Rhizosphere Diazotrophs and Other Bacteria Associated with Native and Encroaching Legumes in the Succulent Karoo Biome in South Africa
Abstract
:1. Introduction
2. Materials and Methods
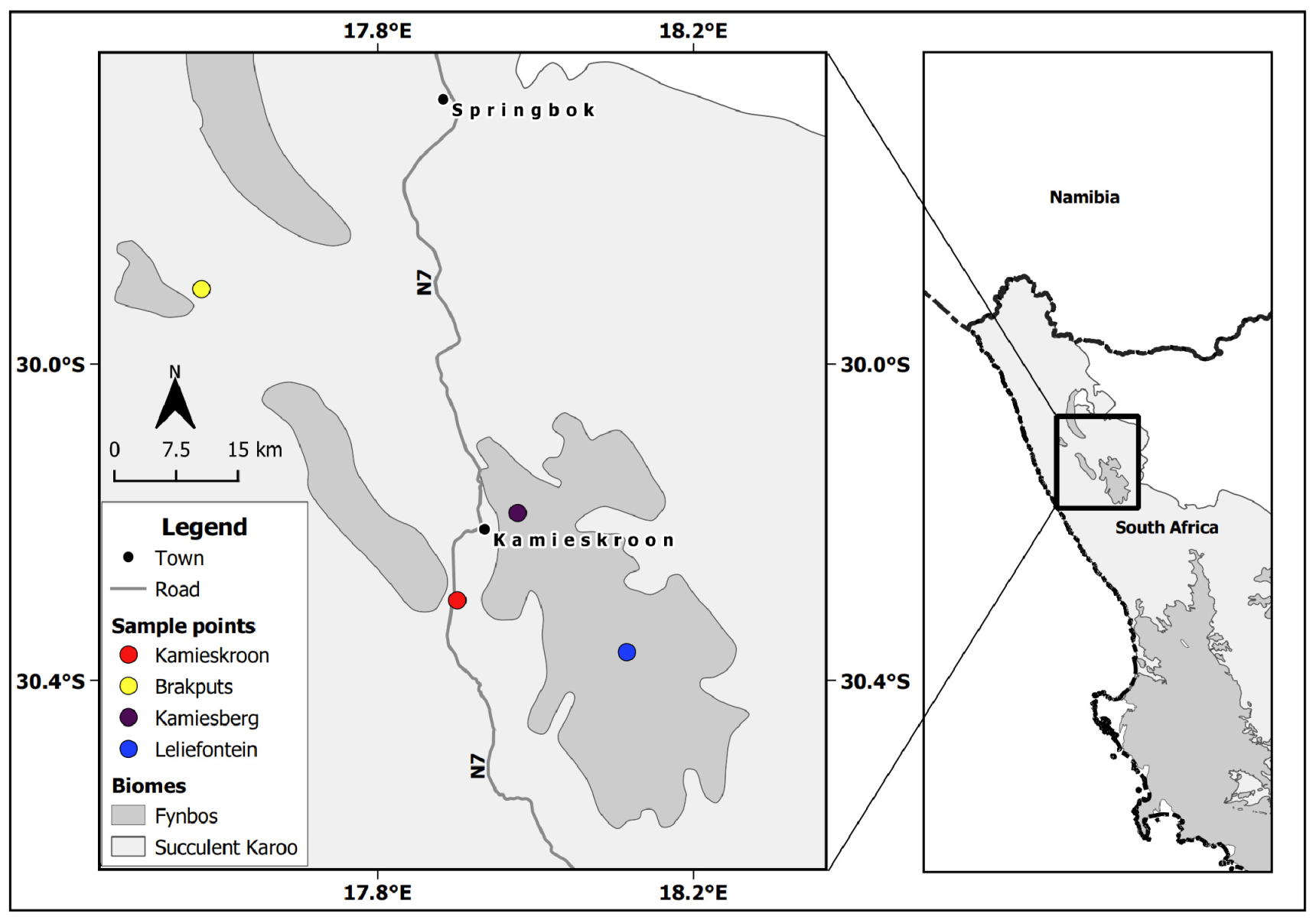
2.1. Study Sites
2.2. Soil Sampling and Physico-Chemical Properties
2.3. Soil Metabarcoding Analysis
2.4. Statistical Analyses of Metabarcoding Data and Soil Physico-Chemical Properties
3. Results
3.1. Physico-Chemical Properties of the Studied Rhizosphere Soils from the Succulent Karoo Biome
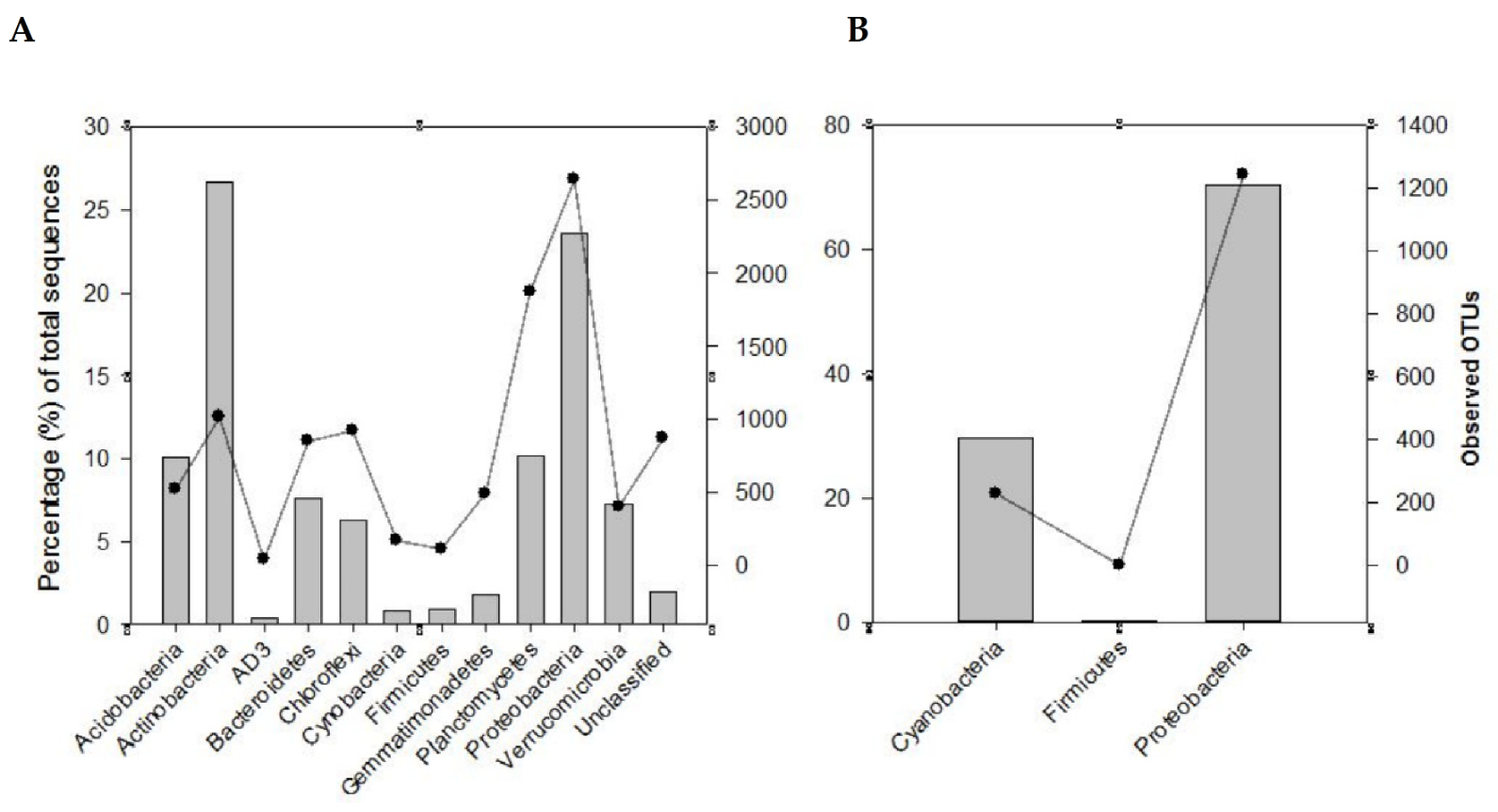
3.2. The Dominant Bacterial Communities in the Rhizosphere Soils Considered
3.3. OTU-Based Diversity of Diazotrophs as Influenced by Legume Species
3.4. Alpha Diversity
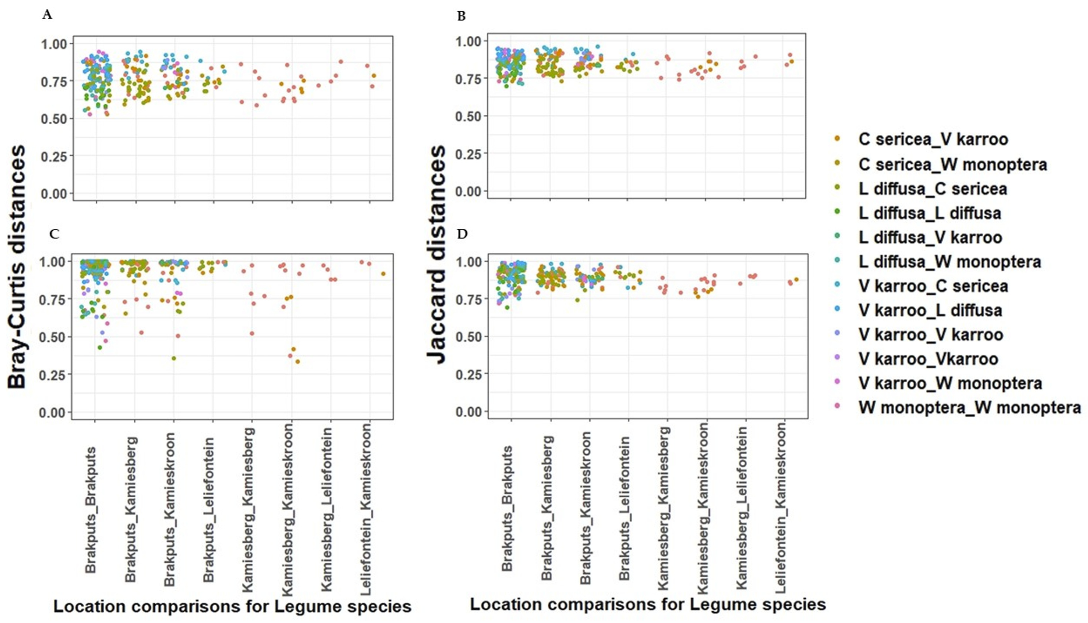
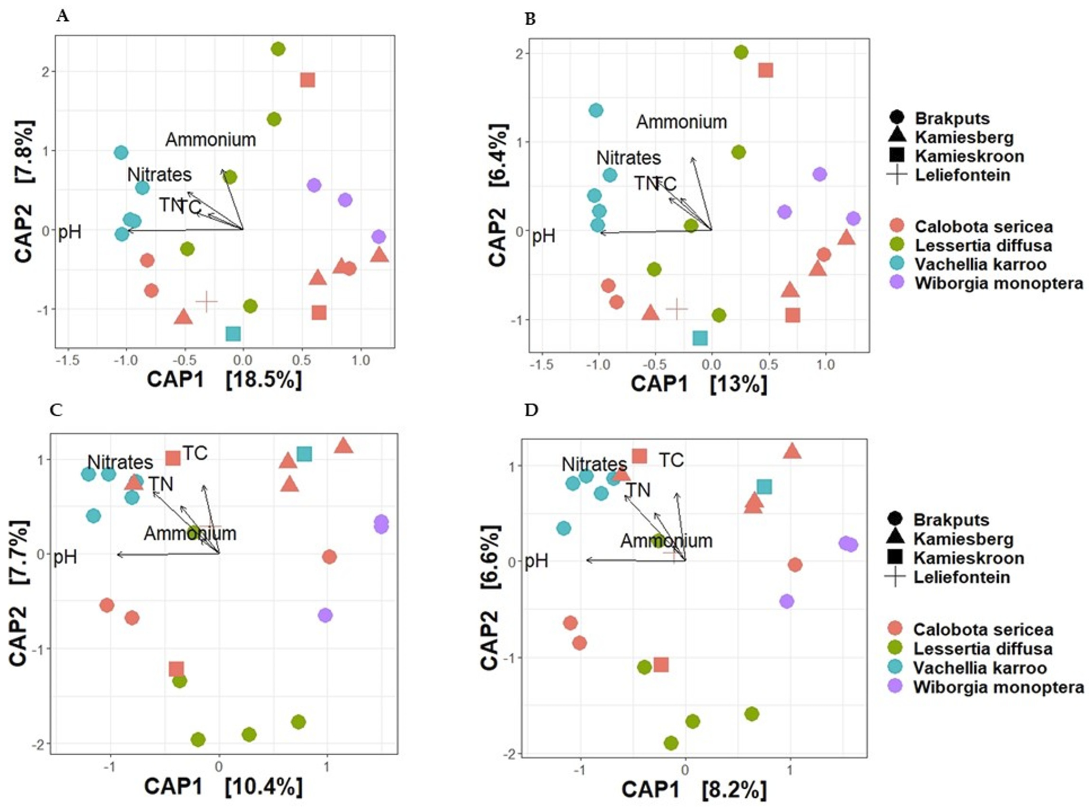
3.5. Beta-Diversity
4. Discussion
4.1. Bacteria Composition in Rhizosphere Soils Used Is Associated with Environmental Conditions
4.2. Legume Species Influence Diazotrophs in the Succulent Karoo Biome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Cowling, R.; Rundel, P.; Desmet, P.; Esler, K. Extraordinary high regional-scale plant diversity in southern African arid lands: Subcontinental and global comparisons. Divers. Distrib. 1998, 4, 27–36. [Google Scholar]
- Mucina, L.; Jürgens, N.; Le Roux, A.; Rutherford, M.C.; Schmiedel, U.; Esler, K.J.; Powrie, L.W.; Desmet, P.G.; Milton, S.J.; Boucher, C. Succulent Karoo Biome. In The Vegetation of South Africa, Lesotho and Swaziland; Strelitzia: Pretoria, South Africa, 2006; Volume 19, pp. 221–299. [Google Scholar]
- Carrick, P.J. Shrub Community Dynamics in a South African Semi-Desert. Ph.D. Thesis, University of Cambridge, Cambridge, UK, 2001. [Google Scholar]
- Hoffman, M.T.; Carrick, P.; Gillson, L.; West, A. Drought, climate change and vegetation response in the succulent karoo, South Africa. S. Afr. J. Sci. 2009, 105, 54–60. [Google Scholar] [CrossRef]
- Mucina, L.; Rutherford, M.C. Biomes and Bioregions of Southern Africa. In The Vegetation of South Africa, Lesotho and Swaziland; Strelitzia: Pretoria, South Africa, 2006; Volume 19, pp. 31–51. [Google Scholar]
- Milton, S.; Dean, W.; Marincowitz, C.; Kerley, G. Effects of the 1990/91 drought on rangeland in the Steytlerville Karoo. S. Afr. J. Sci. 1995, 91, 78–84. [Google Scholar]
- Valverde, A.; De Maayer, P.; Oberholster, T.; Henschel, J.; Louw, M.K.; Cowan, D. Specific microbial communities associate with the rhizosphere of Welwitschia mirabilis, a living fossil. PLoS ONE 2016, 11, e0153353. [Google Scholar]
- Pieterse, Z.; Aveling, T.A.; Jacobs, A.; Cowan, D.A. Diversity and seasonality of fungal communities in soil from the Succulent Karoo biodiversity hotspot, South Africa. J. Arid. Environ. 2020, 172, 104020. [Google Scholar] [CrossRef]
- Lewis, G.P.; Schrire, B. Tribe Indigofereae. In Legumes of the World; Royal Botanic Gardens Kew: Richmond, UK, 2005; pp. 361–365. [Google Scholar]
- Schrire, B. Tribe Millettieae. Legumes of the World; Royal Botanic Gardens Kew: Richmond, UK, 2005; pp. 367–387. [Google Scholar]
- Sprent, J.I.; Ardley, J.; James, E.K. Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol. 2017, 215, 40–56. [Google Scholar] [CrossRef] [Green Version]
