Application of Bioorganic Fertilizer on Panax notoginseng Improves Plant Growth by Altering the Rhizosphere Microbiome Structure and Metabolism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Sample Collection
2.2. Soil Physicochemical Analysis
2.3. Rhizosphere Soil Collection
2.4. Rhizosphere Microbiome Analysis
2.5. Metabolites Profiling from Rhizosphere Samples
2.6. Statistical Analysis and Data Visualization
3. Results
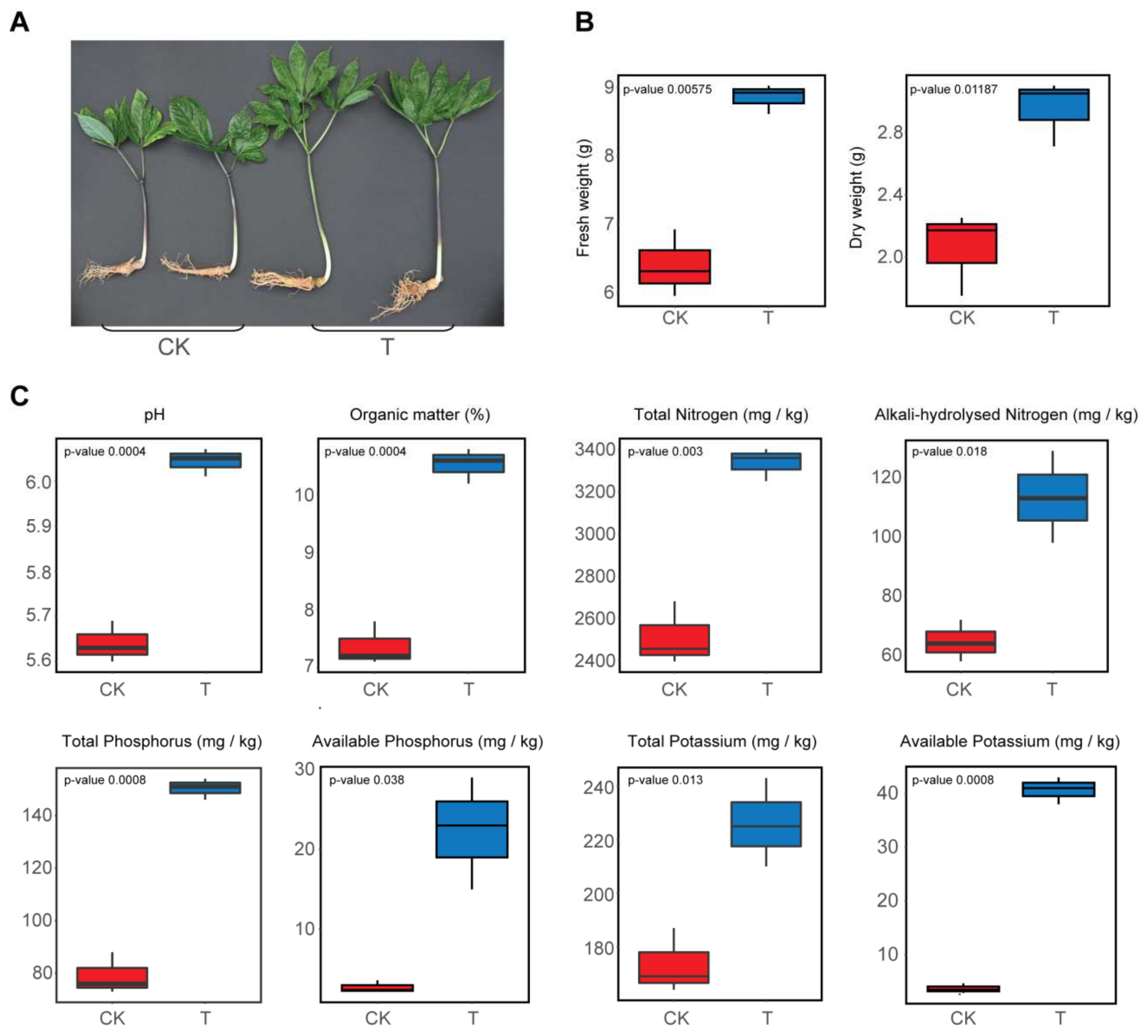
3.1. Impact of a Bioorganic Fertilizer Application on Sanqi Ginseng Growth and Soil Physicochemical Properties
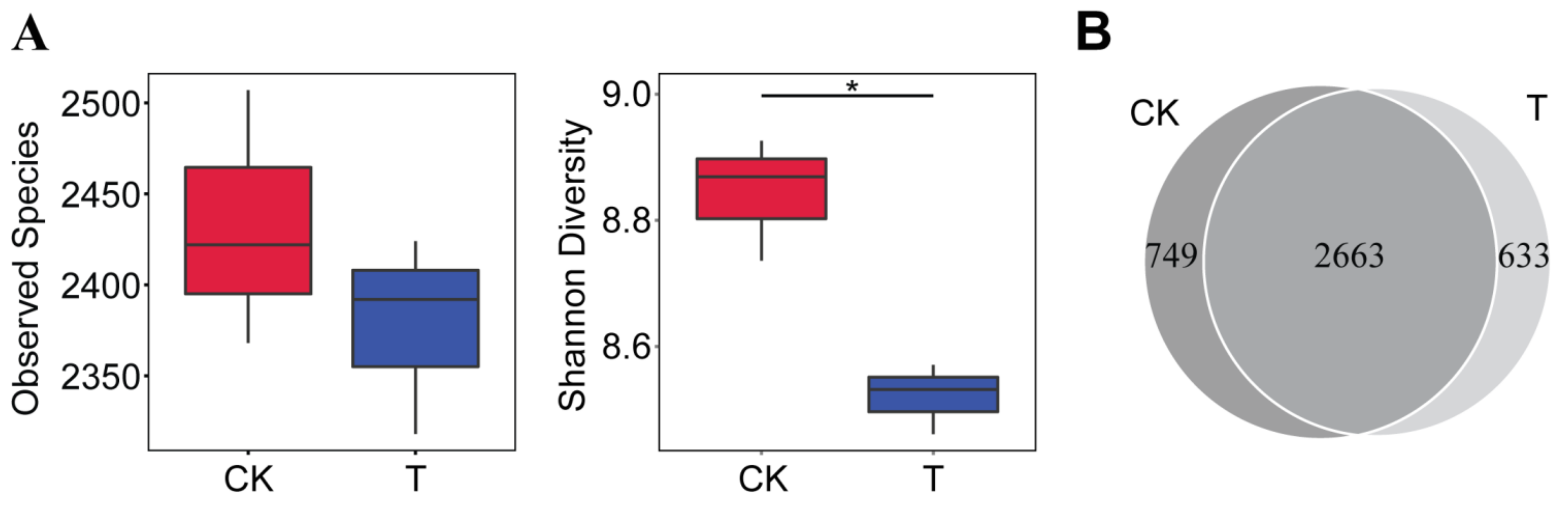
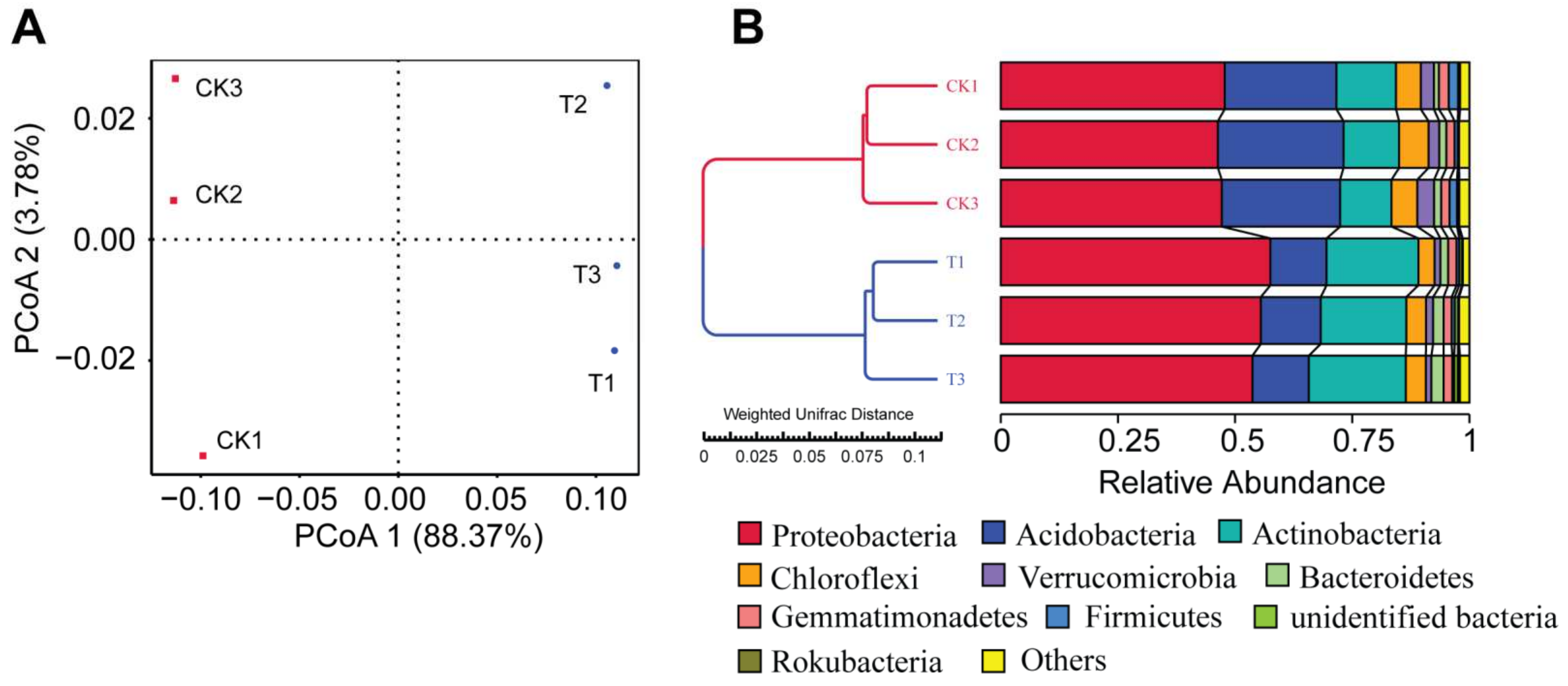
3.2. The Bioorganic Fertilizer Alters P. notoginseng Rhizosphere Bacterial α- and β-Diversity
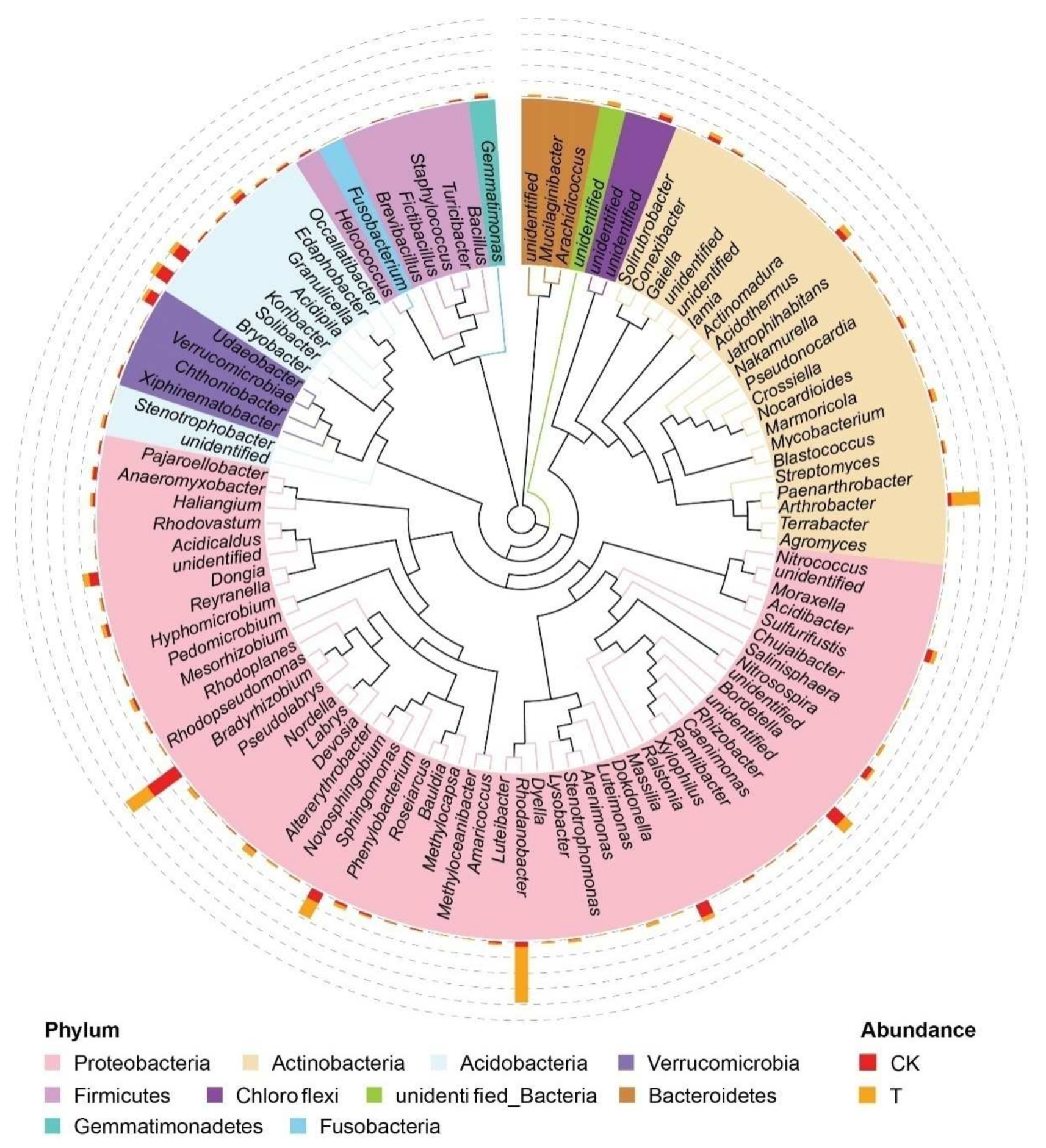
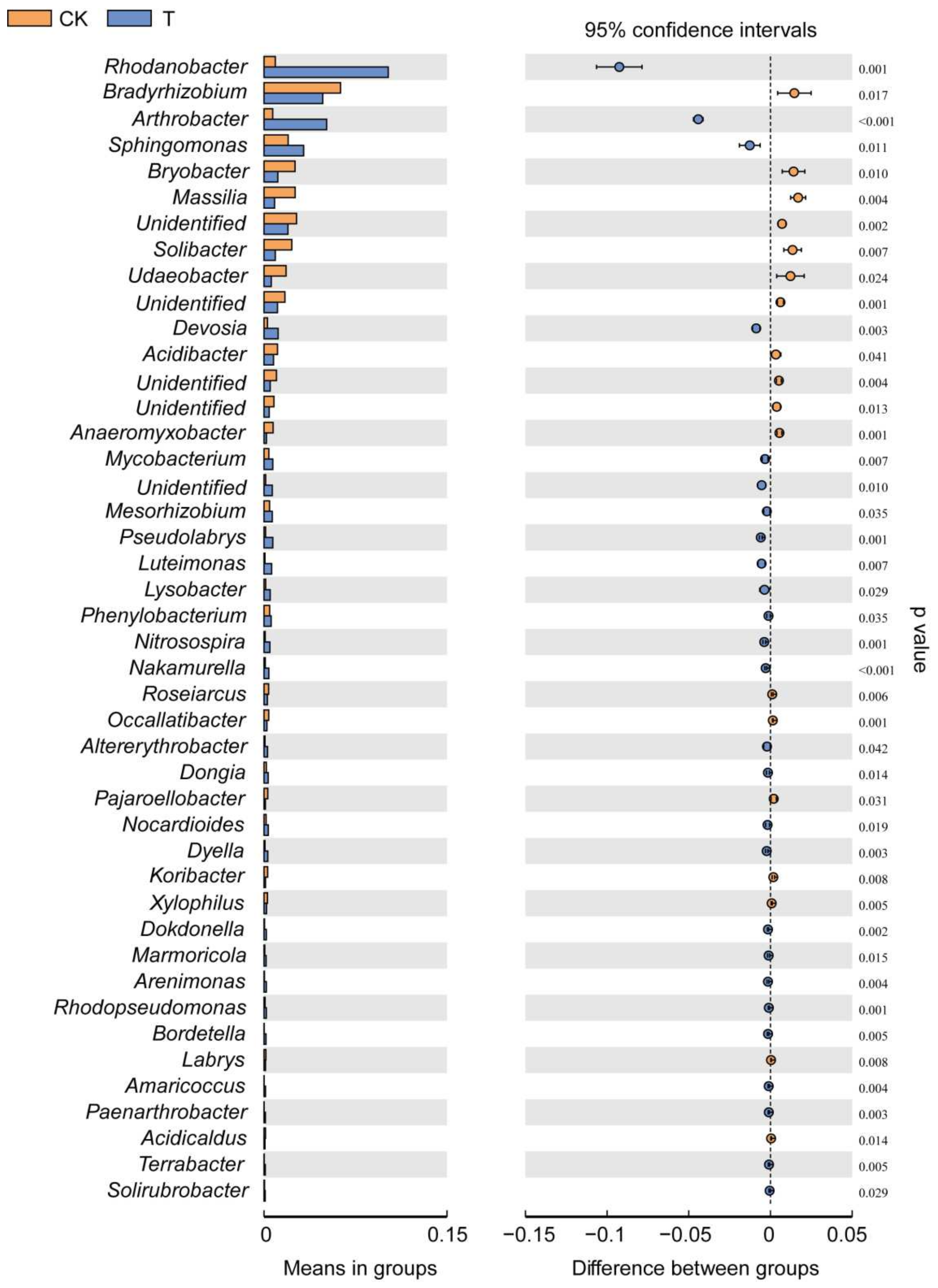
3.3. Effect of a Bioorganic Fertilizer Application on Rhizosphere Bacterial Community Composition
3.4. Changes in Soil Metabolites after Addition of the Bioorganic Fertilizer
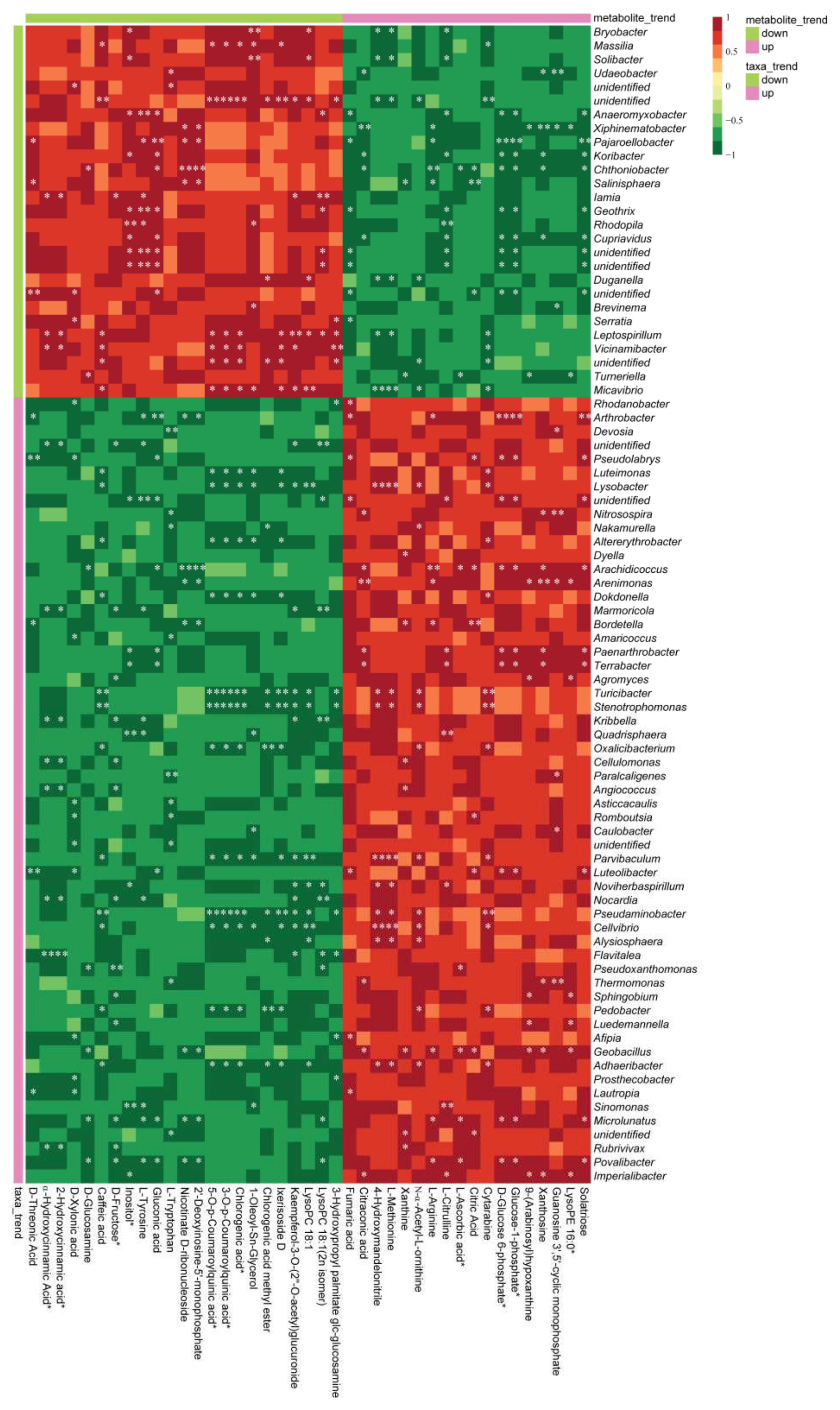
3.5. Correlations between the Bacterial Community and Soil Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Topalović, O.; Hussain, M.; Heuer, H. Plants and associated soil microbiota cooperatively suppress plant-parasitic nematodes. Front. Microbiol. 2020, 11, 313. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Baquerizo, M.; Reich, P.B.; Trivedi, C.; Eldridge, D.J.; Abades, S.; Alfaro, F.D.; Bastida, F.; Berhe, A.A.; Cutler, N.A.; Gallardo, A.; et al. Multiple elements of soil biodiversity drive ecosystem functions across biomes. Nat. Ecol. Evol. 2020, 4, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Hussain, M.; Zhang, W.; Stadler, M.; Liu, X.; Xiang, M. Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi. Mycology 2019, 10, 127–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, M.; Hamid, M.I.; Tian, J.; Hu, J.; Zhang, X.; Chen, J.; Xiang, M.; Liu, X. Bacterial community assemblages in the rhizosphere soil, root endosphere and cyst of soybean cyst nematode-suppressive soil challenged with nematodes. FEMS Microbiol. Ecol. 2018, 94, fiy142. [Google Scholar] [CrossRef] [PubMed]
- Barrios, E. Soil biota, ecosystem services and land productivity. Ecol. Econ. 2007, 64, 269–285. [Google Scholar] [CrossRef]
- Van der Heijden, M.G.; Bardgett, R.D.; van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Cordovez, V.; Dini-Andreote, F.; Carrión, V.J.; Raaijmakers, J.M. Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol. 2019, 73, 69–88. [Google Scholar] [CrossRef]
- Wang, J.; Li, R.; Zhang, H.; Wei, G.; Li, Z. Beneficial bacteria activate nutrients and promote wheat growth under conditions of reduced fertilizer application. BMC Microbiol. 2020, 20, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Wei, Q.; Qi, H.; Wang, X.; Huang, W.; Liu, J.; Liu, S. Optimal schemes and correlation analysis between soil nutrient, pH and microorganism population in orchard of Beijing suburb. J. Fruit Sci. 2011, 28, 15–19. [Google Scholar]
