Predictive Potential of MALDI-TOF Analyses for Wine and Brewing Yeast
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Culture Conditions
2.2. MALDI-TOF MS
2.2.1. Sample Preparation
2.2.2. Mass Spectra Acquisition
2.2.3. Data Analysis
3. Results
3.1. MALDI-TOF Profiles of Strains Cultured on YPD Broth and YPD Agar
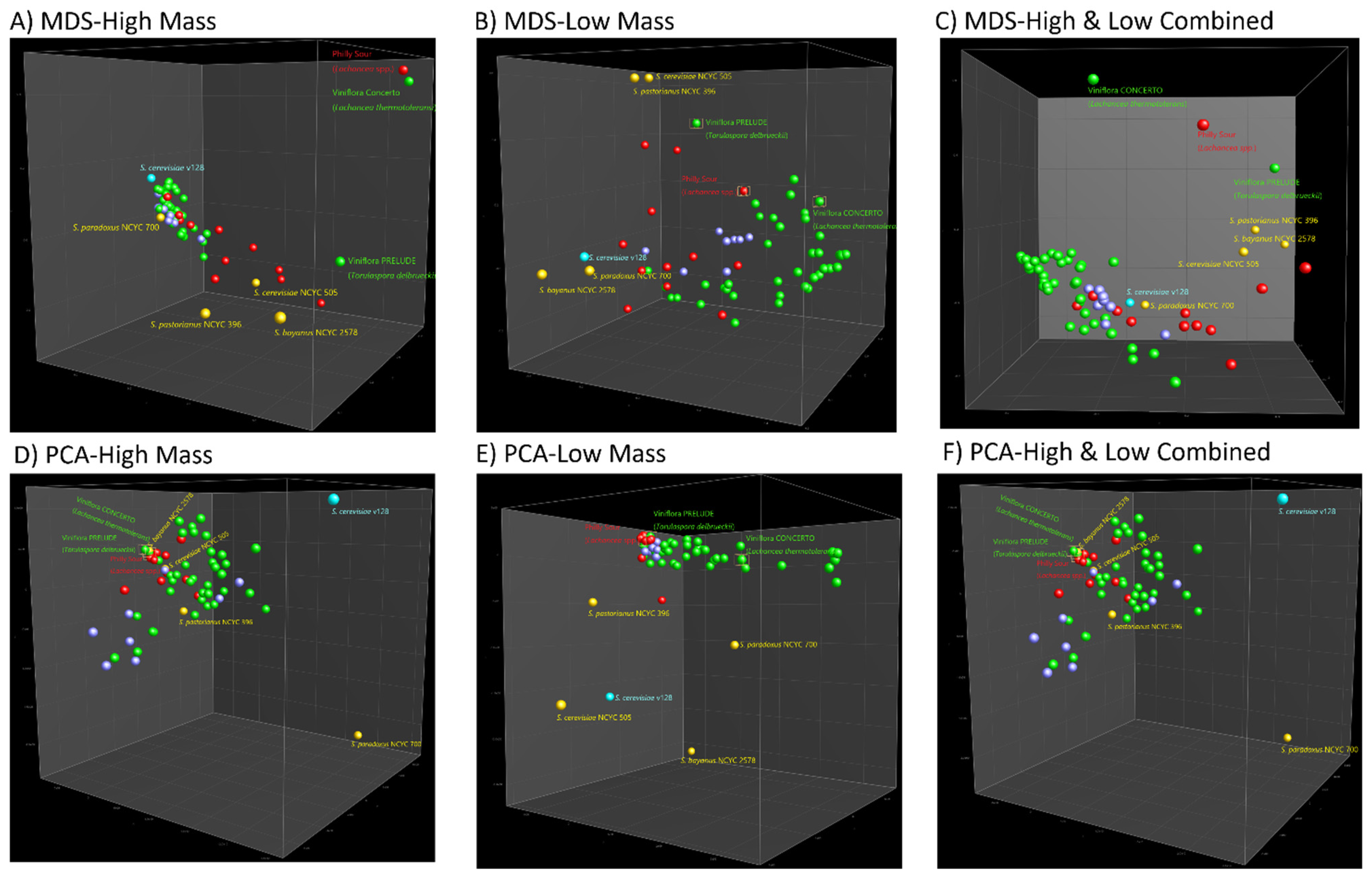
3.2. Strain Classification Using Cluster Analysis and Machine Learning Approaches
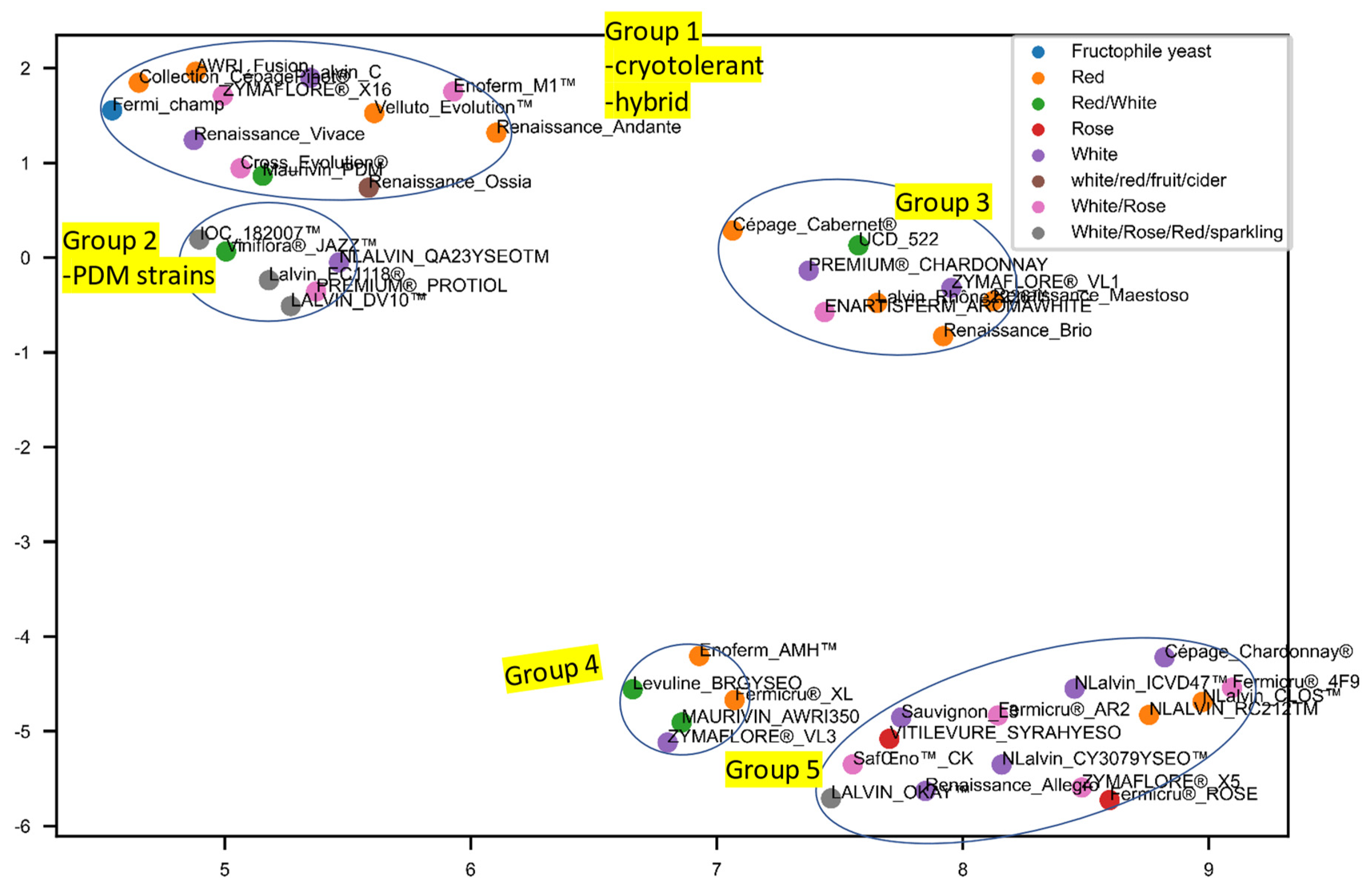
3.3. Separate Analyses Were Undertaken on S. cerevisiae Strains for Which Recommendations Were Extant for Particular Wine Styles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Roullier-Gall, C.; David, V.; Hemmler, D.; Schmitt-Kopplin, P.; Alexandre, H. Exploring yeast sinteractions through metabolic profiling. Sci. Rep. 2020, 10, 6073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbosa, C.; Lage, P.; Vilela, A.; Mendes-Faia, A.; Mendes-Ferreira, A. Phenotypic and metabolic traits of commercial Saccharomyces cerevisiae yeasts. AMB Express 2014, 4, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howell, K.S.; Cozzolino, D.; Bartowsky, E.J.; Fleet, G.H.; Henschke, P.A. Metabolic profiling as a tool for revealing Saccharomyces interactions during wine fermentation. FEMS Yeast Res. 2006, 6, 91–101. [Google Scholar] [CrossRef] [Green Version]
- Legras, J.-L.; Galeote, V.; Bigey, F.; Camarasa, C.; Marsit, S.; Nidelet, T.; Sanchez, I.; Couloux, A.; Guy, J.; Franco-Duarte, R.; et al. Adaptation of S. cerevisiae to Fermented Food Environments Reveals Remarkable Genome Plasticity and the Footprints of Domestication. Mol. Biol. Evol. 2018, 35, 1712–1727. [Google Scholar] [CrossRef] [PubMed]
- Sicard, D.; Legras, J.-L. Bread, beer and wine: Yeast domestication in the Saccharomyces sensu stricto complex. C. R. Biol. 2011, 334, 229–236. [Google Scholar] [CrossRef]
- Gibbons, J.G.; Rinker, D.C. The genomics of microbial domestication in the fermented food environment. Curr. Opin. Genet. Dev. 2015, 35, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Dunn, B.; Levine, R.P.; Sherlock, G. Microarray karyotyping of commercial wine yeast strains reveals shared, as well as unique, genomic signatures. BMC Genom. 2005, 6, 53. [Google Scholar] [CrossRef] [Green Version]