- Azani, N.; Babineau, M.; Bailey, C.D.; Banks, H.; Barbosa, A.R.; Pinto, R.B.; Boatwright, J.S.; Borges, L.M.; Brown, G.K.; Bruneau, A. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny The Legume Phylogeny Working Group (LPWG). Taxon 2017, 66, 44–77. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Hidalgo, P.; Hirsch, A.M. The nodule microbiome: N2-fixing rhizobia do not live alone. Phytobiomes 2017, 1, 70–82. [Google Scholar] [CrossRef] [Green Version]
- Sankhla, I.S.; Tak, N.; Meghwal, R.R.; Choudhary, S.; Tak, A.; Rathi, S.; Sprent, J.I.; James, E.K.; Gehlot, H.S. Molecular characterization of nitrogen fixing microsymbionts from root nodules of Vachellia (Acacia) jacquemontii, a native legume from the Thar Desert of India. Plant Soil 2017, 410, 21–40. [Google Scholar] [CrossRef]
- Lemaire, B.; Dlodlo, O.; Chimphango, S.; Stirton, C.; Schrire, B.; Boatwright, S.; Honnay, O.; Smets, E.; Sprent, J.; James, E.; et al. Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the Core Cape Subregion (South Africa). FEMS Microbiol. Ecol. 2015, 91, 2–17. [Google Scholar] [CrossRef] [Green Version]
- Lucas, L. Post-Fire Response of Botanical and Microbial Communities in the Succulent Karoo; University of the Western Care: Bellville, South Africa, 2018. [Google Scholar]
- Beukes, C.W.; Boshoff, F.S.; Phalane, F.L.; Hassen, A.I.; le Roux, M.M.; Stepkowski, T.; Venter, S.N.; Steenkamp, E.T. Both alpha-and beta-rhizobia occupy the root nodules of Vachellia karroo in South Africa. Front. Microbiol. 2019, 10, 1195. [Google Scholar] [CrossRef]
- Gerding, M.; O’Hara, G.W.; Bräu, L.; Nandasena, K.; Howieson, J.G. Diverse Mesorhizobium spp. with unique nodA nodulating the South African legume species of the genus Lessertia. Plant Soil 2012, 358, 385–401. [Google Scholar] [CrossRef]
- Kamutando, C.N.; Vikram, S.; Kamgan-Nkuekam, G.; Makhalanyane, T.P.; Greve, M.; Le Roux, J.J.; Richardson, D.M.; Cowan, D.A.; Valverde, A. The functional potential of the rhizospheric microbiome of an invasive tree species, Acacia dealbata. Microb. Ecol. 2019, 77, 191–200. [Google Scholar] [CrossRef] [Green Version]
- Lazzaro, L.; Giuliani, C.; Fabiani, A.; Agnelli, A.E.; Pastorelli, R.; Lagomarsino, A.; Benesperi, R.; Calamassi, R.; Foggi, B. Soil and plant changing after invasion: The case of Acacia dealbata in a Mediterranean ecosystem. Sci. Total Environ. 2014, 497, 491–498. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Echeverría, S. Rhizobial hitchhikers from Down Under: Invasional meltdown in a plant-bacteria mutualism? J. Biogeogr. 2010, 37, 1611–1622. [Google Scholar] [CrossRef]
- Crisóstomo, J.A.; Rodríguez-Echeverría, S.; Freitas, H. Co-introduction of exotic rhizobia to the rhizosphere of the invasive legume Acacia saligna, an intercontinental study. Appl. Soil Ecol. 2013, 64, 118–126. [Google Scholar] [CrossRef]
- Le Roux, J.J.; Hui, C.; Keet, J.H.; Ellis, A.G. Co-introduction vs ecological fitting as pathways to the establishment of effective mutualisms during biological invasions. New Phytol. 2017, 215, 1354–1360. [Google Scholar] [CrossRef] [Green Version]
- Dingaan, M.; du Preez, P.J. Vachellia (Acacia) karroo Communities in South Africa: An Overview. In Pure and Applied Biogeography; IntechOpen: London, UK, 2017; pp. 109–141. [Google Scholar]