- Bell, C.W.; Asao, S.; Calderon, F.; Wolk, B.; Wallenstein, M.D. Plant nitrogen uptake drives rhizosphere bacterial community assembly during plant growth. Soil Biol. Biochem. 2015, 85, 170–185. [Google Scholar] [CrossRef]
- Liu, J.; Shu, A.; Song, W.; Shi, W.; Li, M.; Zhang, W.; Li, Z.; Liu, G.; Yuan, F.; Zhang, S.; et al. Long-term organic fertilizer substitution increases rice yield by improving soil properties and regulating soil bacteria. Geoderma 2021, 404, 115287. [Google Scholar] [CrossRef]
- Bai, Y.C.; Chang, Y.Y.; Hussain, M.; Lu, B.; Zhang, J.P.; Song, X.B.; Lei, X.S.; Pei, D. Soil chemical and microbiological properties are changed by long-term chemical fertilizers that limit ecosystem functioning. Microorganisms 2020, 8, 694. [Google Scholar] [CrossRef]
- Wang, C.Z.; McEntee, E.; Wicks, S.; Wu, J.A.; Yuan, C.S. Phytochemical and analytical studies of Panax notoginseng (Burk.) FH chen. J. Nat. Med. 2006, 60, 97–106. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, X.; Xu, Y.; Mei, X.; Jiang, B.; Liao, J.; Yin, Z.; Zheng, J.; Zhao, Z.; Fan, L. Autotoxic ginsenosides in the rhizosphere contribute to the replant failure of Panax notoginseng. PLoS ONE 2015, 10, e0118555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christensen, L.P. Ginsenosides: Chemistry, biosynthesis, analysis, and potential health effects. Adv. Food Nutr. Res. 2008, 55, 1–99. [Google Scholar]
- Sun, S.; Wang, C.Z.; Tong, R.; Li, X.L.; Fishbein, A.; Wang, Q.; He, T.C.; Du, W.; Yuan, C.S. Effects of steaming the root of Panax notoginseng on chemical composition and anticancer activities. Food Chem. 2010, 118, 307e14. [Google Scholar] [CrossRef]
- Xia, P.; Guo, H.; Liang, Z.; Cui, X.; Liu, Y.; Liu, F. Nutritional composition of Sanchi (Panax notoginseng) seed and its potential for industrial use. Genet. Resour. Crop Evol. 2014, 61, 663e7. [Google Scholar] [CrossRef]
- Guo, H.B.; Cui, X.M.; An, N.; Cai, G.P. Sanchi ginseng (Panax notoginseng (Burkill) F. H. Chen) in China: Distribution, cultivation and variations. Genet. Resour. Crop Evol. 2010, 57, 453e60. [Google Scholar] [CrossRef]
- Xia, P.; Guo, H.; Zhao, H.; Jiao, J.; Deyholos, M.K.; Yan, X.; Liu, Y.; Liang, Z. Optimal fertilizer application for Panax notoginseng and effect of soil water on root rot disease and saponin contents. J. Ginseng Res. 2016, 40, 38–46. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, M.H.; Jaafar, H.Z.; Rahmat, A.; Rahman, Z.A. Involvement of nitrogen on flavonoids, glutathione, anthocyanin, ascorbic acid and antioxidant activities of Malaysian medicinal plant Labisia pumila Blume (Kacip Fatimah). Int. J. Mol. Sci. 2012, 13, 393–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awad, M.; Jager, A. Relationships between fruit nutrients and concentrations of flavonoids and chlorogenic acid in ‘Elstar’ apple skin. Sci. Hortic. 2002, 92, 265e76. [Google Scholar] [CrossRef]