- Fay, J.C. The molecular basis of phenotypic variation in yeast. Curr. Opin. Genet. Dev. 2013, 23, 672–677. [Google Scholar] [CrossRef] [Green Version]
- Jadhav, S.; Gulati, V.; Fox, E.M.; Karpe, A.; Beale, D.J.; Sevior, D.; Bhave, M.; Palombo, E.A. Rapid identification and source-tracking of Listeria monocytogenes using MALDI-TOF mass spectrometry. Int. J. Food Microbiol. 2015, 202, 1–9. [Google Scholar] [CrossRef]
- Manukumar, H.M.; Umesha, S. MALDI-TOF-MS based identification and molecular characterization of food associated methicillin-resistant Staphylococcus aureus. Sci. Rep. 2017, 7, 11414. [Google Scholar] [CrossRef]
- Ziino, G.; Marotta, S.M.; Giarratana, F.; Giuffrida, A.; Panebianco, F. Reliability Evaluation of MALDI-TOF MS Associated with SARAMIS Software in Rapid Identification of Thermophilic Campylobacter Isolated from Food. Food Anal. Methods 2019, 12, 1128–1132. [Google Scholar] [CrossRef]
- Mangmee, S.; Reamtong, O.; Kalambaheti, T.; Roytrakul, S.; Sonthayanon, P. MALDI-TOF mass spectrometry typing for predominant serovars of non-typhoidal Salmonella in a Thai broiler industry. Food Control 2020, 113, 107188. [Google Scholar] [CrossRef]
- Posteraro, B.; de Carolis, E.; Vella, A.; Sanguinetti, M. MALDI-TOF mass spectrometry in the clinical mycology laboratory: Identification of fungi and beyond. Exp. Rev. Proteom. 2013, 10, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, J.A.; Miranda, P.; Flores-Félix, J.D.; Sánchez-Juanes, F.; Ageitos, J.M.; González-Buitrago, J.M.; Velázquez, E.; Villa, T.G. Atypical yeasts identified as Saccharomyces cerevisiae by MALDI-TOF MS and gene sequencing are the main responsible of fermentation of chicha, a traditional beverage from Peru. Syst. Appl. Microbiol. 