Isolation and Characterization of Endophytes Bacterial Strains of Momordica charantia L. and Their Possible Approach in Stress Management
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Bacterial Endophytes
2.2. Isolation from Root Bits
2.3. From Transverse Section of Roots
2.4. Isolation from the Crushed Root
3. Characterization of Bacterial Isolates
Genetic Characterization
4. Plant Growth-Promoting (PGP) Traits Analysis
4.1. Production of IAA
4.2. Phosphate Solubilization
4.3. Siderophore Production
4.4. Carbon and Nitrogen Utilization
4.5. Specific Growth Rate
4.6. Hydrolytic Enzyme Assay
4.7. Antibiotic Sensitivity Test
4.8. Stress Tolerance
5. Statistical Analysis
6. Result
6.1. Diversity of Cultivable Endophytic Bacteria
6.2. Colony Morphology and Biochemical Characteristics
6.3. Phylogenetic Analysis
6.4. Plant Growth Promotion (PGP) Trait Analysis
6.5. Carbon and Nitrogen Source Utilization Pattern
6.6. Specific Growth Rate
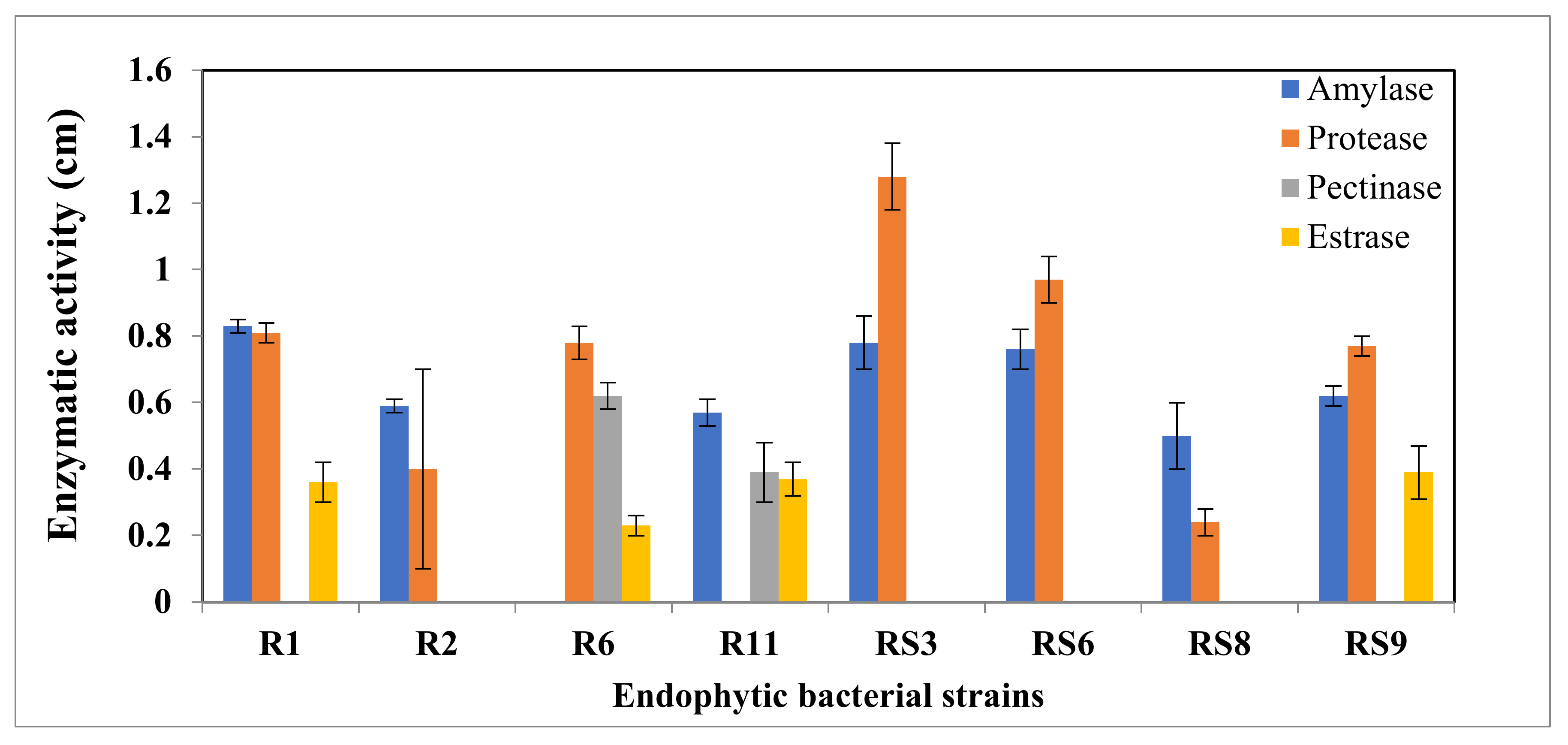
6.7. Hydrolytic Enzyme Assay
6.8. Antibiotic Sensitivity
6.9. Stress Resistance
7. Discussion
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, A.; Zhimo, Y.; Biasi, A.; Salim, S.; Feygenberg, O.; Wisniewski, M.; Droby, S. Endophytic Microbiome in the Carposphere and Its Importance in Fruit Physiology and Pathology. In Postharvest Pathology; Springer: Cham, Switzerland, 2021; pp. 73–88. [Google Scholar] [CrossRef]
- Verma, H.; Kumar, D.; Kumar, V.; Kumari, M.; Singh, S.K.; Sharma, V.K.; Droby, S.; Santoyo, G.; White, J.F.; Kumar, A. The Potential Application of Endophytes in Management of Stress from Drought and Salinity in Crop Plants. Microorganisms 2021, 9, 1729. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Kamrunnahar, A.B.S.; Bhuiyan, M.S.R.; Zeba, N. Morphological characterization, character association and path analysis of bitter gourd (Momordica charantia L.) genotypes. Plant Cell Biotechnol. Mol. Biol. 2021, 22, 53–62. [Google Scholar]
- Jia, S.; Shen, M.; Zhang, F.; Xie, J. Recent Advances in Momordica charantia: Functional Components and Biological Activities. Int. J. Mol. Sci. 2017, 18, 2555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.; Tandon, S.; Semwal, B.; Singh, K. Momordica charantia Linn: A comprehensive review on bitter remedy. J. Pharm. Sci. Opin. 2011, 1, 42–47. [Google Scholar]
- Singh, R.; Kumar, A.; Bhuvaneshwari, K.; Pandey, K.D. Gas Chromatography-Mass Spectrometry Analysis and Phytochemical Screening of Methanolic Fruit Extract of Momordica charantia. J. Recent Adv. Agri. 2012, 1, 122–127. [Google Scholar]
- Yan, J.K.; Wu, L.X.; Qiao, Z.R.; Cai, W.D.; Ma, H. Effect of different drying methods on the product quality and bioactive polysaccharides of bitter gourd (Momordica charantia L.) slices. Food Chem. 2019, 271, 588–596. [Google Scholar] [CrossRef]
