Diatom Biogeography, Temporal Dynamics, and Links to Bacterioplankton across Seven Oceanographic Time-Series Sites Spanning the Australian Continent
Abstract
:1. Introduction
2. Methods
2.1. Sampling Sites, Collection and Processing
2.2. DNA Sequence Processing
2.3. Statistical Analysis
3. Results
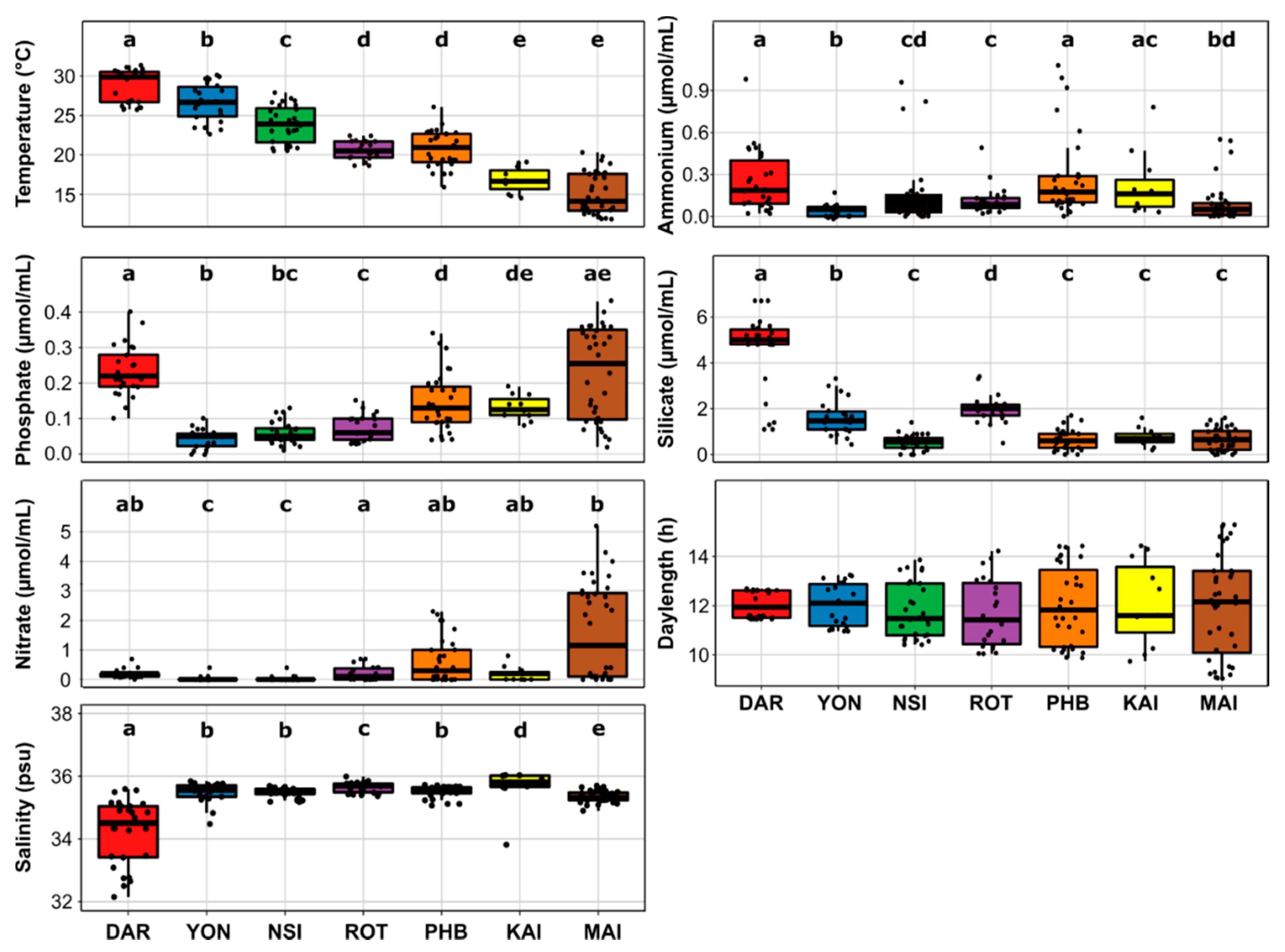
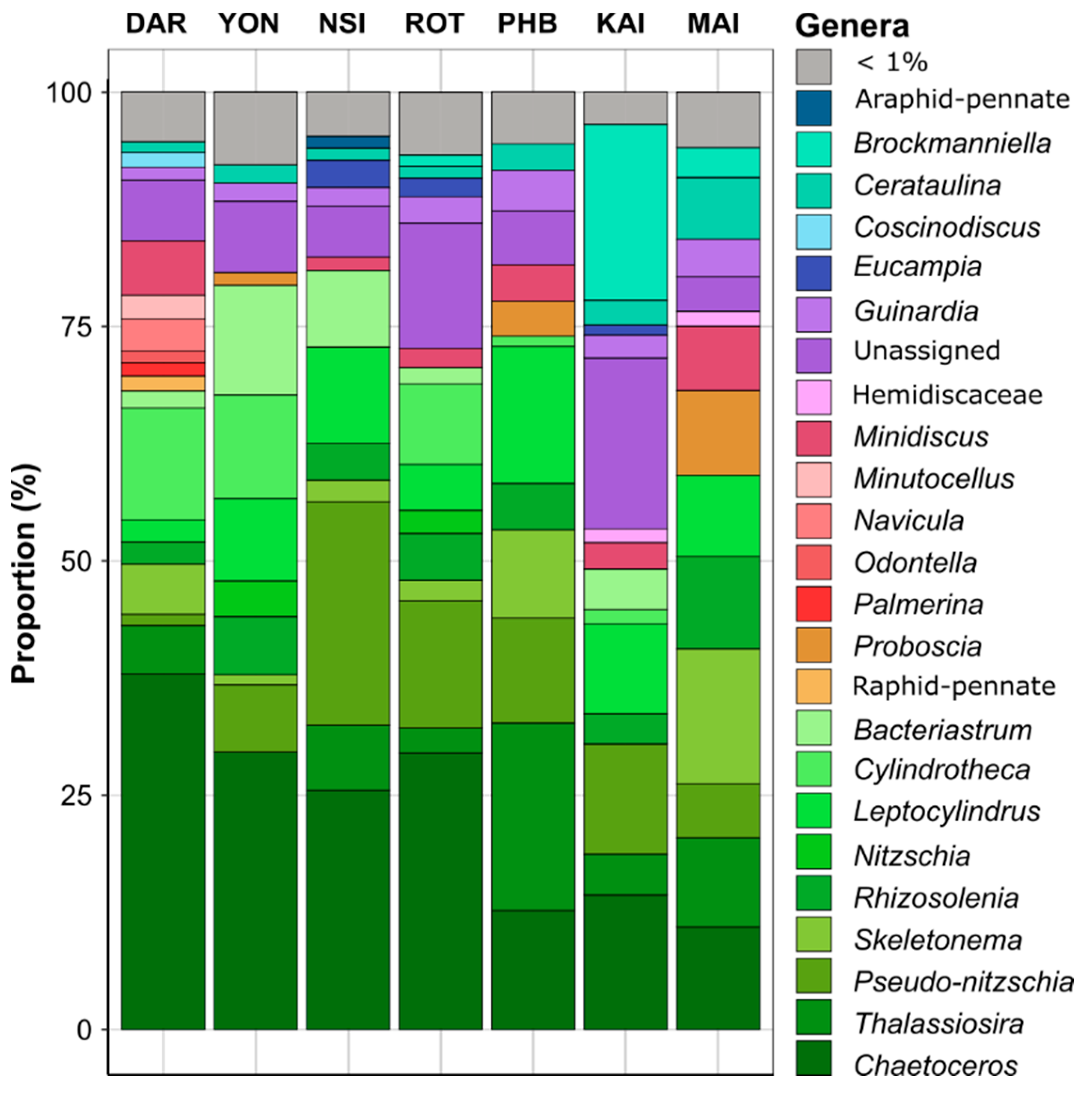
3.1. Regionally Discrete Environmental