2013, 36, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, C.; Gomez-Flechoso, M.A.; Belda, I.; Ruiz, J.; Kayali, N.; Polo, L.; Santos, A. Wine yeasts identification by MALDI-TOF MS: Optimization of the preanalytical steps and development of an extensible open-source platform for processing and analysis of an in-house MS database. Int. J. Food Microbiol. 2017, 254, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Usbeck, J.C.; Kern, C.C.; Vogel, R.F.; Behr, J. Optimization of experimental and modelling parameters for the differentiation of beverage spoiling yeasts by Matrix-Assisted-Laser-Desorption/Ionization–Time-of-Flight Mass Spectrometry (MALDI–TOF MS) in response to varying growth conditions. Food Microbiol. 2013, 36, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Plowman, J.E.; Tian, B.; Clerens, S.; On, S.L. An improved method for MALDI-TOF analysis of wine-associated yeasts. J. Microbiol. Methods 2020, 172, 105904. [Google Scholar] [CrossRef] [PubMed]
- Usbeck, J.C.; Wilde, C.; Bertrand, D.; Behr, J.; Vogel, R.F. Wine yeast typing by MALDI-TOF MS. Appl. Microbiol. Biotechnol. 2014, 98, 3737–3752. [Google Scholar] [CrossRef]
- Moothoo-Padayachie, A.; Kandappa, H.R.; Krishna, S.B.N.; Maier, T.; Govender, P. Biotyping Saccharomyces cerevisiae strains using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Eur. Food Res. Technol. 2013, 236, 351–364. [Google Scholar] [CrossRef]
- Lauterbach, A.; Usbeck, J.C.; Behr, J.; Vogel, R.F. MALDI-TOF MS typing enables the classification of brewing yeasts of the genus Saccharomyces to major beer styles. PLoS ONE 2017, 12, e0181694. [Google Scholar] [CrossRef] [Green Version]
- Lafaye, A.; Junot, C.; Pereira, Y.; Lagniel, G.; Tabet, J.-C.; Ezan, E.; Labarre, J. Combined Proteome and Metabolite-profiling Analyses Reveal Surprising Insights into Yeast Sulfur Metabolism. J. Biol. Chem. 2005, 280, 24723–24730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caglar, M.U.; Hockenberry, A.J.; Wilke, C.O. Predicting bacterial growth conditions from mRNA and protein abundances. PLoS ONE 2018, 13, e0206634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Bruyne, K.; Slabbinck, B.; Waegeman, W.; Vauterin, P.; De Baets, B.; Vandamme, P. Bacterial species identification from MALDI-TOF mass spectra through data analysis and machine learning. Syst. Appl. Microbiol. 