- Ozusaglam, M.A.; Karakoca, K. Antimicrobial and antioxidant activities of Momordica charantia from Turkey. Afr. J. Biotechnol. 2013, 12, 1548–1558. [Google Scholar]
- Hallmann, J.; Quadt-Hallmann, A.; Mahaffee, W.F.; Kloepper, J.W. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997, 43, 895–914. [Google Scholar] [CrossRef]
- Kuklinsky-Sobral, J.; Araújo, W.L.; Mendes, R.; Geraldi, I.O.; Pizzirani-Kleiner, A.A.; Azevedo, J.L. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ. Microbiol. 2004, 6, 1244–1251. [Google Scholar] [CrossRef]
- Conrath, U.; Beckers, G.J.; Flors, V.; García-Agustín, P.; Jakab, G.; Mauch, F.; Newman, M.A.; Pieterse, C.M.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting ready for battle. Mol. Plant-Microbe. Interact. 2006, 19, 1062–1071. [Google Scholar] [CrossRef] [Green Version]
- Pillay, V.K.; Nowak, J. Inoculum density, temperature, and genotype effects on in vitro growth promotion and epiphytic and endophytic colonization of tomato (Lycopersicon esculentum L.) seedlings inoculated with a pseudomonad bacterium. Can. J. Microbiol. 1997, 43, 354–361. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, R.; Yadav, A.; Giri, D.D.; Singh, P.K.; Pandey, K.D. Isolation and characterization of bacterial endophytes of Curcuma longa L. 3 Biotech 2016, 6, 60. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Kumar, A.; Pandey, K.D.; Roy, B.K. Isolation and characterization of bacterial endophytes from the roots of Cassia tora L. Ann. Microbiol. 2015, 65, 1391–1399. [Google Scholar] [CrossRef]
- Lodewyckx, C.; Vangronsveld, J.; Porteous, F.; Moore, E.R.; Taghavi, S.; Mezgeay, M.; der Lelie, D.V. Endophytic bacteria and their potential applications. Crit. Rev. Plant Sci. 2002, 21, 583–606. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, M.; Singh, P.P.; Singh, S.K.; Singh, P.K.; Pandey, K.D. Isolation of plant growth-promoting rhizobacteria and their impact on growth and curcumin content in Curcuma longa L. Biocatal. Agric. Biotechnol. 2016, 8, 1–7. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, R.; Giri, D.D.; Singh, P.K.; Pandey, K.D. Effect of Azotobacter chroococcum CL13 inoculation on growth and curcumin content of turmeric (Curcuma longa L.). Int. J. Curr. Microbiol. App. Sci. 2014, 3, 275–283. [Google Scholar]
- Pikovskaya, R.I. Mobilization of phosphorous in soil in connection with the vital activity of some microbial species. Microbiologia 1948, 17, 362–370. [Google Scholar]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Myers, J.; Kratz, W.A. Relations between pigment content and photosynthetic characteristics in a blue-green alga. J. Gen. Physiol. 1955, 39, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Hankin, L.; Anagnostakis, S.L. The use of solid media for detection of enzyme production by fungi. Mycologia 1975, 67, 597–607. [Google Scholar] [CrossRef]