- Nguyen, P.M.; Niemeyer, E.D. Effects of nitrogen fertilization on the phenolic composition and antioxidant properties of basil (Ocimum basilicum L.). J. Agric. Food Chem. 2008, 56, 8685e91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, J.S.; Gupta, V.K. Soil microbial biomass: A key soil driver in management of ecosystem functioning. Sci. Total Environ. 2018, 634, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wei, L.; Yang, J.; Ahmed, W.; Wang, Y.; Fu, L.; Ji, G. Probiotic consortia: Reshaping the rhizospheric microbiome and its role in suppressing root-rot disease of Panax notoginseng. Front. Microbiol. 2020, 11, 701. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Cao, Y.H.; Cheng, M.H.; Huang, Y.; Mo, M.H.; Wang, Y.; Yang, J.Z.; Yang, F.X. Phylogenetic diversity of bacterial endophytes of Panax notoginseng with antagonistic characteristics towards pathogens of root-rot disease complex. Antonie Van Leeuwenhoek 2013, 103, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yan, K.; Tang, L.; Jia, Z.; Li, Y. Change in deep soil microbial communities due to long-term fertilization. Soil. Biol. Biochem. 2014, 75, 264–272. [Google Scholar] [CrossRef]
- Yu, W.; Guibing, Z.; Liyan, S.; Shanyun, W.; Chengqing, Y. Manure fertilization alters the population of ammonia-oxidizing bacteria rather than ammonia-oxidizing archaea in a paddy soil. J. Basic Microbiol. 2014, 54, 190–197. [Google Scholar]
- Bei, S.; Zhang, Y.L.; Li, T.T.; Christie, P.; Li, X.L.; Zhang, J.L. Response of the soil microbial community to different fertilizer inputs in a wheat-maize rotation on a calcareous soil. Agric. Ecosyst. Environ. 2018, 260, 58–69. [Google Scholar] [CrossRef]
- Tao, C.; Li, R.; Xiong, W.; Shen, Z.; Liu, S.; Wang, B.; Ruan, Y.; Geisen, S.; Shen, Q.; Kowalchuk, G.A. Bioorganic fertilizers stimulate indigenous soil Pseudomonas populations to enhance plant disease suppression. Microbiome 2020, 8, 137. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Y.; Wang, B.; Huang, X.; Yang, L.; Lan, T.; Zhang, J.; Cai, Z. Comparative soil microbial communities and activities in adjacent Sanqi ginseng monoculture and maize-Sanqi ginseng systems. Appl. Soil Ecol. 2017, 120, 89–96. [Google Scholar] [CrossRef]
- Bao, S.D. Soil and Agricultural Chemistry Analysis; Agriculture Publication: Beijing, China, 2000. (In Chinese) [Google Scholar]
- Zeng, Q.; An, S. Identifying the biogeographic patterns of rare and abundant bacterial communities using different primer sets on the loess plateau. Microorganisms 2021, 9, 139. [Google Scholar] [CrossRef]
