Modulation of Streptococcus mutans Adherence to Hydroxyapatite by Engineered Salivary Peptides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Collection of the Parotid Gland Saliva
2.3. Proteins and Peptides Tested
2.4. Hydroxyapatite Disc Preparation
2.5. Acquired Pellicle Formation
2.6. S. mutans Inoculation and Adhesion to the AEP
2.7. Quantitation of Adhered S. mutans
2.8. Extraction of Cell Wall Proteins from S. mutans
2.9. Bacterial Cell Wall Proteome Profile
2.10. Statistical Analysis
3. Results
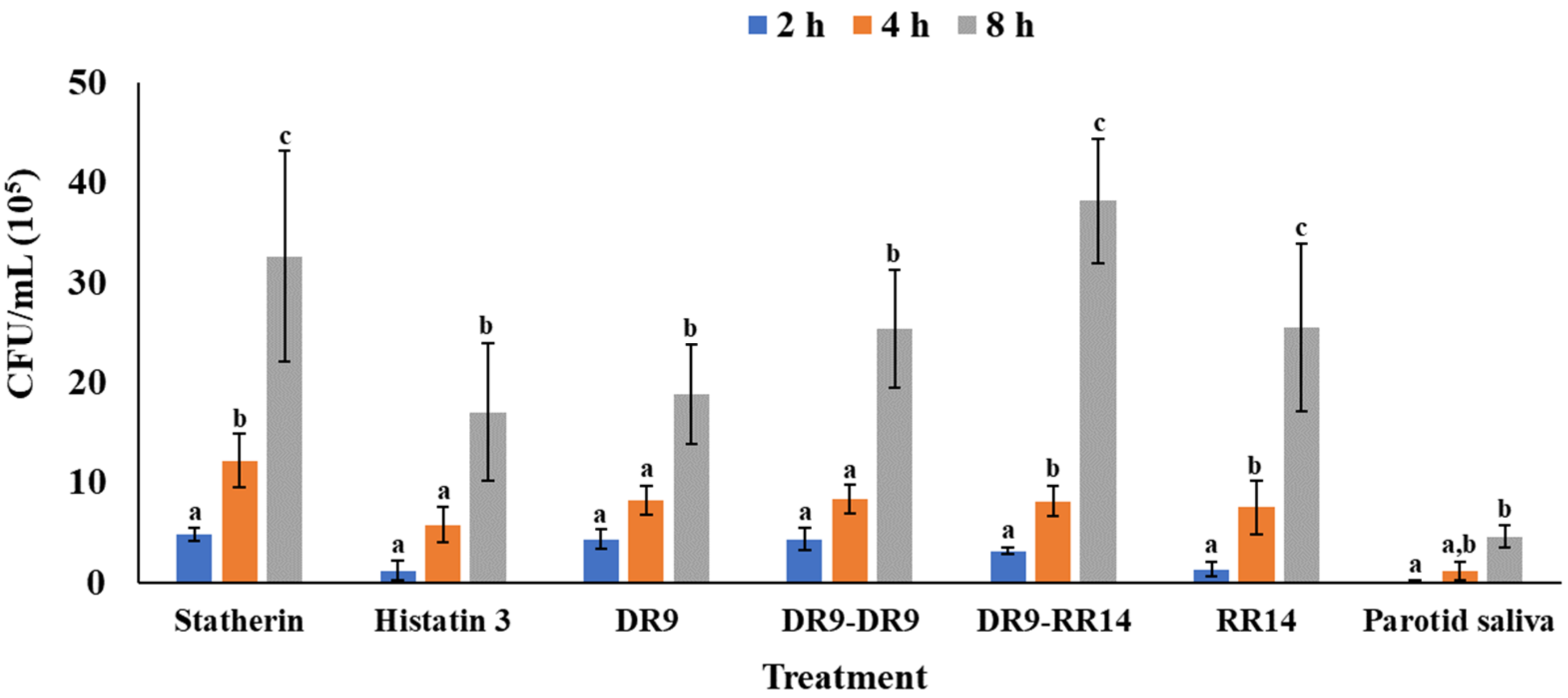
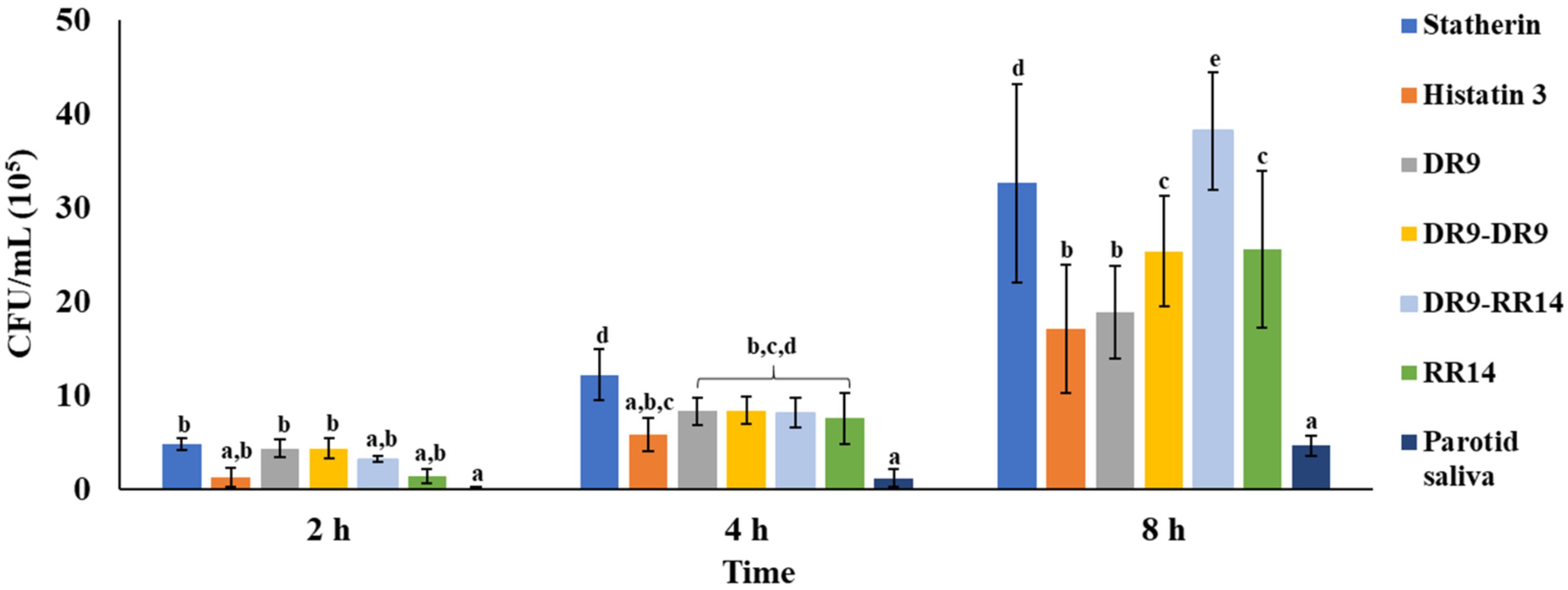
3.1. Quantitation of Adhered S. mutans
3.2. Cell Wall Bacterial Proteome Profiles at Baseline, and in the Adhered and Planktonic Bacteria
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dawes, C. The nomenclature of the integuments of the enamel surface of tooth. Brit. Dent. J. 1963, 115, 65–68. [Google Scholar]
- Siqueira, W.L.; Zhang, W.; Helmerhorst, E.J.; Gygi, S.P.; Oppenheim, F.G. Identification of Protein Components in in vivo Human Acquired Enamel Pellicle Using LC−ESI−MS/MS. J. Proteome Res. 2007, 6, 2152–2160. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, R.; Calheiros-Lobo, M.J.; Williams, J.; Ferrer-Correia, A.J.; Tomer, K.B.; Duarte, J.A.; Domingues, P.M.; Amado, F.M.L. Peptidomic analysis of human acquired enamel pellicle. Biomed. Chromatogr. 2007, 21, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, W.L.; Oppenheim, F.G. Small molecular weight proteins/peptides present in the in vivo formed human acquired enamel pellicle. Arch. Oral Biol. 2009, 54, 437–444. [Google Scholar] [CrossRef] [Green Version]
- Siqueira, W.L.; Custodio, W.; McDonald, E.E. New Insights into the Composition and Functions of the Acquired Enamel Pellicle. J. Dent. Res. 2012, 91, 1110–1118. [Google Scholar] [CrossRef]
- Li, J.; Helmerhorst, E.J.; Leone, C.W.; Troxler, R.F.; Yaskell, T.; Haffajee, A.D.; Socransky, S.S.; Oppenheim, F.G. Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 2004, 97, 1311–1318. [Google Scholar] [CrossRef]
- Marsh, P.D. Dental Plaque as a Microbial Biofilm. Caries Res. 2004, 38, 204–211. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Marsh, P.D.; Zaura, E. Dental biofilm: Ecological interactions in health and disease. J. Clin. Periodontol. 2017, 44, S12–S22. [Google Scholar] [CrossRef]
- Shimotoyodome, A.; Kobayashi, H.; Tokimitsu, I.; Matsukubo, T.; Takaesu, Y. Statherin and Histatin 1 Reduce Parotid Saliva-Promoted Streptococcus mutans Strain MT8148 Adhesion to Hydroxyapatite Surfaces. Caries Res. 2006, 40, 403–411. [Google Scholar] [CrossRef]