- Vieira, J.D.G. Purificação e caracterização de uma a-amilase de Streptomyces sp. Ph.D. Thesis, University of São Paulo, Saul Palo, Brasil, 1999; p. 116. [Google Scholar]
- Hankin, L.; Zucker, M.; Sands, D.C. Improved solid medium for the detection and enumeration of pectolytic bacteria. Appl. Environ. Microbiol. 1971, 22, 205–209. [Google Scholar] [CrossRef]
- Sierra, G. A simple method for the detection of lipolytic activity of micro-organisms and some observations on the influence of the contact between cells and fatty substrates. Antonie Van Leeuwenhoek 1957, 23, 15–22. [Google Scholar] [CrossRef]
- Shukla, P.N. Diversity and Characteristics of Methylotrophs from a Degraded Coal Mine Area; Banaras Hindu University: Varanasi, India, 2009; p. 34. [Google Scholar]
- Visalakchi, S.; Muthumary, J. Taxol (anticancer drug) producing endophytic fungi: An overview. Int. J. Pharm. Biol. Sci. 2010, 1, 1–9. [Google Scholar]
- Bugbee, W.M.; Cole, D.F.; Nielsen, G. Microflora and invert sugars in juice from healthy tissue of stored sugarbeets. Appl. Environ. Microbiol. 1975, 29, 780–781. [Google Scholar] [CrossRef]
- Gardner, J.M.; Feldman, A.W.; Zablotowicz, R.M. Identity and behavior of xylem-residing bacteria in rough lemon roots of Florida citrus trees. Appl. Environ. Microbiol. 1982, 43, 1335–1342. [Google Scholar] [CrossRef] [Green Version]
- Gagné, S.; Richard, C.; Rousseau, H.; Antoun, H. Xylem-residing bacteria in alfalfa roots. Can. J. Microbiol. 1987, 33, 996–1000. [Google Scholar] [CrossRef]
- Boer, S.D.; Copeman, R.J. Endophytic bacterial flora in Solanum tuberosum and its significance in bacterial ring rot diagnosis. Can. J. Plant Sci. 1974, 54, 115–122. [Google Scholar] [CrossRef]
- Fox, G.E.; Wisotzkey, J.D.; Jurtshuk, P., Jr. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Evol. Microbiol. 1992, 42, 166–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandamme, P.; Pot, B.; Gillis, M.; De Vos, P.; Kersters, K.; Swings, J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Mol. Biol. Rev. 1996, 60, 407–438. [Google Scholar] [CrossRef] [PubMed]
- Lapage, S.P.; Sneath, P.H.A.; Lessel, E.F.; Skerman, V.B.D.; Seeliger, H.P.R.; Clark, W.A. International Code of Nomenclature of Bacteria (1990 Revision); American Society for Microbiology: Washington, DC, USA, 1992. [Google Scholar]
- Stackbrandt, E. Defining taxonomic ranks. In The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackbrandt, E., Eds.; Springer: New York, NY, USA, 2000. [Google Scholar]