- Gondwe, R.L.; Kinoshita, R.; Suminoe, T.; Aiuchi, D.; Palta, J.P.; Tani, M. Available soil nutrients and NPK application impacts on yield, quality, and nutrient composition of potatoes growing during the main season in Japan. Am. J. Potato Res. 2020, 97, 234–245. [Google Scholar] [CrossRef]
- Wang, J.H.; Wang, X.J.; Xu, M.G.; Feng, G.; Zhang, W.J.; Lu, C.A. Crop yield and soil organic matter after long-term straw return to soil in China. Nutr. Cycl. Agroecosyst. 2015, 102, 371–381. [Google Scholar] [CrossRef]
- Blair, N.; Faulkner, R.D.; Till, A.R.; Korschens, M.; Schulz, E. Long-term management impacts on soil C, N and physical fertility: Part II: Bad Lauchstadt static and extreme FYM experiments. Soil Till. Res. 2006, 91, 39–47. [Google Scholar] [CrossRef]
- Liang, Q.; Chen, H.Q.; Gong, Y.S.; Fan, M.S.; Yang, H.F.; Lal, R.; Kuzyakov, Y. Effects of 15 years of manure and inorganic fertilizers on soil organic carbon fractions in a wheat-maize system in the North China plain. Nutr. Cycl. Agroecosyst. 2012, 92, 21–33. [Google Scholar] [CrossRef]
- Ozlu, E.; Kumar, S. Response of soil organic carbon, ph, electrical conductivity, and water stable aggregates to long-term annual manure and inorganic fertilizer. Soil Sci. Soc. Am. J. 2018, 82, 1243–1251. [Google Scholar] [CrossRef]
- Bhattacharyya, R.; Ghosh, B.N.; Mishra, P.K.; Mandal, B.; Rao, C.S.; Sarkar, D.; Das, K.; Anil, K.S.; Lalitha, M.; Hati, K.M.; et al. Soil degradation in India: Challenges and potential solutions. Sustainability 2015, 7, 3528–3570. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Xia, F.; Liu, X.M.; He, Y.; Xu, J.M.; Brookes, P.C. Effects of nitrogen fertilizer on the acidification of two typical acid soils in South China. J. Soils Sediments 2014, 14, 415–422. [Google Scholar] [CrossRef]
- Yang, X.D.; Ni, K.; Shi, Y.Z.; Yi, X.Y.; Zhang, Q.F.; Fang, L.; Ma, L.F.; Ruan, J.Y. Effects of long-term nitrogen application on soil acidification and solution chemistry of a tea plantation in China. Agric. Ecosyst. Environ. 2018, 252, 74–82. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, C.; Chen, Z.; Zhang, G.; Chen, L.; Wu, Z. Stoichiometric analyses of soil nutrients and enzymes in a Cambisol soil treated with inorganic fertilizers or manures for 26 years. Geoderma 2019, 353, 382–390. [Google Scholar] [CrossRef]
- Van Bruggen, A.H.C.; Sharma, K.; Kaku, E.; Karfopoulos, S.; Zelenev, V.V.; Blok, W.J. Soil health indicators and Fusarium wilt suppression in organically and conventionally managed greenhouse soils. Appl. Soil Ecol. 2015, 86, 192–201. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Smit, E.; Leeflang, P.; Gommans, S.; van den Broek, J.; van Mil, S.; Wernars, K. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 2001, 67, 2284–2291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Yu, Z.; Yao, Q.; Hu, X.; Zhang, W.; Mi, G.; Wang, G. Distinct soil bacterial communities in response to the cropping system in a Mollisol of northeast China. App. Soil Ecol. 2017, 119, 407–416. [Google Scholar] [CrossRef]
