The Identification of Multidrug-Resistant Microorganisms including Bergeyella zoohelcum Acquired from the Skin/Prosthetic Interface of Amputees and Their Susceptibility to Medihoney™ and Garlic Extract (Allicin)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Swab Collection
2.2. Isolate Recovery
2.3. Microbiological Identification Methods
2.4. Bacterial Standardisation
2.5. Preparation of Allicin (Garlic Extract)
2.6. Preparation of Medihoney™ Stock
2.7. Bacterial Inhibition Assays
2.7.1. Determination of Minimum Inhibitory Concentration with Allicin
2.7.2. Determination of Minimum Inhibitory Concentration with Manuka Honey
2.8. Bacterial Inoculation
Determination of Minimum Bactericidal Concentration
2.9. Antibiotic Sensitivity Testing
3. Results
3.1. Participants
3.2. Bacterial Identification
3.2.1. Cultural Appearance
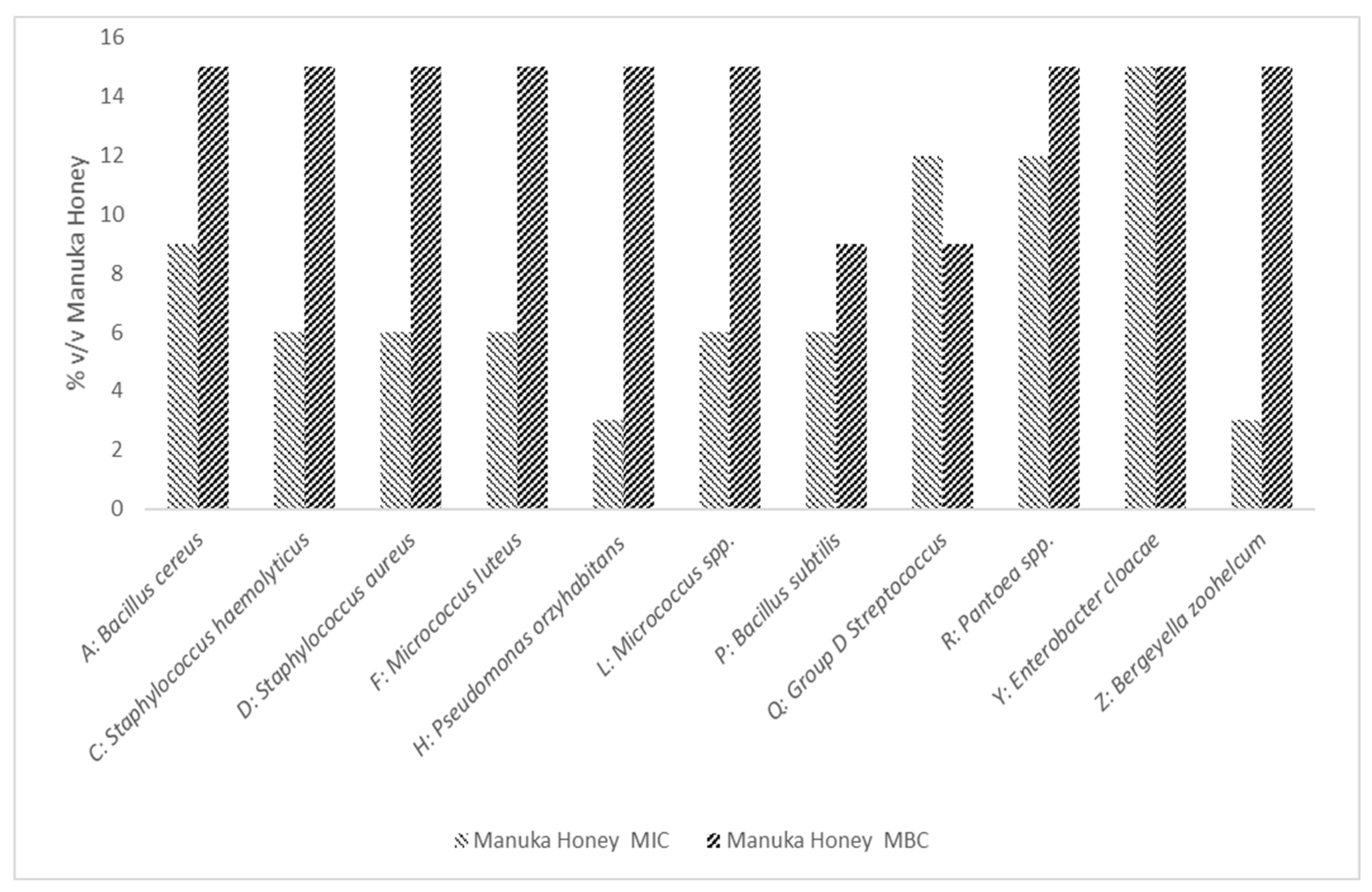
3.2.2. MIC and MBC Determination of Manuka Honey
3.2.3. MIC and MBC Determination of Allicin
3.3. Antibiotic Susceptibility
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Diabetes UK. 26,378 Diabetes-Related Lower Limb Amputations in the Last Three Years. 2018. Available online: https://www.diabetes.org.uk/about_us/news/lower-limb-amputations (accessed on 9 August 2021).