- Guo, L.D. A method to promote sporulation in palm endophytic fungi. Fungal Divers. 1998, 1, 109–113. [Google Scholar]
- Rivera, M.H.; López-Munguía, A.; Soberón, X.; Saab-Rincón, G. α-Amylase from Bacillus licheniformis mutants near to the catalytic site: Effects on hydrolytic and transglycosylation activity. Protein Eng. 2003, 16, 505–514. [Google Scholar] [CrossRef] [Green Version]
- Reinhold-Hurek, B.; Hurek, T. Life in grasses: Diazotrophic endophytes. Trend Microbiol. 1998, 6, 139–144. [Google Scholar] [CrossRef]
- Elbeltagy, A.; Nishioka, K.; Suzuki, H.; Sato, T.; Sato, Y.I.; Morisaki, H.; Mitsui, H.; Minamisawa, K. Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. Soil Sci. Plant Nutr. 2000, 46, 617–629. [Google Scholar] [CrossRef]
- Here, T.; Reinhold-Hurek, B.; Van Montagu, M.; Kellenberger, E. Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J. Bacteriol. 1994, 176, 1913–1923. [Google Scholar]
- Quadt-Hallmann, A.; Kloepper, J.W. Immunological detection and localization of the cotton endophyte Enterobacter asburiae JM22 in different plant species. Can. J. Microbiol. 1996, 42, 1144–1154. [Google Scholar] [CrossRef]
- Jung, Y.J.; Lee, J.K.; Sung, C.G.; Oh, T.K.; Kim, H.K. Nonionic detergent-induced activation of an esterase from Bacillus megaterium 20-1. J. Mol. Catal. B Enzym. 2003, 26, 223–229. [Google Scholar] [CrossRef]
- Litzner, B.R.; Caton, T.M.; Schneegurt, M.A. Carbon substrate utilization, antibiotic sensitivity, and numerical taxonomy of bacterial isolates from the Great Salt Plains of Oklahoma. Arch. Microbiol. 2006, 185, 286–296. [Google Scholar] [CrossRef]
- Le, T.H.; Ng, C.; Chen, H.; Yi, X.Z.; Koh, T.H.; Barkham, T.M.S.; Zhou, Z.; Gin, K.Y.H. Occurrences and characterization of antibiotic-resistant bacteria and genetic determinants of hospital wastewater in a tropical country. Antimicrob. Agents Chemother. 2016, 60, 7449–7456. [Google Scholar] [CrossRef] [Green Version]
- Sziderics, A.H.; Rasche, F.; Trognitz, F.; Sessitsch, A.; Wilhelm, E. Bacterial endophytes contribute to abiotic stress adaptation in pepper plants (Capsicum annuum L.). Can. J. Microbiol. 2007, 53, 1195–1202. [Google Scholar] [CrossRef]
- Arora, S.; Patel, P.N.; Vanza, M.J.; Rao, G.G. Isolation and characterization of endophytic bacteria colonizing halophyte and other salt-tolerant plant species from coastal Gujarat. Afr. J. Microbiol. Res. 2014, 8, 1779–1788. [Google Scholar]



| Characteristics | R1 | R2 | R6 | R11 | RS3 | RS6 | RS8 | RS9 |
|---|---|---|---|---|---|---|---|---|
| Cell Shape | R | R | R | R | R | R | C | R |
| Gram’s Reaction | + | + | − | − | + | + | − | + |
| Motility | + | + | + | + | − | + | + | + |
| Oxidase | − | + | + | − | − | + | + | − |