- Bratthall, D.; Petersen, P.E.; Stjernswärd, J.R.; Brown, L.J. Oral and Craniofacial Diseases and Disorders. In Disease Control Priorities in Developing Countries, 2nd ed.; Jamison, D.T., Breman, J.G., Measham, A.R., Alleyne, G., Claeson, M., Evans, D.B., Jha, P., Mills, A., Musgrove, P., Eds.; The International Bank for Reconstruction and Development/The World Bank: Washington DC, USA, 2006. [Google Scholar]
- Hamada, S.; Koga, T.; Ooshima, T. Virulence Factors of Streptococcus mutans and Dental Caries Prevention. J. Dent. Res. 1984, 63, 407–411. [Google Scholar] [CrossRef]
- Hannig, C.; Hannig, M.; Attin, T. Enzymes in the acquired enamel pellicle. Eur. J. Oral Sci. 2005, 113, 2–13. [Google Scholar] [CrossRef]
- Basiri, T.; Johnson, N.D.; Moffa, E.B.; Mulyar, Y.; Serra Nunes, P.L.; Machado, M.; Siqueira, W.L. Duplicated or Hybridized Peptide Functional Domains Promote Oral Homeostasis. J. Dent. Res. 2017, 96, 1162–1167. [Google Scholar] [CrossRef]
- Valente, M.T.; Moffa, E.B.; Crosara, K.T.B.; Xiao, Y.; De Oliveira, T.M.; Machado, M.A.D.A.M.; Siqueira, W.L. Acquired Enamel Pellicle Engineered Peptides: Effects on Hydroxyapatite Crystal Growth. Sci. Rep. 2018, 8, 3766. [Google Scholar] [CrossRef]
- Xiao, Y.; Karttunen, M.; Jalkanen, J.; Mussi, M.C.; Liao, Y.; Grohe, B.; Lagugné-Labarthet, F.; Siqueira, W.L. Hydroxyapatite Growth Inhibition Effect of Pellicle Statherin Peptides. J. Dent. Res. 2015, 94, 1106–1112. [Google Scholar] [CrossRef]
- Siqueira, W.L.; Margolis, H.C.; Helmerhorst, E.J.; Mendes, F.M.; Oppenheim, F.G. Evidence of Intact Histatins in the in vivo Acquired Enamel Pellicle. J. Dent. Res. 2010, 89, 626–630. [Google Scholar] [CrossRef]
- Helmerhorst, E.J.; Alagl, A.S.; Siqueira, W.L.; Oppenheim, F.G. Oral fluid proteolytic effects on histatin 5 structure and function. Arch. Oral Biol. 2006, 51, 1061–1070. [Google Scholar] [CrossRef]
- Marin, L.M.; Cury, J.A.; Siqueira, W.L. Validation of a cariogenic biofilm model by evaluating the effect of fluoride on enamel demineralization. J. Microbiol. Methods 2021, 192, 106386. [Google Scholar] [CrossRef]
- Azzopardi, P.V.; O’Young, J.; Lajoie, G.; Karttunen, M.; Goldberg, H.A.; Hunter, G.K. Roles of Electrostatics and Conformation in Protein-Crystal Interactions. PLoS ONE 2010, 5, e9330. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, Y.; Washio, J.; Sasaki, K.; Takahashi, N. A high-sensitive and non-radioisotopic fluorescence dye method for evaluating bacterial adhesion to denture materials. Dent. Mater. J. 2013, 32, 585–591. [Google Scholar] [CrossRef] [Green Version]