- Dai, Z.; Su, W.; Chen, H.; Barberán, A.; Zhao, H.; Yu, M.; Yu, L.; Brookes, P.C.; Schadt, C.W.; Chang, S.X.; et al. Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of Actinobacteria and Proteobacteria in agro-ecosystems across the globe. Glob. Change Biol. 2018, 24, 3452–3461. [Google Scholar] [CrossRef]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [Green Version]
- Madhaiyan, M.; Poonguzhali, S.; Saravanan, V.S.; Kwon, S.W. Rhodanobacter glycinis sp. nov., a yellow-pigmented gammaproteobacterium isolated from the rhizoplane of field-grown soybean. Int. J. Syst. Evol. Microbiol. 2014, 64, 2023–2028. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Liu, X.; Zhang, Q. Effects of combined biochar and organic fertilizer on nitrous oxide fluxes and the related nitrifier and denitrifier communities in a saline-alkali soil. Sci. Total Environ. 2019, 686, 199–211. [Google Scholar] [CrossRef]
- De Clercq, D.; Van Trappen, S.; Cleenwerck, I.; Ceustermans, A.; Swings, J.; Coosemans, J.; Ryckeboer, J. Rhodanobacter spathiphylli sp. nov., a gammaproteobacterium isolated from the roots of Spathiphyllum plants grown in a compost-amended potting mix. Int. J. Syst. Evol. Microbiol. 2006, 56, 1755–1759. [Google Scholar] [CrossRef]
- Du, J.; Singh, H.; Ngo, H.T.; Won, K.; Kim, K.Y.; Yi, T.H. Lysobacter tyrosinelyticus sp. nov. isolated from gyeryongsan national park soil. J. Microbiol. 2015, 53, 365–370. [Google Scholar] [CrossRef]
- Wu, L.; Jiang, Y.; Zhao, F.; He, X.; Liu, H.; Yu, K. Increased organic fertilizer application and reduced chemical fertilizer application affect the soil properties and bacterial communities of grape rhizosphere soil. Sci. Rep. 2020, 10, 9568. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Yu, X.; Gao, N.; Ota, S.; Shiratori, Y.; Nishizawa, T.; Isobe, K.; He, X.; Senoo, K. Genome Sequence of Arthrobacter sp. UKPF54-2, a Plant Growth-Promoting Rhizobacterial Strain Isolated from Paddy Soil. Microbiol. Resour. Announc. 2019, 8, e01005-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busse, H.J.R. Review of the taxonomy of the genus Arthrobacter, emendation of the genus Arthrobacter sensu lato, proposal to reclassify selected species of the genus Arthrobacter in the novel genera Glutamicibacter gen. nov., Paeniglutamicibacter gen. nov., Pseudoglutamicibacter gen. nov., Paenarthrobacter gen. nov. and Pseudarthrobacter gen. nov., and emended description of Arthrobacter roseus. Int. J. Syst. Evol. Microbiol. 2016, 66, 9–37. [Google Scholar]