| Citrate | + | + | + | + | − | + | + | + |
| Indole | − | − | − | − | − | − | − | − |
| H2S production | − | − | − | − | − | − | − | − |
| Phenylalanine deaminase | − | − | − | − | − | − | − | − |
| Carbohydrate fermentation | + | + | + | − | − | + | − | − |
| Starch hydrolysis | − | + | + | − | + | + | + | + |
| Urease | − | − | − | − | + | − | − | − |
| Nitrate reductase | − | + | + | + | + | + | + | − |
| Catalase | + | + | + | + | + | + | + | + |
| S.N | Strains | Accession Number | Nearest Phylogenetic Neighbour | Phylogenetic Domain |
|---|---|---|---|---|
| 1. | R1 | JX500429 | Bacillus licheniformis strain YC3-C (HQ698961.1) | Firmicutes |
| 2. | R2 | JX472914 | Bacillus sp. NM03 (JX303661.1) | Firmicutes |
| 3. | R6 | JX472915 | Agrobacterium tumefaciens strain X9, (JX002661) | β-proteobacteria |
| 4. | R11 | JX472917 | Uncultured bacterium clone KWB120 (JX047140) | Uncultured bacteria clone |
| 5. | RS3 | JX472920 | Bacillus subtilis strain BVC21 (JQ660604) | Firmicutes |
| 6. | RS6 | JX472923 | Bacillus subtilis strain N11 (JQ973708) | Firmicutes |
| 7. | RS8 | JX472924 | Uncultured bacterium clone BULK-114 (JN161941) | Uncultured bacterial strain |
| 8. | RS9 | JX472925 | Lysinibacillus fusiformis strain TL (JQ991004) | Firmicutes |
| Carbon Source | R1 | R2 | R6 | R11 | RS3 | RS6 | RS8 | RS9 |
|---|---|---|---|---|---|---|---|---|
| Glucose | + | + | + | + | + | + | + | + |
| Sucrose | + | + | + | + | + | + | + | + |
| Sodium citrate | + | + | + | + | − | − | + | − |
| Sodium acetate | − | − | + | − | − | − | + | − |
| Sodium formate | − | − | + | − | − | − | + | − |
| Mannitol | − | − | + | − | − | − | + | − |
| Malic acid | − | − | − | + | − | − | − | − |
| Methanol | − | − | − | + | − | − | + | + |
| Nitrogen Source | ||||||||
| Yeast extract | + | + | + | + | + | + | + | + |
| Potassium Nitrate | + | + | + | + | + | + | − | + |
| Sodium Nitrite | + | − | + | − | + | + | − | + |
| Ammonium acetate | + | − | + | − | + | + | + | + |
| Ammonium sulphate | + | + | + | + | + | + | + | + |
| Ammonium chloride | + | + | + | + | + | + | + | + |
| Alanine | + | + | + | + | + | + | + | − |
| Lysine | + | + | + | + | + | + | + | − |
| Glycine | + | + | + | + | + | + | + | + |
| Glutamine | + | + | + | + | + | + | + | + |
| Isoleucine | + | + | + | + | + | + | + | + |
| Arginine | − | + | + | + | + | + | + | + |
| Cysteine | + | + | + | − | + | + | + | + |
| Aspartic acid | − | + | + | − | + | − | − | − |
| Glutamic acid | − | + | + | − | + | − | − | − |
| Proline | + | + | + | + | + | + | − | − |
| Strains | Antibiotic Sensitivity (Antibiotics Inhibition Zone in mm) | |||||
|---|---|---|---|---|---|---|