- Müller, F.L.; Raitt, L.M.; Chimphango, S.B.; Samuels, M.I.; Cupido, C.F.; Boatwright, J.S.; Knight, R.; Trytsman, M. Prioritisation of native legume species for further evaluation as potential forage crops in water-limited agricultural systems in South Africa. Environ. Monit. Assess. 2017, 189, 512. [Google Scholar] [CrossRef]
- Moiloa, N.; Chimphango, S.; Muasya, A. A phylogenetic study of the genus Wiborgia (Crotalarieae, Fabaceae). S. Afr. J. Bot. 2018, 115, 179–193. [Google Scholar] [CrossRef]
- Moiloa, N. Phylogenetic Relationships and the Effects of Edaphic Heterogeneity on the Distribution of Wiborgia (Fabaceae) in the Greater Cape Floristic Region. Ph.D. Thesis, University of Cape Town, Cape Town, South Africa, 2016. [Google Scholar]
- Esler, K.; Von Staden, L.; Midgley, G. Determinants of the Fynbos/Succulent Karoo biome boundary: Insights from a reciprocal transplant experiment. S. Afr. J. Bot. 2015, 101, 120–128. [Google Scholar] [CrossRef]
- Van Wyk, A.E.; Smith, G.F. Regions of Floristic Endemism in Southern Africa: A Review with Emphasis on Succulents; Umdaus Press: Pretoria, South Africa, 2001; pp. 1–160. [Google Scholar]
- Rebelo, A.G.; Boucher, C.; Helme, N.; Mucina, L.; Rutherford, M.C. Fynbos Biome. In The Vegetation of South Africa, Lesotho and Swaziland; Strelitzia: Pretoria, South Africa, 2006; Volume 19, pp. 53–219. [Google Scholar]
- Pavan-Kumar, A.; Gireesh-Babu, P.; Lakra, W. DNA metabarcoding: A new approach for rapid biodiversity assessment. J. Cell Sci. Mol. Biol. 2015, 2, 111. [Google Scholar]
- Flores, M.; Morales, L.; Avila, A.; González, V.; Bustos, P.; García, D.; Mora, Y.; Guo, X.; Collado-Vides, J.; Pinero, D. Diversification of DNA sequences in the symbiotic genome of Rhizobium etli. J. Bacteriol. 2005, 187, 7185–7192. [Google Scholar] [CrossRef] [Green Version]
- Pichler, M.; Coskun, Ö.K.; Ortega-Arbulú, A.S.; Conci, N.; Wörheide, G.; Vargas, S.; Orsi, W.D. A 16S rRNA gene sequencing and analysis protocol for the Illumina MiniSeq platform. Microbiol. Open 2018, 7, e00611. [Google Scholar] [CrossRef]
- Poly, F.; Ranjard, L.; Nazaret, S.; Gourbière, F.; Monrozier, L.J. Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl. Environ. Microbiol. 2001, 67, 2255–2262. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Welch, D.M.; Morrison, H.G.; Sogin, M.L. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 2010, 12, 1889–1898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaby, J.C.; Buckley, D.H. A comprehensive aligned nifH gene database: A multipurpose tool for studies of nitrogen-fixing bacteria. Database 2014, 2014, bau001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaby, J.C.; Rishishwar, L.; Valderrama-Aguirre, L.C.; Green, S.J.; Valderrama-Aguirre, A.; Jordan, I.K.; Kostka, J.E. Diazotroph community characterization via a high-throughput nifH amplicon sequencing and analysis pipeline. Appl. Environ. Microbiol. 2018, 84, e01512–e01517. [Google Scholar] [CrossRef] [Green Version]
- Swanson, K.S.; Dowd, S.E.; Suchodolski, J.S.; Middelbos, I.S.; Vester, B.M.; Barry, K.A.; Nelson, K.E.; Torralba, M.; Henrissat, B.; Coutinho, P.M. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 2011, 5, 639–649. [Google Scholar] [CrossRef]