- He, W.J.; Zhang, L.; Yi, S.Y.; Tang, X.L.; Yuan, Q.S.; Guo, M.W.; Wu, A.B.; Qu, B.; Li, H.P.; Liao, Y.C. An aldo-keto reductase is responsible for Fusarium toxin-degrading activity in a soil Sphingomonas strain. Sci. Rep. 2017, 7, 9549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mwaheb, M.A.; Hussain, M.; Tian, J.; Zhang, X.; Hamid, M.I.; El-Kassim, N.A.; Hassan, G.M.; Xiang, M.; Liu, X. Synergetic suppression of soybean cyst nematodes by chitosan and Hirsutella minnesotensis via the assembly of the soybean rhizosphere microbial communities. Biol. Control 2017, 115, 85–94. [Google Scholar] [CrossRef]
- Hamid, M.I.; Hussain, M.; Wu, Y.; Zhang, X.; Xiang, M.; Liu, X. Successive soybean-monoculture cropping assembles rhizosphere microbial communities for the soil suppression of soybean cyst nematode. FEMS Microbiol. Ecol. 2017, 93, fiw222. [Google Scholar] [CrossRef] [Green Version]
- Rivas, R.; Velázquez, E.; Willems, A.; Vizcaíno, N.; Subba-Rao, N.S.; Mateos, P.F.; Gillis, M.; Dazzo, F.B.; Martínez-Molina, E. A new species of Devosia that forms a unique nitrogen-fixing root-nodule symbiosis with the aquatic legume Neptunia natans (Lf) Druce. Appl. Environ. Microbiol. 2002, 68, 5217–5222. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Chen, D.; Zhang, R.; Hang, X.; Li, R.; Shen, Q. Amino acids hydrolyzed from animal carcasses are a good additive for the production of bioorganic fertilizer. Front. Microbiol. 2016, 7, 1290. [Google Scholar]
- Eo, J.; Park, K.C. Long-term effects of imbalanced fertilization on the composition and diversity of soil bacterial community. Agric. Ecosyst. Environ. 2016, 231, 176–182. [Google Scholar] [CrossRef]
- Romanenko, L.A.; Tanaka, N.; Svetashev, V.I.; Kurilenko, V.V.; Mikhailov, V.V. Luteimonas vadosa sp. nov., isolated from seashore sediment. Int. J. Syst. Evol. Microbiol. 2012, 63, 1261–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Hong, Q.; Zhang, C.F.; Yang, H.X.; Hu, G.; Zhu, S.J.; Zhang, Y.K.; Liu, X.W.; Zhang, H.; Zhao, C.R. Luteimonas soli sp. nov., isolated from farmland soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 4809–4815. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, X.; Yu, M.; Qiao, Y.; Zhang, X.H. Genomic analysis of Luteimonas abyssi XH031T: Insights into its adaption to the subseafloor environment of South Pacific Gyre and ecological role in biogeochemical cycle. BMC Genomics 2015, 16, 1092. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Liu, W.; Zhu, C.; Luo, G.; Kong, Y.; Ling, N.; Wang, M.; Dai, J.; Shen, Q.; Guo, S. Bacterial rather than fungal community composition is associated with microbial activities and nutrient-use efficiencies in a paddy soil with short-term organic amendments. Plant Soil 2018, 424, 335–349. [Google Scholar] [CrossRef]