- Busscher, H.J.; Van Der Mei, H.C. Physico-Chemical Interactions in Initial Microbial Adhesion and Relevance for Biofilm Formation. Adv. Dent. Res. 1997, 11, 24–32. [Google Scholar] [CrossRef]
- Whittaker, C.J.; Klier, C.M.; Kolenbrander, P.E. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 1996, 50, 513–552. [Google Scholar] [CrossRef]
- Gorbunoff, M.J.; Timasheff, S.N. The interaction of proteins with hydroxyapatite. Anal. Biochem. 1984, 136, 440–445. [Google Scholar] [CrossRef]
- Rölla, G.; Robrish, S.A.; Bowen, W.H. Interaction of hydroxyapatite and protein-coated hydroxyapatite with Streptococcus mutans and Streptococcus sanguis. Acta Pathol. Microbiol. Scand. Sect. B Microbiol. 1977, 85B, 341–346. [Google Scholar] [CrossRef]
- Goobes, G.; Goobes, R.; Schueler-Furman, O.; Baker, D.; Stayton, P.S.; Drobny, G.P. Folding of the C-terminal bacterial binding domain in statherin upon adsorption onto hydroxyapatite crystals. Proc. Natl. Acad. Sci. USA 2006, 103, 16083–16088. [Google Scholar] [CrossRef] [Green Version]
- Simonson, L.G.; Reiher, D.A. Effect of human saliva and various compounds on the adsorption of the bacterium Streptococcus mutans to hydroxyapatite. Arch. Oral Biol. 1981, 26, 143–146. [Google Scholar] [CrossRef]
- Reynolds, E.C.; Wong, A. Effect of adsorbed protein on hydroxyapatite zeta potential and Streptococcus mutans adherence. Infect. Immun. 1983, 39, 1285–1290. [Google Scholar] [CrossRef] [Green Version]
- Rölla, G. Formation of dental integuments--some basic chemical considerations. Swed. Dent. J. 1977, 1, 241–251. [Google Scholar]
- Heffernan, R.; Dehzangi, A.; Lyons, J.; Paliwal, K.; Sharma, A.; Wang, J.; Sattar, A.; Zhou, Y.; Yang, Y. Highly accurate sequence-based prediction of half-sphere exposures of amino acid residues in proteins. Bioinformatics 2015, 32, 843–849. [Google Scholar] [CrossRef]
- Makrodimitris, K.; Masica, D.L.; Kim, E.T.; Gray, J.J. Structure Prediction of Protein−Solid Surface Interactions Reveals a Molecular Recognition Motif of Statherin for Hydroxyapatite. J. Am. Chem. Soc. 2007, 129, 13713–13722. [Google Scholar] [CrossRef]
- Raj, P.A.; Johnsson, M.; Levine, M.J.; Nancollas, G.H. Salivary statherin. Dependence on sequence, charge, hydrogen bonding potency, and helical conformation for adsorption to hydroxyapatite and inhibition of mineralization. J. Biol. Chem. 1992, 267, 5968–5976. [Google Scholar] [CrossRef]