- Dowd, S.E.; Callaway, T.R.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Hagevoort, R.G.; Edrington, T.S. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 2008, 8, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowd, S.E.; Sun, Y.; Secor, P.R.; Rhoads, D.D.; Wolcott, B.M.; James, G.A.; Wolcott, R.D. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008, 8, 43. [Google Scholar] [CrossRef] [Green Version]
- Pinto, A.J.; Schroeder, J.; Lunn, M.; Sloan, W.; Raskin, L. Spatial-temporal survey and occupancy-abundance modeling to predict bacterial community dynamics in the drinking water microbiome. mBio 2014, 5, e01135-14. [Google Scholar] [CrossRef] [Green Version]
- Benito, X.; Fritz, S.C.; Steinitz-Kannan, M.; Vélez, M.I.; McGlue, M.M. Lake regionalization and diatom metacommunity structuring in tropical South America. Ecol. Evol. 2018, 8, 7865–7878. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.J.; Willis, T.J. Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology 2003, 84, 511–525. [Google Scholar] [CrossRef]
- Makhalanyane, T.P.; Valverde, A.; Gunnigle, E.; Frossard, A.; Ramond, J.-B.; Cowan, D.A. Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 2015, 39, 203–221. [Google Scholar] [CrossRef]
- Dojani, S.; Kauff, F.; Weber, B.; Büdel, B. Genotypic and phenotypic diversity of cyanobacteria in biological soil crusts of the Succulent Karoo and Nama Karoo of southern Africa. Microb. Ecol. 2014, 67, 286–301. [Google Scholar] [CrossRef]
- Op den Camp, H.J.; Islam, T.; Stott, M.B.; Harhangi, H.R.; Hynes, A.; Schouten, S.; Jetten, M.S.; Birkeland, N.K.; Pol, A.; Dunfield, P.F. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 2009, 1, 293–306. [Google Scholar] [CrossRef]
- Kovaleva, O.; Merkel, A.Y.; Novikov, A.; Baslerov, R.; Toshchakov, S.; Bonch-Osmolovskaya, E. Tepidisphaera mucosa gen. nov., sp. nov., a moderately thermophilic member of the class Phycisphaerae in the phylum Planctomycetes, and proposal of a new family, Tepidisphaeraceae fam. nov., and a new order, Tepidisphaerales ord. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 549–555. [Google Scholar] [CrossRef]
- Mhete, M.; Eze, P.N.; Rahube, T.O.; Akinyemi, F.O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Sci. Afr. 2020, 7, e00246. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Dutta, A.; Sarkar, J.; Panigrahi, M.K.; Sar, P. Low-abundance members of the Firmicutes facilitate bioremediation of soil impacted by highly acidic mine drainage from the Malanjkhand copper project, India. Front. Microbiol. 2018, 9, 2882. [Google Scholar] [CrossRef] [PubMed]
- Kamutando, C.N.; Vikram, S.; Kamgan-Nkuekam, G.; Makhalanyane, T.P.; Greve, M.; Le Roux, J.J.; Richardson, D.M.; Cowan, D.; Valverde, A. Soil nutritional status and biogeography influence rhizosphere microbial communities associated with the invasive tree Acacia dealbata. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muema, E.K.; Cadisch, G.; Röhl, C.; Vanlauwe, B.; Rasche, F. Response of ammonia-oxidizing bacteria and archaea to biochemical quality of organic inputs combined with mineral nitrogen fertilizer in an arable soil. Appl. Soil Ecol. 2015, 95, 128–139. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitter, E.K.; Tosi, M.; Obregón, D.; Dunfield, K.E.; Germida, J.J. Rethinking crop nutrition in times of modern microbiology: Innovative biofertilizer technologies. Front. Sustain. Food Syst. 2021, 5, 29. [Google Scholar] [CrossRef]