- Lin, Y.; Ye, G.; Luo, J.; Di, H.J.; Liu, D.; Fan, J.; Ding, W. Nitrosospira cluster 8a plays a predominant role in the nitrification process of a subtropical Ultisol under long-term inorganic and organic fertilization. Appl. Environ. Microbiol. 2018, 84, e01031-18. [Google Scholar] [CrossRef] [Green Version]
- Cheng, N.; Peng, Y.; Kong, Y.; Li, J.; Sun, C. Combined effects of biochar addition and nitrogen fertilizer reduction on the rhizosphere metabolomics of maize Zea mays L. seedlings. Plant Soil 2018, 433, 19–35. [Google Scholar] [CrossRef]
- White III, R.A.; Rivas-Ubach, A.; Borkum, M.I.; Köberl, M.; Bilbao, A.; Colby, S.M.; Hoyt, D.W.; Bingol, K.; Kim, Y.M.; Wendler, J.P. The state of rhizospheric science in the era of multi-omics, a practical guide to omics technologies. Rhizosphere 2017, 3, 212–221. [Google Scholar] [CrossRef]
- Fisk, L.M.; Barton, L.; Jones, D.L.; Glanville, H.C.; Murphy, D.V. Root exudate carbon mitigates nitrogen loss in a semi-arid soil. Soil Biol. Biochem. 2015, 88, 380–389. [Google Scholar] [CrossRef] [Green Version]
- Gul, S.; Whalen, J.K.; Thomas, B.W.; Sachdeva, V.; Deng, H. Physico-chemical properties and microbial responses in biochar-amended soils: Mechanisms and future directions. Agric. Ecosyst. Environ. 2015, 206, 46–59. [Google Scholar] [CrossRef]
- Coskun, D.; Britto, D.T.; Shi, W.M.; Kronzucker, H.J. How plant root exudates shape the nitrogen cycle. Trends Plant Sci. 2017, 22, 661–673. [Google Scholar] [CrossRef]
- Salvador, V.H.; Lima, R.B.; dos Santos, W.D.; Soares, A.R.; Böhm, P.A.F.; Marchiosi, R.; Ferrarese, M.L.L.; Ferrarese-Filho, O. Cinnamic acid increases lignin production and inhibits soybean root growth. PLoS ONE 2013, 8, e69105. [Google Scholar] [CrossRef] [Green Version]
- Whitehead, D.C. Identification of p-hydroxybenzoic, vanillic, p-coumaric and ferulic acids in soils. Nature 1964, 202, 417–418. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Wei, Z.; Wang, X.; Friman, V.P.; Huang, J.; Wang, X.; Mei, X.; Xu, Y.; Shen, Q.; Jousset, A. Pathogen invasion indirectly changes the composition of soil microbiome via shifts in root exudation profile. Biol. Fertil. Soils 2016, 52, 997–1005. [Google Scholar] [CrossRef] [Green Version]
- Toor, R.K.; Savage, G.P.; Heeb, A. Influence of different types of fertilisers on the major antioxidant components of tomatoes. J. Food Compos. Anal. 2006, 19, 20–27. [Google Scholar] [CrossRef]
- Mitchell, A.E.; Hong, Y.J.; Koh, E.; Barrett, D.M.; Bryant, D.E.; Denison, R.F.; Kaffka, S. Ten-year comparison of the influence of organic and conventional crop management practices on the content of flavonoids in tomatoes. J. Agric. Food Chem. 2007, 55, 6154–6159. [Google Scholar] [CrossRef]
- Haichar, F.E.; Santaella, C.; Heulin, T.; Achouak, W. Root exudates mediated interactions belowground. Soil Biol. Biochem. 2014, 77, 69–80. [Google Scholar] [CrossRef]