- Long, J.R.; Shaw, W.J.; Stayton, P.S.; Drobny, G.P. Structure and Dynamics of Hydrated Statherin on Hydroxyapatite As Determined by Solid-State NMR. Biochemistry 2001, 40, 15451–15455. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.L.; Xu, T.; Lamkin, M.S.; Brodin, P.; Aars, H.; Berg, T.; Oppenheim, F.G. Physiological Regulation of the Secretion of Histatins and Statherins in Human Parotid Saliva. J. Dent. Res. 1994, 73, 1811–1817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacKay, B.J.; Denepitiya, L.; Iacono, V.J.; Krost, S.B.; Pollock, J.J. Growth-inhibitory and bactericidal effects of human parotid salivary histidine-rich polypeptides on Streptococcus mutans. Infect. Immun. 1984, 44, 695–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payne, J.B.; Iacono, V.J.; Crawford, I.T.; Lepre, B.M.; Bernzweig, E.; Grossbard, B.L. Selective effects of histidine-rich polypeptides on the aggregation and viability of Streptococcus mutans and Streptococcus sanguis. Oral Microbiol. Immunol. 1991, 6, 169–176. [Google Scholar] [CrossRef]
- Gibbons, R.J.; Hay, D.I. Adsorbed Salivary Acidic Proline-rich Proteins Contribute to the Adhesion of Streptococcus mutans JBP to Apatitic Surfaces. J. Dent. Res. 1989, 68, 1303–1307. [Google Scholar] [CrossRef]
- Roger, V.; Tenovuo, J.; Lenander-Lumikari, M.; Söderling, E.; Vilja, P. Lysozyme and Lactoperoxidase Inhibit the Adherence of Streptococcus mutans NCTC 10449 (Serotype c) to Saliva-Treated Hydroxyapatite in vitro. Caries Res. 1994, 28, 421–428. [Google Scholar] [CrossRef]
- Ajdić, D.; McShan, W.M.; McLaughlin, R.E.; Savić, G.; Chang, J.; Carson, M.B.; Primeaux, C.; Tian, R.; Kenton, S.; Jia, H.; et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 2002, 99, 14434–14439. [Google Scholar] [CrossRef] [Green Version]
- Banas, J.A.; Biswas, S.; Zhu, M. Effects of DNA Methylation on Expression of Virulence Genes in Streptococcus mutans. Appl. Environ. Microbiol. 2011, 77, 7236–7242. [Google Scholar] [CrossRef] [Green Version]
- Klein, M.I.; DeBaz, L.; Agidi, S.; Lee, H.; Xie, G.; Lin, A.H.-M.; Hamaker, B.R.; Lemos, J.; Koo, H. Dynamics of Streptococcus mutans Transcriptome in Response to Starch and Sucrose during Biofilm Development. PLoS ONE 2010, 5, e13478. [Google Scholar] [CrossRef]
- Mattos-Graner, R.O.; Jin, S.; King, W.F.; Chen, T.; Smith, D.J.; Duncan, M.J. Cloning of the Streptococcus mutans Gene Encoding Glucan Binding Protein B and Analysis of Genetic Diversity and Protein Production in Clinical Isolates. Infect. Immun. 2001, 69, 6931–6941. [Google Scholar] [CrossRef] [Green Version]