- World Health Organization (WHO). Guidelines for Training Personnel in Developing countries for Prosthetics and Orthotics Services. 2005. Available online: https://apps.who.int/iris/handle/10665/43127 (accessed on 7 September 2020).
- Frölke, J.P.; van de Meent, H. The endo-exo prosthesis for patients with a problematic amputation stump. Ned. Tijdschr. Geneeskd. 2010, 154, A2010. [Google Scholar] [PubMed]
- Gholizadeh, H.; Abu Osman, N.A.; Eshraghi, A.; Ali, S.; Yahyavi, E.S. Satisfaction and Problems Experienced with Transfemoral Suspension Systems: A Comparison Between Common Suction Socket and Seal-In Liner. Arch. Phys. Med. Rehabil. 2013, 94, 1584–1589. [Google Scholar] [CrossRef] [PubMed]
- Beyaz, S.; Güler, Ü.Ö.; Bağır, G.Ş. Factors affecting lifespan following below-knee amputation in diabetic patients. Acta Orthop. Traumatol. Turc. 2017, 51, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Z.M.; Ng, N.S.L.; Thomas, C. Prevention and treatment of diabetic foot ulcers. J. R. Soc. Med. 2017, 110, 104–109. [Google Scholar] [CrossRef] [Green Version]
- Weledji, E.P.; Fokam, P. Treatment of the diabetic foot—To amputate or not? BMC Surg. 2014, 14, 83. [Google Scholar] [CrossRef] [Green Version]
- Ghoseiri, K.; Zheng, Y.P.; Leung, A.K.; Rahgozar, M.; Aminian, G.; Lee, T.H.; Safari, M.R. Temperature measurement and control system for transtibial prostheses: Functional evaluation. Assist. Technol. 2018, 30, 16–23. [Google Scholar] [CrossRef]
- Cooper, S. Bacterial Growth and Division: Biochemistry and Regulation of Prokaryotic and Eukaryotic Division Cycles; Elsevier: Amsterdam, The Netherlands, 2012; p. 528. [Google Scholar]
- OPEDGE.COM. Responses to Amputee Odor Question. Available online: https://opedge.com/OANDPL/ViewMessage/6539CC54-6905-428D-9A4B-982C9E674B37 (accessed on 25 June 2020).
- Campbell, L.; Zirwas, M.J. Triclosan. Dermatitis 2006, 17, 204–207. [Google Scholar]
- Westfall, C.; Flores-Mireles, A.L.; Robinson, J.I.; Lynch, A.J.; Hultgren, S.; Henderson, J.P.; Levin, P.A. The Widely Used Antimicrobial Triclosan Induces High Levels of Antibiotic Tolerance In Vitro and Reduces Antibiotic Efficacy up to 100-Fold In Vivo. Antimicrob. Agents Chemother. 2019, 63, e02312–e02318. [Google Scholar] [CrossRef] [Green Version]
- Carey, D.E.; McNamara, P.J. The impact of triclosan on the spread of antibiotic resistance in the environment. Front. Microbiol. 2015, 5, 780. [Google Scholar] [CrossRef] [Green Version]
- Pattamayutanon, P.; Angeli, S.; Thakeow, P.; Abraham, J.; Disayathanoowat, T.; Chantawannakul, P. Biomedical Activity and Related Volatile Compounds of Thai Honeys from 3 Different Honeybee Species. J. Food Sci. 2015, 80, M2228-40. [Google Scholar] [CrossRef]
- Comvita® UK. The Immense Benefits of Medihoney. Available online: https://www.comvita.co.uk/ingredients/manuka-honey/the-science-behind-manuka-honey.list?gclid=EAIaIQobChMI4peeyabP9QIVFIfVCh2JywcBEAAYASACEgIUwPD_BwE&gclsrc=aw.ds (accessed on 26 October 2020).