2011, 34, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Mazher, A. Visualization Framework for High-Dimensional Spatio-Temporal Hydrological Gridded Datasets using Machine-Learning Techniques. Water 2020, 12, 590. [Google Scholar] [CrossRef] [Green Version]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Zhang, J.; Plowman, J.E.; Tian, B.; Clerens, S.; On, S.L. The influence of growth conditions on MALDI-TOF MS spectra of winemaking yeast: Implications for industry applications. J. Microbiol. Methods 2021, 188, 106280. [Google Scholar] [CrossRef]
- Borneman, A.R.; Forgan, A.H.; Kolouchova, R.; Fraser, J.; Schmidt, S. Whole Genome Comparison Reveals High Levels of Inbreeding and Strain Redundancy Across the Spectrum of Commercial Wine Strains of Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2016, 6, 957–971. [Google Scholar] [CrossRef] [Green Version]
- DDunn, B.; Richter, C.; Kvitek, D.J.; Pugh, T.; Sherlock, G. Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 2012, 22, 908–924. [Google Scholar] [CrossRef] [Green Version]
- Vranckx, K.; De Bruyne, K.; Pot, B. Analysis of MALDI-TOF MS Spectra using the BioNumerics Software. In MALDI-TOF and Tandem MS for Clinical Microbiology; Shah, H.N., Gharbia, S.E., Eds.; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2017. [Google Scholar]
- Bisson, L.F. Yeast Hybrids in Winemaking. Catal. Discov. Pract. 2017, 1, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Reich, M.; Bosshard, P.P.; Stark, M.; Beyser, K.; Borgmann, S. Species Identification of Bacteria and Fungi from Solid and Liquid Culture Media by MALDI-TOF Mass Spectrometry. J. Bacteriol. Parasitol. 2013, 10, 5. [Google Scholar] [CrossRef] [Green Version]
- Ryzhov, V.; Fenselau, C. Characterization of the Protein Subset Desorbed by MALDI from Whole Bacterial Cells. Anal. Chem. 2001, 73, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Richter, C.L.; Dunn, B.; Sherlock, G.; Pugh, T. Comparative metabolic footprinting of a large number of commercial wine yeast strains in Chardonnay fermentations. FEMS Yeast Res. 2013, 13, 394–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monerawela, C.; Bond, U. Brewing up a storm: The genomes of lager yeasts and how they evolved. Biotechnol. Adv. 2017, 35, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Legras, J.