- Fujita, K.; Matsumoto-Nakano, M.; Inagaki, S.; Ooshima, T. Biological functions of glucan-binding protein B of Streptococcus mutans. Oral Microbiol. Immunol. 2007, 22, 289–292. [Google Scholar] [CrossRef]
- Crowley, P.J.; Brady, L.J.; Piacentini, D.A.; Bleiweis, A.S. Identification of a salivary agglutinin-binding domain within cell surface adhesin P1 of Streptococcus mutans. Infect. Immun. 1993, 61, 1547–1552. [Google Scholar] [CrossRef] [Green Version]
- Yin, A.; Margolis, H.C.; Yao, Y.; Grogan, J.; Oppenheim, F.G. Multi-component adsorption model for pellicle formation: The influence of salivary proteins and non-salivary phospho proteins on the binding of histatin 5 onto hydroxyapatite. Arch. Oral Biol. 2005, 51, 102–110. [Google Scholar] [CrossRef]
- Marin, L.M. Effect of Histatin- and Statherin-Derived Engineered Salivary Peptides on Streptococcus mutans Adhesion and on Enamel Demineralization Provoked by Cariogenic Biofilms. Ph.D. Thesis, The University of Western Ontario, London, ON, Canada, 2019. [Google Scholar]
- Marin-Gallon, L.M. Effect of Histatin- and Statherin- Derived Engineered Salivary Peptides on Streptococcus mutans Adhesion and on Enamel Demineralization Provoked by Cariogenic Biofilms. Ph.D. Thesis, University of Campinas, Piracicaba, SP, Brazil, 2019. [Google Scholar]

| Peptide/Protein Name | Peptide Sequence | pI |
|---|---|---|
| Statherin | DSpSpEEKFLRRIGRFGYGYGPYQPVPEQPLYPQPYQPQYQQYTF | 4.4 |
| Histatin 3 | DSHAKRHHGYKRKFHEKHHSHRGYRSNYLYDN | 10.4 |
| DR9 | DSpSpEEKFLR | 3.6 |
| DR9-DR9 | DSpSpEEKFLRDSpSpEEKFLR | 3.4 |
| DR9-RR14 | DSpSpEEKFLRRKFHEKHHSHRGYR | 7.1 |
| RR14 | RKFHEKHHSHRGYR | 11.0 |
| Treatment | Time (h) | |||
|---|---|---|---|---|
| n | 2 | 4 | 8 | |
| Mean of CFU/Disc*105 (±S.D.) | ||||
| Statherin | 6 | 4.8 (±0.7) | 12.2 (±2.7) | 32.6 (±10.6) |
| Histatin 3 | 6 | 1.2 (±1.0) | 5.8 (±1.7) | 17.0 (±6.9) |
| DR9 | 6 | 4.3 (±1.0) | 8.3 (±1.4) | 18.8 (±5.0) |
| DR9-DR9 | 6 | 4.3 (±1.1) | 8.3 (±1.4) | 25.3 (±5.9) |
| DR9-RR14 | 6 | 3.2 (±0.3) | 8.1 (±1.5) | 38.1 (±6.2) |
| RR14 | 6 | 1.3 (±0.7) | 7.5 (±2.7) | 25.5 (±8.4) |
| Parotid saliva | 6 | 0.0 (±0.2) | 1.1 (±1.0) | 4.6 (±1.1) |
| Gene Name | Protein Name | Protein Function |
|---|---|---|
| adhE | Aldehyde-alcohol dehydrogenase | Alcohol metabolic process |
| ilvC * | Ketol-acid reductoisomerase (NADP(+)) | Amino acid biosynthesis |
| gapC * | Glyceraldehyde-3-phosphate dehydrogenase | Carbohydrate metabolic process |
| spaP * | Cell surface antigen I/II | Cell wall antigen |
| hup * | DNA-binding protein HU | Chromosome condensation |
| eno * | Enolase | Glycolytic process |
| pgk * | Phosphoglycerate kinase | Glycolytic process |