- Song, J.; Min, L.; Wu, J.; He, Q.; Chen, F.; Wang, Y. Response of the microbial community to phosphate-solubilizing bacterial inoculants on Ulmus chenmoui Cheng in Eastern China. PLoS ONE 2021, 16, e0247309. [Google Scholar] [CrossRef]
- Zeng, Y.; Koblížek, M. Phototrophic Gemmatimonadetes: A New “Purple” Branch on the Bacterial Tree of Life. In Modern Topics in the Phototrophic Prokaryotes; Springer: Basel, Switzerland, 2017; pp. 163–192. [Google Scholar]
- Xia, Y.; Kong, Y.; Thomsen, T.R.; Nielsen, P.H. Identification and ecophysiological characterization of epiphytic protein-hydrolyzing Saprospiraceae (“Candidatus Epiflobacter” spp.) in activated sludge. Appl. Environ. Microbiol. 2008, 74, 2229–2238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felestrino, É.B.; Santiago, I.F.; Freitas, L.d.S.; Rosa, L.H.; Ribeiro, S.P.; Moreira, L.M. Plant growth promoting bacteria associated with langsdorffia hypogaea-rhizosphere-host biological interface: A neglected model of bacterial prospection. Front. Microbiol. 2017, 8, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, V.M.; Teixeira, K.R.d.S. Nitrogen fixing bacteria in the family Acetobacteraceae and their role in agriculture. J. Basic Microbiol. 2015, 55, 931–949. [Google Scholar] [CrossRef] [PubMed]
- Andrews, M.; Andrews, M.E. Specificity in legume-rhizobia symbioses. Int. J. Mol. Sci. 2017, 18, 705. [Google Scholar] [CrossRef] [Green Version]
- Hegyi, A.; Nguyen, T.B.K.; Posta, K. Metagenomic Analysis of Bacterial Communities in Agricultural Soils from Vietnam with Special Attention to Phosphate Solubilizing Bacteria. Microorganisms 2021, 9, 1796. [Google Scholar] [CrossRef] [PubMed]
- Jorquera, M.A.; Shaharoona, B.; Nadeem, S.M.; de la Luz Mora, M.; Crowley, D.E. Plant growth-promoting rhizobacteria associated with ancient clones of creosote bush (Larrea tridentata). Microb. Ecol. 2012, 64, 1008–1017. [Google Scholar] [CrossRef]
- Khadem, A.F.; Pol, A.; Jetten, M.S.; den Camp, H.J.O. Nitrogen fixation by the verrucomicrobial methanotroph ‘Methylacidiphilum fumariolicum’SolV. Microbiology 2010, 156, 1052–1059. [Google Scholar] [CrossRef] [Green Version]
- Kielak, A.M.; Cipriano, M.A.; Kuramae, E.E. Acidobacteria strains from subdivision 1 act as plant growth-promoting bacteria. Arch. Microbiol. 2016, 198, 987–993. [Google Scholar] [CrossRef] [Green Version]
- Glick, B.R.; Todorovic, B.; Czarny, J.; Cheng, Z.; Duan, J.; McConkey, B. Promotion of plant growth by bacterial ACC deaminase. Crit. Rev. Plant Sci. 2007, 26, 227–242. [Google Scholar] [CrossRef]
- Saleem, M.; Arshad, M.; Hussain, S.; Bhatti, A.S. Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. J. Ind. Microbiol. Biotechnol. 2007, 34, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Mellado, J.; Onofre-Lemus, J.; Estrada-De Los Santos, P.; Martínez-Aguilar, L. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 2007, 73, 5308–5319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, P.N.; Ardley, J.K. Review of the genus Methylobacterium and closely related organisms: A proposal that some Methylobacterium species be reclassified into a new genus, Methylorubrum gen. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 2727–2748. [Google Scholar] [CrossRef] [PubMed]