| Chloramphenicol (30 mcg/disc) | Polymixin-B (300 unit/disc) | Erythromycin (15 mcg/disc) | Rifampicin (5 mcg/disc) | Spectinomycin (30 mcg/disc) | Kannamycin (10 mcg/disc) | |
| R1 | 10.3 ± 0.05 a(I) | 14.6 ± 1.5 a(I) | 12.6 ± 1.5 a(I) | 19 ± 1 a(S) | 40.6 ± 3.2 a(S) | 23 ± 1 a (S) |
| R2 | 42.6 ± 1.15 b(S) | 11 ± 1 a (I) | 26.3 ± 2.08 b(S) | R | 30 ± 1 b (S) | 37 ± 2 b (S) |
| R6 | 28 ± 2 c(S) | 13 ± 1 a(I) | 19 ± 1 b(S) | 28 ± 2.6 b(S) | 23.6 ± 1.5 c(S) | 22.6 ± 1.5 a(S) |
| R11 | 26.6 ± 1.5 c(S) | R | R | 10 ± 1 c(I) | 10 ± 1 d(I) | 24 ± 1 a (S) |
| RS3 | R | 12 a (I) | 15 ± 1 a(S) | 11 ± 1 c(I) | 30.6 ± 1.5 b(S) | 30 ± 1 c (S) |
| RS6 | R | 9.6 ± 2 a(I) | 24 ± 1 b(S) | 17.6 ± 1.5 d(S) | 20 ± 1 c(S) | 26.6 ± 1.15 a |
| RS8 | 39 ± 2 d(S) | 12.6 ± 0.5 a(I) | 39.73 c(S) | 16 ± 2 d(S) | 43 ± 2 a(S) | 40 ± 2.64 b (S) |
| RS9 | 26.3 ± 1.5 e(S) | 10 ± 1.54 a(I) | R | 14 ± 2 d(I) | 23 ± 1.7 c(S) | R |
| Strains | concn | R1 | R2 | R6 | R11 | RS3 | RS6 | RS8 | RS9 |
|---|---|---|---|---|---|---|---|---|---|
| NaCl | 4% | + | + | + | + | + | + | + | + |
| 6% | + | − | − | + | + | + | + | − | |
| 8% | + | − | − | − | + | + | − | − | |
| 10% | + | − | − | − | − | + | − | − | |
| 12% | − | − | − | − | − | − | − | − | |
| KOH | 3% | − | − | − | − | − | − | − | − |
| NaN3 | 0.02% | + | − | − | − | + | + | + | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, R.; Pandey, K.D.; Singh, M.; Singh, S.K.; Hashem, A.; Al-Arjani, A.-B.F.; Abd_Allah, E.F.; Singh, P.K.; Kumar, A. Isolation and Characterization of Endophytes Bacterial Strains of Momordica charantia L. and Their Possible Approach in Stress Management. Microorganisms 2022, 10, 290. https://doi.org/10.3390/microorganisms10020290
Singh R, Pandey KD, Singh M, Singh SK, Hashem A, Al-Arjani A-BF, Abd_Allah EF, Singh PK, Kumar A. Isolation and Characterization of Endophytes Bacterial Strains of Momordica charantia L. and Their Possible Approach in Stress Management. Microorganisms. 2022; 10(2):290. https://doi.org/10.3390/microorganisms10020290
Chicago/Turabian StyleSingh, Ritu, Kapil Deo Pandey, Monika Singh, Sandeep Kumar Singh, Abeer Hashem, Al-Bandari Fahad Al-Arjani, Elsayed Fathi Abd_Allah, Prashant Kumar Singh, and Ajay Kumar. 2022. "Isolation and Characterization of Endophytes Bacterial Strains of Momordica charantia L. and Their Possible Approach in Stress Management" Microorganisms 10, no. 2: 290. https://doi.org/10.3390/microorganisms10020290
APA StyleSingh, R., Pandey, K. D., Singh, M., Singh, S. K., Hashem, A., Al-Arjani, A.-B. F., Abd_Allah, E. F., Singh, P. K., & Kumar, A. (2022). Isolation and Characterization of Endophytes Bacterial Strains of Momordica charantia L. and Their Possible Approach in Stress Management. Microorganisms, 10(2), 290. https://doi.org/10.3390/microorganisms10020290