- Van Dam, N.M.; Bouwmeester, H.J. Metabolomics in the rhizosphere: Tapping into belowground chemical communication. Trends Plant Sci. 2016, 21, 256–265. [Google Scholar] [CrossRef]
- Adeleke, R.; Nwangburuka, C.; Oboirien, B. Origins, roles and fate of organic acids in soils: A review. S. Afr. J. Bot. 2017, 108, 393–406. [Google Scholar] [CrossRef]
- Baudoin, E.; Benizri, E.; Guckert, A. Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil Biol. Biochem. 2003, 35, 1183–1192. [Google Scholar] [CrossRef]
- Zhu, S.S.; Vivanco, J.M.; Manter, D.K. Nitrogen fertilizer rate affects root exudation, the rhizosphere microbiome and nitrogen-use-efficiency of maize. Appl. Soil Ecol. 2016, 107, 324–333. [Google Scholar] [CrossRef] [Green Version]
- Haase, S.; Neumann, G.; Kania, A.; Kuzyakov, Y.; Römheld, V.; Kandeler, E. Elevation of atmospheric CO2 and N-nutritional status modify nodulation, nodule-carbon supply, and root exudation of Phaseolus vulgaris L. Soil Biol. Biochem. 2007, 39, 2208–2221. [Google Scholar] [CrossRef]
- Carvalhais, L.C.; Dennis, P.G.; Dmitri, F.; Fedoseyenko, D.; Hajirezaei, M.R.; Borriss, R.; von Wiren, N. Root exudation of sugars, amino acids, and organic acids by maize as affected by nitrogen, phosphorus, potassium, and iron deficiency. J. Plant Nutr. Soil Sci. 2011, 174, 3–11. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, R.; Wang, S.; Xiong, B.; Gu, H.; Wang, H.; Ji, C.; Jia, W.; Horowitz, A.R.; Zhen, W.; Asher, J.B.; et al. Application of Bioorganic Fertilizer on Panax notoginseng Improves Plant Growth by Altering the Rhizosphere Microbiome Structure and Metabolism. Microorganisms 2022, 10, 275. https://doi.org/10.3390/microorganisms10020275
Shi R, Wang S, Xiong B, Gu H, Wang H, Ji C, Jia W, Horowitz AR, Zhen W, Asher JB, et al. Application of Bioorganic Fertilizer on Panax notoginseng Improves Plant Growth by Altering the Rhizosphere Microbiome Structure and Metabolism. Microorganisms. 2022; 10(2):275. https://doi.org/10.3390/microorganisms10020275
Chicago/Turabian StyleShi, Rui, Shu Wang, Bingjie Xiong, Haiyan Gu, Huiling Wang, Chao Ji, Weijia Jia, Abraham Rami Horowitz, Wenjie Zhen, Jiftah Ben Asher, and et al. 2022. "Application of Bioorganic Fertilizer on Panax notoginseng Improves Plant Growth by Altering the Rhizosphere Microbiome Structure and Metabolism" Microorganisms 10, no. 2: 275. https://doi.org/10.3390/microorganisms10020275
APA StyleShi, R., Wang, S., Xiong, B., Gu, H., Wang, H., Ji, C., Jia, W., Horowitz, A. R., Zhen, W., Asher, J. B., & He, X. (2022). Application of Bioorganic Fertilizer on Panax notoginseng Improves Plant Growth by Altering the Rhizosphere Microbiome Structure and Metabolism. Microorganisms, 10(2), 275. https://doi.org/10.3390/microorganisms10020275