- Sy, A.; Giraud, E.; Samba, R.; Gillis, M.; Dreyfus, B. Nodulation of certain legumes of the genus Crotalaria by the new species Methylobacterium. Can. J. Microbiol. 2001, 47, 503–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selao, T. Regulation of Nitrogen Fixation in Rhodospirillum Rubrum: Through Proteomics and Beyond. Ph.D. Thesis, Stockholm University, Stovkholm, Sweden, 2010. [Google Scholar]
- Bao, Z.; Sasaki, K.; Okubo, T.; Ikeda, S.; Anda, M.; Hanzawa, E.; Kakizaki, K.; Sato, T.; Mitsui, H.; Minamisawa, K. Impact of Azospirillum sp. B510 inoculation on rice-associated bacterial communities in a paddy field. Microbes Environ. 2013, 28, 487–490. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, R.A.; Klopprogge, K.; Grabbe, R. Regulation of nitrogen fixation in Klebsiella pneumoniae and Azotobacter vinelandii: NifL, transducing two environmental signals to the nif transcriptional activator NifA. J. Mol. Microbiol. Biotechnol. 2002, 4, 235–242. [Google Scholar]
- Jourand, P.; Giraud, E.; Bena, G.; Sy, A.; Willems, A.; Gillis, M.; Dreyfus, B.; de Lajudie, P. Methylobacterium nodulans sp. nov., for a group of aerobic, facultatively methylotrophic, legume root-nodule-forming and nitrogen-fixing bacteria. Int. J. Syst. Evol. Microbiol. 2004, 54, 2269–2273. [Google Scholar] [CrossRef]
- Liu, X.Y.; Wang, E.T.; Li, Y.; Chen, W.X. Diverse bacteria isolated from root nodules of Trifolium, Crotalaria and Mimosa grown in the subtropical regions of China. Arch. Microbiol. 2007, 188, 1–14. [Google Scholar] [CrossRef]
- Phalane, F.L.; Steenkamp, E.; Law, I.; Botha, W. The Diversity of Root Nodule Bacteria Associated with Lebeckia Species in South Africa. Master’s Thesis, University of Pretoria, Pretoria, South Africa, 2008. [Google Scholar]
- Madigan, M.; Cox, S.S.; Stegeman, R.A. Nitrogen fixation and nitrogenase activities in members of the family Rhodospirillaceae. J. Bacteriol. 1984, 157, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Alef, K.; Kleiner, D. Regulatory aspects of inorganic nitrogen metabolism in the Rhodospirillaceae. Arch. Microbiol. 1982, 133, 239–241. [Google Scholar] [CrossRef]
- Bae, H.-S.; Rash, B.A.; Rainey, F.A.; Nobre, M.F.; Tiago, I.; da Costa, M.S.; Moe, W.M. Description of Azospira restricta sp. nov., a nitrogen-fixing bacterium isolated from groundwater. Int. J. Syst. Evol. Microbiol. 2007, 57, 1521–1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, B.T.; Yagi, J.M.; Jeon, C.O.; Madsen, E.M. Role of nitrogen fixation in the autecology of Polaromonas naphthalenivorans in contaminated sediments. Environ. Microbiol. 2012, 14, 1544–1557. [Google Scholar] [CrossRef] [PubMed]
- Schmalenberger, A.; Hodge, S.; Bryant, A.; Hawkesford, M.J.; Singh, B.K.; Kertesz, M.A. The role of Variovorax and other Comamonadaceae in sulfur transformations by microbial wheat rhizosphere communities exposed to different sulfur fertilization regimes. Environ. Microbiol. 2008, 10, 1486–1500. [Google Scholar] [CrossRef] [PubMed]
- Tsavkelova, E.A.; Cherdyntseva, T.A.; Klimova, S.Y.; Shestakov, A.I.; Botina, S.G.; Netrusov, A.I. Orchid-associated bacteria produce indole-3-acetic acid, promote seed germination, and increase their microbial yield in response to exogenous auxin. Arch. Microbiol. 2007, 188, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Coats, V.C.; Pelletreau, K.N.; Rumpho, M.E. Amplicon pyrosequencing reveals the soil microbial diversity associated with invasive J apanese barberry (B erberis thunbergii DC.). Mol. Ecol. 2014, 23, 1318–1332. [Google Scholar] [CrossRef]