- Robson, V.; Dodd, S.; Thomas, S. Standardized antibacterial honey (MedihoneyTM) with standard therapy in wound care: Randomized clinical trial. J. Adv. Nurs. 2009, 65, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Biglari, B.; Linden, P.H.; Simon, A.; Aytac, S.; Gerner, H.J.; Moghaddam, A. Use of Medihoney as a non-surgical therapy for chronic pressure ulcers in patients with spinal cord injury. Spinal Cord. 2012, 50, 165–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alhashim, M.; Lombardo, J. Mechanism of Action of Topical Garlic on Wound Healing. Dermatol. Surg. 2018, 44, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.; Levina, N.; van der Linden, M.; Gruhlke, M.; Martin, C.; Slusarenko, A.J. Diallylthiosulfinate (Allicin), a Volatile Antimicrobial from Garlic (Allium sativum), Kills Human Lung Pathogenic Bacteria, Including MDR Strains, as a Vapor. Molecules 2017, 22, 1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, L.; Bazin, T.; Truchetet, M.-E.; Schaeverbeke, T.; Delhaes, L.; Pradeu, T. Protective Microbiota: From Localized to Long-Reaching Co-Immunity. Front. Immunol. 2017, 8, 1678. [Google Scholar] [CrossRef]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef] [Green Version]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [Green Version]
- Stevens, D.L.; Bryant, A.E. Impetigo, Erysipelas and Cellulitis. In Streptococcus pyogenes: Basic Biology to Clinical Manifestations; Ferretti, J.J., Stevens, D.L., Fischetti, V.A., Eds.; University of Oklahoma Health Sciences Center: Oklahoma City, OK, USA, 2016. [Google Scholar]
- Kühbacher, A.; Burger-Kentischer, A.; Rupp, S. Interaction of Candida Species with the Skin. Microorganisms 2017, 5, 32. [Google Scholar] [CrossRef] [Green Version]
- Rajkumari, N.; Mathur, P.; Misra, M.C. Soft Tissue and Wound Infections Due to Enterococcus spp. Among Hospitalized Trauma Patients in a Developing Country. J. Glob. Infect. Dis. 2014, 6, 189–193. [Google Scholar]
- George, E.K.; De Jesus, O.; Vivekanandan, R. Clostridium Tetani; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Bottone, E.J. Bacillus cereus, a Volatile Human Pathogen. Clin. Microbiol. Rev. 2010, 23, 382–398. [Google Scholar] [CrossRef] [Green Version]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Public Health England. UK Standards for Microbiology Investigations: Introduction to the Preliminary Identification of Med-ically Important Bacteria and Fungi from Culture Standards Unit. 2017. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1020471/ID_1i3.pdf (accessed on 30 May 2019).
- Public Health England. UK Standards for Microbiology Investigations: Staining Procedures. 2019. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/802769/TP_39i3.pdf (accessed on 30 May 2019).
- Public Health England. UK Standards for Microbiology Investigations: Catalase Test 2019. Available online: https://www.gov.uk/government/publications/smi-tp-8-catalase-test (accessed on 30 May 2020).
- Public Health England. UK Standards for Microbiology Investigations: Oxidase Test. 2019. Available online: https://www.gov.uk/government/publications/smi-tp-26-oxidase-test (accessed on 20 July 2020).
- Public Health England. UK Standards for Microbiology Investigations: Inoculation of Culture Media for Bacteriology. 2017. Available online: https://www.gov.uk/government/publications/smi-q-5-inoculation-of-culture-media-for-bacteriology (accessed on 20 July 2020).
- Jung, B.; Hoilat, G.J. MacConkey Medium; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Payment, P.; Coffin, E.; Paquette, G. Blood agar to detect virulence factors in tap water heterotrophic bacteria. Appl. Environ. Microbiol. 1994, 60, 1179–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Griethuysen, A.; Bes, M.; Etienne, J.; Zbinden, R.; Kluytmans, J. International Multicenter Evaluation of Latex Agglutination Tests for Identification of Staphylococcus aureus. J. Clin. Microbiol. 2001, 39, 86–89. [Google Scholar] [CrossRef] [Green Version]
- Thermo Fisher Scientific. Remel. Staphaurex. Available online: https://www.fishersci.co.uk/shop/products/remel-staphaurex-diagnostic-tests/p-4529287 (accessed on 20 August 2021).