| fbaA * | Fructose-1,6-biphosphate aldolase | Glycolytic process |
| gpmA * | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | Glycolytic process |
| lplA | Lipoate--protein ligase | Protein biosynthesis |
| groL * | 60 kDa chaperonin | Protein folding |
| dnaK * | Chaperone protein DnaK | Protein folding |
| clp * | Putative Clp-like ATP-dependent protease, ATP-binding subunit | Transcription |
| tuf * | Elongation factor Tu | Translation |
| rplL * | 50S ribosomal protein L7/L12 | Translation |
| rplE * | 50S ribosomal protein L5 | Translation |
| rpsC * | 30S ribosomal protein S3 | Translation |
| rpsE * | 30S ribosomal protein S5 | Translation |
| rplK * | 50S ribosomal protein L11 | Translation |
| rpsB * | 30S ribosomal protein S2 | Translation |
| rpsJ * | 30S ribosomal protein S10 | Translation |
| rplU * | 50S ribosomal protein L21 | Translation |
| rplA * | 50S ribosomal protein L1 | Translation |
| rpsD * | 30S ribosomal protein S4 | Translation |
| rpsG | 30S ribosomal protein S7 | Translation |
| rplO * | 50S ribosomal protein L15 | Translation |
| rs1 * | Putative ribosomal protein S1 sequence-specific DNA-binding protein | Translation |
| rpsH * | 30S ribosomal protein S8 | Translation |
| rplJ * | 50S ribosomal protein L10 | Translation |
| fusA * | Elongation factor G | Translation |
| rplF * | 50S ribosomal protein L6 | Translation |
| rplX * | 50S ribosomal protein L24 | Translation |
| rpsS | 30S ribosomal protein S19 | Translation |
| tsf * | Elongation factor Ts | Translation |
| oppA * | Putative oligopeptide ABC transporter, substrate-binding protein OppA | Transport |
| livK * | Putative ABC transporter, branched chain amino acid-binding protein | Transport |
| SMU_1641c * | Uncharacterized protein | Uncharacterized |
| SMU_63c | Uncharacterized protein | Uncharacterized |
| Time (h) | Protein Function | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 4 | 8 | |||||||||||||||||||||
| Treatment | A | B | C | D | E | F | G | A | B | C | D | E | F | G | A | B | C | D | E | F | G | ||
| Gene Name | |||||||||||||||||||||||
| carB * | x | x | Amino acid biosynthesis | ||||||||||||||||||||
| SMU_241c | x | x | x | x | Amino acid transport | ||||||||||||||||||
| gbpB | x | x | x | Biofilm formation | |||||||||||||||||||
| gtfB * | x | x | x | EPS biosynthesis | |||||||||||||||||||
| eno | x | x | Glycolytic process | ||||||||||||||||||||
| pgk | x | x | x | x | x | x | x | x | Glycolytic process | ||||||||||||||
| recG * | x | x | Metabolic processes | ||||||||||||||||||||