- Berthrong, S.T.; Yeager, C.M.; Gallegos-Graves, L.; Steven, B.; Eichorst, S.A.; Jackson, R.B.; Kuske, C.R. Nitrogen fertilization has a stronger effect on soil nitrogen-fixing bacterial communities than elevated atmospheric CO2. Appl. Environ. Microbiol. 2014, 80, 3103–3112. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, J.N.; Bombar, D.; Paerl, R.W.; Riemann, L. Diazotrophs and N2-fixation associated with particles in coastal estuarine waters. Front. Microbiol. 2018, 9, 2759. [Google Scholar] [CrossRef]
- Sarria-Guzmán, Y.; Chávez-Romero, Y.; Gómez-Acata, S.; Montes-Molina, J.A.; Morales-Salazar, E.; Dendooven, L.; Navarro-Noya, Y.E. Bacterial communities associated with different Anthurium andraeanum L. plant tissues. Microbes Environ. 2016, 31, 321–328. [Google Scholar] [CrossRef] [Green Version]
- Bertics, V.J.; Löscher, C.; Salonen, I.; Dale, A.W.; Gier, J.; Schmitz, R.; Treude, T. Occurrence of benthic microbial nitrogen fixation coupled to sulfate reduction in the seasonally hypoxic Eckernförde Bay, Baltic Sea. Biogeosciences 2013, 10, 1243–1258. [Google Scholar] [CrossRef] [Green Version]
- Tourova, T.P.; Spiridonova, E.M.; Berg, I.A.; Slobodova, N.V.; Boulygina, E.S.; Sorokin, D.Y. Phylogeny and evolution of the family Ectothiorhodospiraceae based on comparison of 16S rRNA, cbbL and nifH gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2387–2398. [Google Scholar] [CrossRef]
- Holmes, D.E.; Nevin, K.P.; Lovley, D.R. In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl. Environ. Microbiol. 2004, 70, 7251–7259. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yang, Q.; Ling, J.; Van Nostrand, J.D.; Shi, Z.; Zhou, J.; Dong, J. Diversity and structure of diazotrophic communities in mangrove rhizosphere, revealed by high-throughput sequencing. Front. Microbiol. 2017, 8, 2032. [Google Scholar] [CrossRef]
- Xiang, Q.; Luo, L.; Liang, Y.; Chen, Q.; Zhang, X.; Gu, Y. The diversity, growth promoting abilities and anti-microbial activities of bacteria isolated from the fruiting body of Agaricus bisporus. Pol. J. Microbiol. 2017, 66, 201–207. [Google Scholar] [CrossRef] [Green Version]
- Li, H.-B.; Singh, R.K.; Singh, P.; Song, Q.-Q.; Xing, Y.-X.; Yang, L.-T.; Li, Y.-R. Genetic diversity of nitrogen-fixing and plant growth promoting Pseudomonas species isolated from sugarcane rhizosphere. Front. Microbiol. 2017, 8, 1268. [Google Scholar] [CrossRef]
- Ahemad, M.; Khan, M.S. Effect of fungicides on plant growth promoting activities of phosphate solubilizing Pseudomonas putida isolated from mustard (Brassica compestris) rhizosphere. Chemosphere 2012, 86, 945–950. [Google Scholar] [CrossRef]
- Ahemad, M.; Khan, M.S. Evaluation of plant-growth-promoting activities of rhizobacterium Pseudomonas putida under herbicide stress. Ann. Microbiol. 2012, 62, 1531–1540. [Google Scholar] [CrossRef]
- Gardner, T.; Acosta-Martinez, V.; Senwo, Z.; Dowd, S.E. Soil rhizosphere microbial communities and enzyme activities under organic farming in Alabama. Diversity 2011, 3, 308–328. [Google Scholar] [CrossRef]