- Ayeni, F.A.; Andersen, C.; Nørskov-Lauritsen, N. Comparison of growth on mannitol salt agar, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, VITEK® 2 with partial sequencing of 16S rRNA gene for identification of coagulase-negative staphylococci. Microb. Pathog. 2017, 105, 255–259. [Google Scholar] [CrossRef]
- Bixler-Forell, E.; Martin, W.J.; Moody, M.D. Clinical evaluation of the improved streptex method for grouping streptococci. Diagn. Microbiol. Infect. Dis. 1984, 2, 113–118. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific. Remel. Streptex Rapid Remel Europe. 2016. Available online: https://www.thermofisher.com/order/catalog/product/R30950555 (accessed on 12 July 2020).
- BioMérieux. API Staph 2013. Available online: https://www.mediray.co.nz/media/15784/om_biomerieux_test-kits_ot-20500_package_insert-20500.pdf (accessed on 12 July 2020).
- Kloos, W.E.; Wolfshohl, J.F. Identification of Staphylococcus species with the API STAPH-IDENT system. J. Clin. Microbiol. 1982, 16, 509–516. [Google Scholar] [CrossRef] [Green Version]
- BioMérieux. API 20 E 2002. Available online: http://biomanufacturing.org/uploads/files/587872707301898351-api20einstructions.pdf (accessed on 14 July 2020).
- Holmes, B.; Willcox, W.R.; Lapage, S.P. Identification of Enterobacteriaceae by the API 20E system. J. Clin. Pathol. 1978, 31, 22–30. [Google Scholar] [CrossRef]
- BioMérieux. API Candida 2011. Available online: https://www.mediray.co.nz/media/15780/om_biomerieux_test-kits_ot-10500_package_insert_-10500.pdf (accessed on 3 August 2021).
- Campbell, C.K.; Davey, K.G.; Holmes, A.D.; Szekely, A.; Warnock, D.W. Comparison of the API Candida System with the AUXACOLOR System for Identification of Common Yeast Pathogens. J. Clin. Microbiol. 1999, 37, 821–823. [Google Scholar] [CrossRef] [Green Version]
- BD Advancing the World of Health. BBL CrystalTM Identification Systems Gram-Positive ID Kit 2002. Available online: https://legacy.bd.com/ds/technicalCenter/clsi/clsi-Crysgp.pdf (accessed on 20 July 2020).
- BD Advancing the World of Health. BBL CrystalTM Identification Systems Enteric/Nonfermenter ID Kit 2001. Available online: https://legacy.bd.com/ds/technicalCenter/clsi/clsi-CrysE_nf.pdf (accessed on 20 July 2020).
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol; American Society for Microbiology: Washington, DC, USA, 2009. [Google Scholar]
- Dutkiewicz, J.; Mackiewicz, B.; Kinga Lemieszek, M.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part III. Deleterious effects: Infections of humans, animals and plants. Ann. Agric. Environ. Med. 2016, 23, 197–205. [Google Scholar] [CrossRef]
- Geng, L.; Xu, M.; Yu, L.; Li, J.; Zhou, Y.; Wang, Y.; Chen, J. Risk factors and the clinical and surgical features of fungal prosthetic joint infections: A retrospective analysis of eight cases. Exp. Ther. Med. 2016, 12, 991–999. [Google Scholar] [CrossRef] [Green Version]
- O&P Virtual Library. 26: Skin Problems of the Amputee. Available online: http://www.oandplibrary.org/alp/chap26-01.asp (accessed on 4 November 2020).