-L.; Merdinoglu, D.; Cornuet, J.-M.; Karst, F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 2007, 16, 2091–2102. [Google Scholar] [CrossRef]
- Krogerus, K.; Preiss, R.; Gibson, B. A Unique Saccharomyces cerevisiae × Saccharomyces uvarum Hybrid Isolated From Norwegian Farmhouse Beer: Characterization and Reconstruction. Front. Microbiol. 2018, 9, 2253. [Google Scholar] [CrossRef] [Green Version]
- Fay, J.C.; Benavides, J.A. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005, 1, 66–71. [Google Scholar] [CrossRef]
- Stefanini, I.; Dapporto, L.; Berná, L.; Polsinelli, M.; Turillazzi, S.; Cavalieri, D. Social wasps are a Saccharomyces mating nest. Proc. Natl. Acad. Sci. USA 2016, 113, 2247–2251. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Través, L.; Lopes, C.A.; Querol, A.; Barrio, E. On the Complexity of the Saccharomyces bayanus Taxon: Hybridization and Potential Hybrid Speciation. PLoS ONE 2014, 9, e93729. [Google Scholar] [CrossRef] [Green Version]
- Silhavy-Richter, K.; Hack, R.; Regner, F.; Mandl, K. Differentiation of commercial wine yeast strains by molecular markers. Mitt Klosterneubg. 2020, 70, 28–43. [Google Scholar]
- Eglinton, J.; Francis, I.; Henschke, P. Selection and potential of Australian Saccharomyces bayanus yeast for increasing the diversity of red and white wine sensory properties. In: Yeast’s contribution to the sensory profile of wine: Maintaining typicity and biodiversity in the context of globalization. In Proceedings of the Les XVIIes Entretiens Scientifiques Lallemand: Blagnac Cedex, France: Lallemand, La Rioja, Argentina, 27–28 April 2005; pp. 5–12. Available online: https://www.lallemandwine.com/wp-content/uploads/2014/10/ESL-2005-La-Rioja-Yeasts-contribution-to-the-sensory-profile-of-wine.pdf (accessed on 9 December 2021).
