [CrossRef]
- Li, H.-B.; Singh, R.K.; Singh, P.; Song, Q.-Q.; Xing, Y.-X.; Yang, L.-T.; Li, Y.-R. Genetic diversity of nitrogen-fixing and plant growth promoting Pseudomonas species isolated from sugarcane rhizosphere. Front. Microbiol. 2017, 8, 1268. [Google Scholar] [CrossRef]
- Ahemad, M.; Khan, M.S. Effect of fungicides on plant growth promoting activities of phosphate solubilizing Pseudomonas putida isolated from mustard (Brassica compestris) rhizosphere. Chemosphere 2012, 86, 945–950. [Google Scholar] [CrossRef]
- Ahemad, M.; Khan, M.S. Evaluation of plant-growth-promoting activities of rhizobacterium Pseudomonas putida under herbicide stress. Ann. Microbiol. 2012, 62, 1531–1540. [Google Scholar] [CrossRef]
- Gardner, T.; Acosta-Martinez, V.; Senwo, Z.; Dowd, S.E. Soil rhizosphere microbial communities and enzyme activities under organic farming in Alabama. Diversity 2011, 3, 308–328. [Google Scholar] [CrossRef]
- Räsänen, L.A.; Sprent, J.I.; Lindström, K. Symbiotic properties of sinorhizobia isolated from Acacia and Prosopis nodules in Sudan and Senegal. Plant Soil 2001, 235, 193–210. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, H.; Fu, S.; Yao, Q. Variation in soil microbial community structure associated with different legume species is greater than that associated with different grass species. Front. Microbiol. 2017, 8, 1007. [Google Scholar] [CrossRef] [PubMed]
- Kambai, C.; Joshua, V.; Olatidoye, O.; Yakubu, C.; Adaaja, B.; Olaniyi, J. Comparative Study of Soil Bacteria from the Rhizosphere of Two Selected Tree Species (Anogeissus leiocarpa and Pterocarpus erinaceus) in Shere Hills, Plateau State, Nigeria. J. Appl. Sci. Environ. Manag. 2021, 25, 1147–1153. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).