- Davin-Regli, A.; Pagès, J.-M. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 2015, 18, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okwundu, N.; Mercer, J. Pantoea agglomerans cutaneous infection. J. Dermatol. Dermatol. Surg. 2019, 23, 41. [Google Scholar] [CrossRef]
- Tena, D.; Fernández, C. Pseudomonas oryzihabitans: An unusual cause of skin and soft tissue infection. Infect. Dis. 2015, 47, 820–824. [Google Scholar]
- Barros, E.M.; Ceotto, H.; Bastos, M.C.F.; dos Santos, K.R.N.; Giambiagi-deMarval, M. Staphylococcus haemolyticus as an Important Hospital Pathogen and Carrier of Methicillin Resistance Genes. J. Clin. Microbiol. 2012, 50, 166–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbull, P. Medical Microbiology, 4th ed.; NCBI Bookshelf; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Bowler, P.G.; Duerden, B.I.; Armstrong, D.G. Wound Microbiology and Associated Approaches to Wound Management. Clin. Microbiol. Rev. 2001, 14, 244–269. [Google Scholar] [CrossRef] [Green Version]
- Russell, A.D. 4-Mechanisms of bacterial resistance to antibiotics and biocides. In Progress in Medicinal Chemistry; Ellis, G.P., Luscombe, D.K., Oxford, A.W., Eds.; Elsevier: Amsterdam, The Netherlands, 1998; pp. 133–197. [Google Scholar]
- Lin, W.-R.; Chen, Y.-S.; Liu, Y.-C. Cellulitis and Bacteremia Caused by Bergeyella zoohelicum. J. Formos. Med. Assoc. 2007, 106, 573–576. [Google Scholar] [CrossRef]
- Muramatsu, Y.; Haraya, N.; Horie, K.; Uchida, L.; Kooriyama, T.; Suzuki, A.; Horiuchi, M. Bergeyella zoohelicum isolated from oral cavities of therapy dogs. Zoonoses Public Health 2019, 66, 936–942. [Google Scholar] [CrossRef]
- Sharma, S.; Salazar, H.; Sharma, S.; Nasser, M.F.; Dahdouh, M. Bergeyella zoohelicum Bacteremia from Therapy Dog Kisses. Cureus 2019, 11, e4494. [Google Scholar]
- Goldstein, E.J.C.; Citron, D.M.; Merriam, C.V.; Warren, Y.; Tyrell, K. Comparative In Vitro Activities of GAR-936 against Aerobic and Anaerobic Animal and Human Bite Wound Pathogens. Anitmicrobial. Agents Chemother. 2000, 44, 2747–2751. [Google Scholar] [CrossRef] [Green Version]
- Carter, D.A.; Blair, S.E.; Cokcetin, N.N.; Bouzo, D.; Brooks, P.; Schothauer, R.; Harry, E.J. Therapeutic Manuka Honey: No Longer So Alternative. Front. Microbiol. 2016, 7, 569. [Google Scholar] [CrossRef] [Green Version]
- Shukla, S.K.; Paustian, D.L.; Stockwell, P.J.; Morey, R.E.; Jordan, J.G.; Levett, P.N.; Frank, D.N.; Reed, K.D. Isolation of a Fastidious bergeyella Species Associated with Cellulitis after a Cat Bite and a Phylogenetic Comparison with Bergeyella zoohelicum Strains. J. Clin. Microbiol. 2004, 42, 290–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Isolate ID | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Susceptibility Profile | B. cereus | S. haemolyticus | S. aureus | M. luteus | Micrococcus spp. | B. subtilis | Group D Streptococcus | P. orzyhabitans | Pantoea spp. | E. cloacae | B. zoohelcum |
| Penicillin (1 unit) | R | R | R | R | R | R | R | R | R | R | R |
| Erythromycin (10 ug) | S | S | S | S | S | S | S | S | R | R | R |
| Ampicillin (10 ug) | R | R | R | R | R | R | R | R | R | R | R |
| Vancomycin (30 ug) | S | S | S | S | S | S | S | S | R | R | S |
| Ceftazidime (10 μg) | R | R | R | R | R | R | R | R | S | S | R |
| Ciprofloxacin (10 μg) | S | S | S | S | S | S | S | S | S | S | S |
| Gentamicin (10 μg) | S | S | S | S | S | S | S | S | S | S | R |
| Colistin-sulphate (50 μg) | S | S | S | S | S | S | S | S | S | S | S |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harsent, R.; Macleod, J.; Rowlands, R.S.; Smith, P.M.; Rushmere, N.; Blaxland, J. The Identification of Multidrug-Resistant Microorganisms including Bergeyella zoohelcum Acquired from the Skin/Prosthetic Interface of Amputees and Their Susceptibility to Medihoney™ and Garlic Extract (Allicin). Microorganisms 2022, 10, 299. https://doi.org/10.3390/microorganisms10020299
Harsent R, Macleod J, Rowlands RS, Smith PM, Rushmere N, Blaxland J. The Identification of Multidrug-Resistant Microorganisms including Bergeyella zoohelcum Acquired from the Skin/Prosthetic Interface of Amputees and Their Susceptibility to Medihoney™ and Garlic Extract (Allicin). Microorganisms. 2022; 10(2):299. https://doi.org/10.3390/microorganisms10020299
Chicago/Turabian StyleHarsent, Ruby, Joshua Macleod, Richard S. Rowlands, Paul M. Smith, Neil Rushmere, and James Blaxland. 2022. "The Identification of Multidrug-Resistant Microorganisms including Bergeyella zoohelcum Acquired from the Skin/Prosthetic Interface of Amputees and Their Susceptibility to Medihoney™ and Garlic Extract (Allicin)" Microorganisms 10, no. 2: 299. https://doi.org/10.3390/microorganisms10020299