- Räsänen, L.A.; Sprent, J.I.; Lindström, K. Symbiotic properties of sinorhizobia isolated from Acacia and Prosopis nodules in Sudan and Senegal. Plant Soil 2001, 235, 193–210. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, H.; Fu, S.; Yao, Q. Variation in soil microbial community structure associated with different legume species is greater than that associated with different grass species. Front. Microbiol. 2017, 8, 1007. [Google Scholar] [CrossRef] [PubMed]
- Kambai, C.; Joshua, V.; Olatidoye, O.; Yakubu, C.; Adaaja, B.; Olaniyi, J. Comparative Study of Soil Bacteria from the Rhizosphere of Two Selected Tree Species (Anogeissus leiocarpa and Pterocarpus erinaceus) in Shere Hills, Plateau State, Nigeria. J. Appl. Sci. Environ. Manag. 2021, 25, 1147–1153. [Google Scholar] [CrossRef]

| Alpha Diversity Indices 1 | |||
|---|---|---|---|
| Legume Species | Richness (Observed Taxa) | Shannon | Shannoneven |
| 16S rRNA | |||
| C. sericea | 546 ± 25.8 a | 5.69 ± 0.07 a | 0.905 ± 0.005 a |
| L. diffusa | 496 ± 34.7 a | 5.63 ± 0.09 a | 0.908 ± 0.007 ab |
| V. karroo | 687 ± 31.6 b | 6.09 ± 0.08 b | 0.932 ± 0.006 b |
| W. monoptera | 482 ± 44.8 a | 5.45 ± 0.11 a | 0.883 ± 008 a |
| nifH | |||
| C. sericea | 65.5 ± 10.0 a | 2.06± 0.21 ab | 0.50 ± 0.04 ab |
| L. diffusa | 45.2 ± 13.6 a | 1.53 ± 0.28 a | 0.40 ± 0.06 a |
| V. karroo | 115.0 ± 12.1 b | 2.91 ± 0.25 b | 0.62 ± 0.05 b |
| W. monoptera | 72.9 ± 17.2 ab | 2.83 ± 0.36 b | 0.67 ± 0.08 b |
| Alpha Diversity Indices | Soil pH | TN [%] | TC [%] | NH4+ [mg kg−1] | NO3− [mg kg−1] | |
|---|---|---|---|---|---|---|
| Richness (Observed taxa) | ||||||
| 16S rRNA | r | 0.70 *** | 0.54 ** | 0.52 * | ns | ns |
| r2 | 0.46 *** | 0.21 * | 0.20 * | ns | ns | |
| nifH | r | ns | 0.62 ** | 0.66 *** | 0.42 * | 0.46 * |
| r2 | ns | 0.38 ** | 0.43 *** | 0.18 * | 0.21 * | |
| Shannon Index | ||||||
| 16S rRNA | r | 0.77 *** | 0.53 ** | 0.50 * | ns | ns |
| r2 | 0.56 *** | 0.22 ** | 0.18 * | ns | ns | |
| nifH | r | ns | 0.55 ** | 0.59 ** | ns | ns |
| r2 | ns | 0.30 ** | 0.35 ** | ns | ns | |
| Shannoneven Index | ||||||
| 16S rRNA | r | 0.79 *** | 0.49 * | 0.42 * | ns | ns |
| r2 | 0.59 *** | 0.21 * | 0.15 * | ns | ns | |
| nifH | r | ns | ns | 0.45 * | ns | ns |
| r2 | ns | ns | 0.20 * | ns | ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muema, E.K.; Steenkamp, E.T.; Venter, S.N. Rhizosphere Diazotrophs and Other Bacteria Associated with Native and Encroaching Legumes in the Succulent Karoo Biome in South Africa. Microorganisms 2022, 10, 216. https://doi.org/10.3390/microorganisms10020216
Muema EK, Steenkamp ET, Venter SN. Rhizosphere Diazotrophs and Other Bacteria Associated with Native and Encroaching Legumes in the Succulent Karoo Biome in South Africa. Microorganisms. 2022; 10(2):216. https://doi.org/10.3390/microorganisms10020216
Chicago/Turabian StyleMuema, Esther K., Emma T. Steenkamp, and Stephanus N. Venter. 2022. "Rhizosphere Diazotrophs and Other Bacteria Associated with Native and Encroaching Legumes in the Succulent Karoo Biome in South Africa" Microorganisms 10, no. 2: 216. https://doi.org/10.3390/microorganisms10020216
APA StyleMuema, E. K., Steenkamp, E. T., & Venter, S. N. (2022). Rhizosphere Diazotrophs and Other Bacteria Associated with Native and Encroaching Legumes in the Succulent Karoo Biome in South Africa. Microorganisms, 10(2), 216. https://doi.org/10.3390/microorganisms10020216