- Coi, A.L.; Bigey, F.; Mallet, S.; Marsit, S.; Zara, G.; Gladieux, P.; Galeote, V.; Budroni, M.; Dequin, S.; Legras, J.L. Genomic signatures of adaptation to wine biological ageing conditions in biofilm-forming flor yeasts. Mol. Ecol. 2017, 26, 2150–2166. [Google Scholar] [CrossRef]
- Hart, R.; Jolly, N.; Mohamed, G.; Booyse, M.; Ndimba, B. Characterisation of Saccharomyces cerevisiae hybrids selected for low volatile acidity formation and the production of aromatic Sauvignon blanc wine. Afr. J. Biotechnol. 2016, 15, 2068–2081. [Google Scholar] [CrossRef] [Green Version]
- Novo, M.; Bigey, F.; Beyne, E.; Galeote, V.; Gavory, F.; Mallet, S.; Cambon, B.; Legras, J.-L.; Wincker, P.; Casaregola, S.; et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. USA 2009, 106, 16333–16338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamero, A.; Tronchoni, J.; Querol, A.; Belloch, C. Production of aroma compounds by cryotolerant Saccharomyces species and hybrids at low and moderate fermentation temperatures. J. Appl. Microbiol. 2013, 114, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Gamero, A.; Orte, M.P.H.; Querol, A.; Ferreira, V. Effect of aromatic precursor addition to wine fermentations carried out with different Saccharomyces species and their hybrids. Int. J. Food Microbiol. 2011, 147, 33–44. [Google Scholar] [CrossRef]
- Liang, H.-Y.; Chen, J.; Reeves, M.; Han, B.-Z. Aromatic and sensorial profiles of young Cabernet Sauvignon wines fermented by different Chinese autochthonous Saccharomyces cerevisiae strains. Food Res. Int. 2013, 51, 855–865. [Google Scholar] [CrossRef]
- Kim, J.; Darlington, A.; Salvador, M.; Utrilla, J.; Jiménez, J.I. Trade-offs between gene expression, growth and phenotypic diversity in microbial populations. Curr. Opin. Biotechnol. 2019, 62, 29–37. [Google Scholar] [CrossRef]
- Batistote, M.; da Cruz, S.H.; Ernandes, J.R. Altered Patterns of Maltose and Glucose Fermentation by Brewing and Wine Yeasts Influenced by the Complexity of Nitrogen Source. J. Inst. Brew. 2006, 112, 84–91. [Google Scholar] [CrossRef]
- Carrau, F.M.; Medina, K.; Farina, L.; Boido, E.; Henschke, P.A.; Dellacassa, E. Production of fermentation aroma compounds by Saccharomyces cerevisiae wine yeasts: Effects of yeast assimilable nitrogen on two model strains. FEMS Yeast Res. 2008, 8, 1196–1207. [Google Scholar] [CrossRef] [Green Version]


| Commercial Strains | Genetic Background |
|---|---|
| Wine strains | |
| AWRI Fusion * | S. cerevisiae × S. cariocanus |
| Cepage Cabernet | S. cerevisiae |
| Cepage Chardonnay | S. cerevisiae-Strain n° LW05 |
| Collection Cepage Pinot | S. cerevisiae |
| Cross Evolution | S. cerevisiae var. cerevisiae |
| Enartisferm Aroma White | S. cerevisiae |
| Enoferm AMH™ | S. cerevisiae var. cerevisiae |
| Enoferm M1 | S. cerevisiae |
| Fermi champ | S. cerevisiae (ex bayanus) |
| Fermicru 4F9 * | S. cerevisiae-Strain n° 4F9 |