| SMU_1367c * | x | x | x | Metabolic processes | |||||||||||||||||||
| SMU_546 | x | x | Metabolic processes | ||||||||||||||||||||
| dnaK | x | x | Protein folding | ||||||||||||||||||||
| groL | x | x | x | Protein folding | |||||||||||||||||||
| SMU_488 * | x | x | x | x | x | x | x | x | x | x | Protein folding | ||||||||||||
| SMU_1779 * | x | x | x | x | x | x | RNA methylation | ||||||||||||||||
| Rny * | x | x | x | x | x | RNA processing | |||||||||||||||||
| ciaR * | x | x | x | x | Transcription | ||||||||||||||||||
| mtlR | x | x | x | x | Transcription | ||||||||||||||||||
| rpoC | x | x | x | x | x | x | x | Transcription | |||||||||||||||
| rplL | x | x | Translation | ||||||||||||||||||||
| pacL * | x | x | x | x | x | x | x | x | x | x | x | x | Transport | ||||||||||
| SMU_1093 * | x | x | Transport | ||||||||||||||||||||
| SMU_723 * | x | x | Transport | ||||||||||||||||||||
| SMU_946 * | x | Transport | |||||||||||||||||||||
| SMU_1116c * | x | x | x | Uncharacterized | |||||||||||||||||||
| SMU_1140c * | x | x | Uncharacterized | ||||||||||||||||||||
| SMU_1641c | x | x | x | x | Uncharacterized | ||||||||||||||||||
| SMU_1979c * | x | x | x | x | x | x | x | x | x | x | x | x | x | x | Uncharacterized | ||||||||
| SMU_2073c * | x | x | x | x | x | x | x | Uncharacterized | |||||||||||||||
| SMU_252 * | x | x | Uncharacterized | ||||||||||||||||||||
| SMU_49 * | x | x | Uncharacterized | ||||||||||||||||||||
| SMU_631 * | x | x | Uncharacterized | ||||||||||||||||||||
| satE | x | x | x | Unknown | |||||||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marin, L.M.; Xiao, Y.; Cury, J.A.; Siqueira, W.L. Modulation of Streptococcus mutans Adherence to Hydroxyapatite by Engineered Salivary Peptides. Microorganisms 2022, 10, 223. https://doi.org/10.3390/microorganisms10020223
Marin LM, Xiao Y, Cury JA, Siqueira WL. Modulation of Streptococcus mutans Adherence to Hydroxyapatite by Engineered Salivary Peptides. Microorganisms. 2022; 10(2):223. https://doi.org/10.3390/microorganisms10020223
Chicago/Turabian StyleMarin, Lina Maria, Yizhi Xiao, Jaime Aparecido Cury, and Walter Luiz Siqueira. 2022. "Modulation of Streptococcus mutans Adherence to Hydroxyapatite by Engineered Salivary Peptides" Microorganisms 10, no. 2: 223. https://doi.org/10.3390/microorganisms10020223
APA StyleMarin, L. M., Xiao, Y., Cury, J. A., & Siqueira, W. L. (2022). Modulation of Streptococcus mutans Adherence to Hydroxyapatite by Engineered Salivary Peptides. Microorganisms, 10(2), 223. https://doi.org/10.3390/microorganisms10020223