| Fermicru AR2 | S. cerevisiae-Strain n° L0122 |
| Fermicru Rose * | S. cerevisiae-Strain n°LW10 |
| Fermicru XL | S. cerevisiae-Strain n° CECTA 11947 |
| IOC 18-2007 * | S. cerevisiae var. bayanus |
| Lalvin C | S. cerevisiae var. bayanus |
| Lalvin CLOS | S. cerevisiae var. cerevisiae |
| Lalvin CY 3079 | S. cerevisiae var. cerevisiae |
| Lalvin DV10 * | S. cerevisiae var. bayanus |
| Lalvin EC1118 * | S. cerevisiae var. bayanus |
| Lalvin ICV D47 | S. cerevisiae var. cerevisiae |
| Lalvin OKAY | S. cerevisiae var. cerevisiae |
| Lalvin RC212 | S. cerevisiae var. cerevisiae |
| Lalvin Rhone 2226 | S. cerevisiae |
| LalvinQA 23 * | S. cerevisiae |
| Levuline BRG | S. cerevisiae |
| Maurivin AWRI 350 | S. cerevisiae |
| Maurivin PDM * | S. cerevisiae (var. bayanus) |
| Premium Chardonnay | S. cerevisiae |
| PREMIUM® PROTIOL | S. cerevisiae |
| Renaissance Allegro | S. cerevisiae bayanus |
| Renaissance Andante | S. cerevisiae |
| Renaissance Brio (Brioso) | S. cerevisiae |
| Renaissance Maestoso | S. cerevisiae |
| Renaissance Vivace | S. cerevisiae bayanus |
| Rennaissance Ossia | S. cerevisiae |
| SafoenoTM CK | S. cerevisiae |
| Sauvignon L3 | S. cerevisiae |
| UCD522 | S. cerevisiae |
| Velluto Evolution™ | S. cerevisiae/uvarum |
| Viniflora Jazz | S. cerevisiae |
| Viniflora® PRELUDE™ | Torulaspora delbrueckii |
| Viniflora® CONCERTO™ | Lachancea thermotolerans |
| Vitilevure Syrah | S. cerevisiae |
| Zymaflore VL1 | S. cerevisiae |
| Zymaflore VL3 | S. cerevisiae |
| Zymaflore X5 | S. cerevisiae |
| ZYMAFLORE® X16 | S. cerevisiae |
| Brewing strains | |
| BRY-97 American West Coast Yeast | S. cerevisiae |
| LalBrew KÖln | S. cerevisiae |
| Belle Saison | S. cerevisiae var. diastaticus |
| Mangrove Jack’s New World Strong Ale Yeast | S. cerevisiae |
| Philly Sour | Lachancea spp. |
| LalBrew Verdant IPA | S. cerevisiae |
| Mangrove Jack’s Californian Lager Yeast | S. cerevisiae |
| Mangrove Jack’s Bohemian Lager Yeast | S. cerevisiae |
| Saflager S-23 Yeast | S. cerevisiae |
| Mangrove Jack’s Bavarian Wheat Yeast | S. cerevisiae |
| Mangrove Jack’s Belgian Wit Yeast | S. cerevisiae |
| Safbrew WB-06 Wheat Yeast | S. cerevisiae var. diastaticus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Plowman, J.E.; Tian, B.; Clerens, S.; On, S.L.W. Predictive Potential of MALDI-TOF Analyses for Wine and Brewing Yeast. Microorganisms 2022, 10, 265. https://doi.org/10.3390/microorganisms10020265
Zhang J, Plowman JE, Tian B, Clerens S, On SLW. Predictive Potential of MALDI-TOF Analyses for Wine and Brewing Yeast. Microorganisms. 2022; 10(2):265. https://doi.org/10.3390/microorganisms10020265
Chicago/Turabian StyleZhang, Junwen, Jeffrey E. Plowman, Bin Tian, Stefan Clerens, and Stephen L. W. On. 2022. "Predictive Potential of MALDI-TOF Analyses for Wine and Brewing Yeast" Microorganisms 10, no. 2: 265. https://doi.org/10.3390/microorganisms10020265