Patterns and Diatom Communities
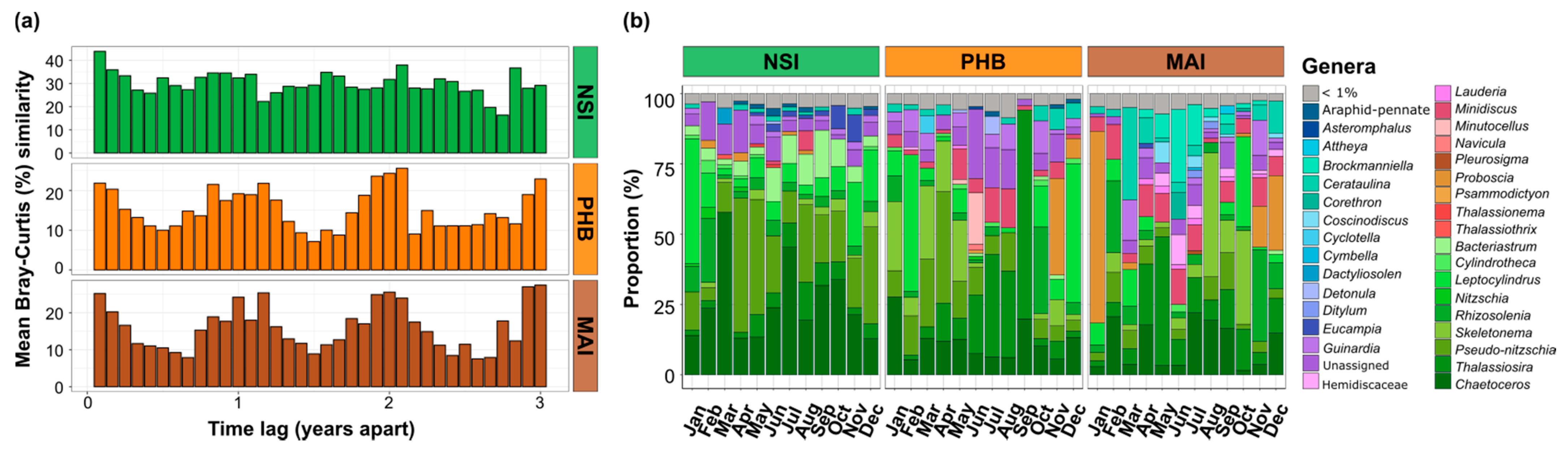
3.2. Diatom Communities Exhibit Dissimilar Levels of Seasonality According to Location
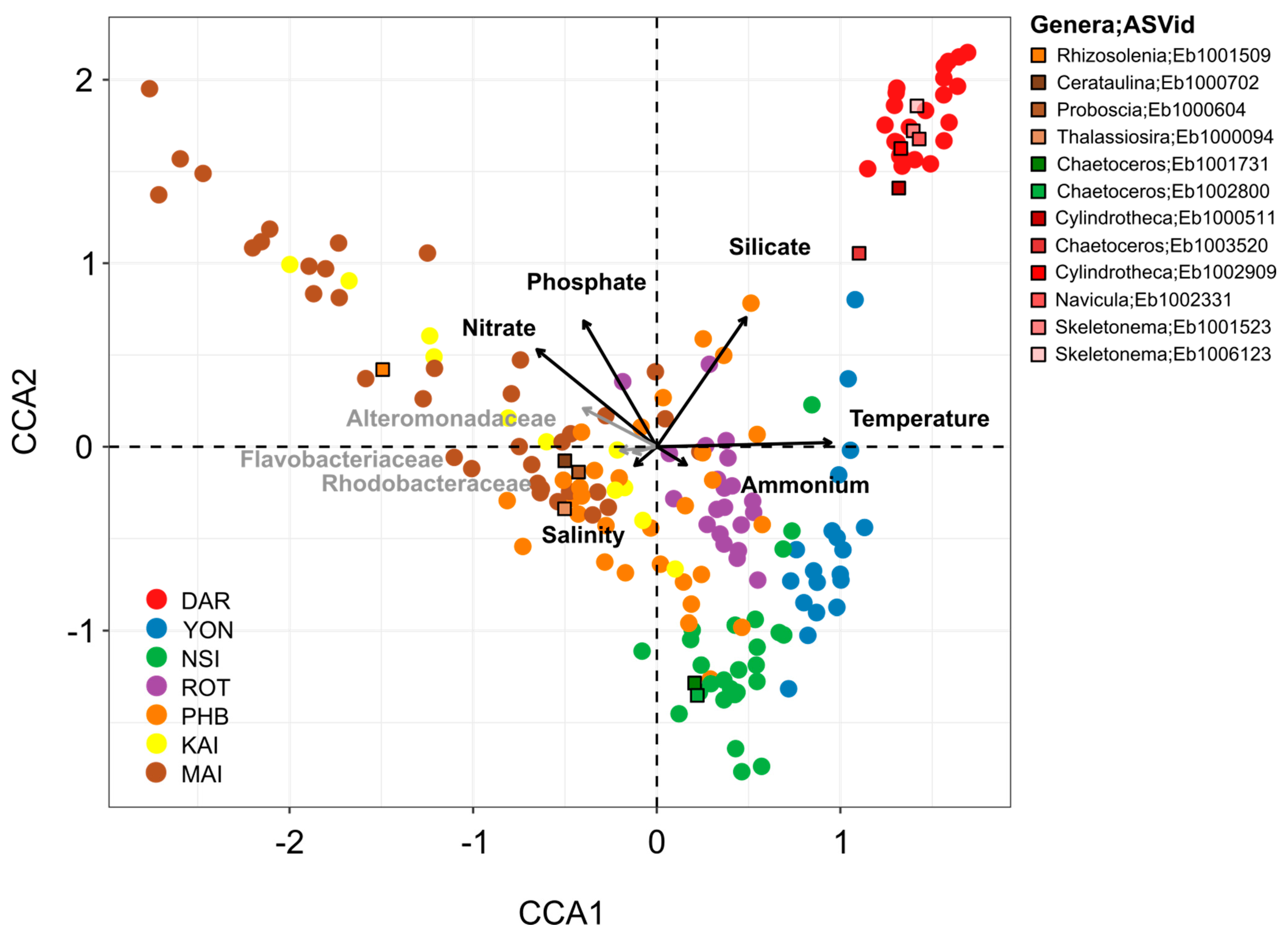
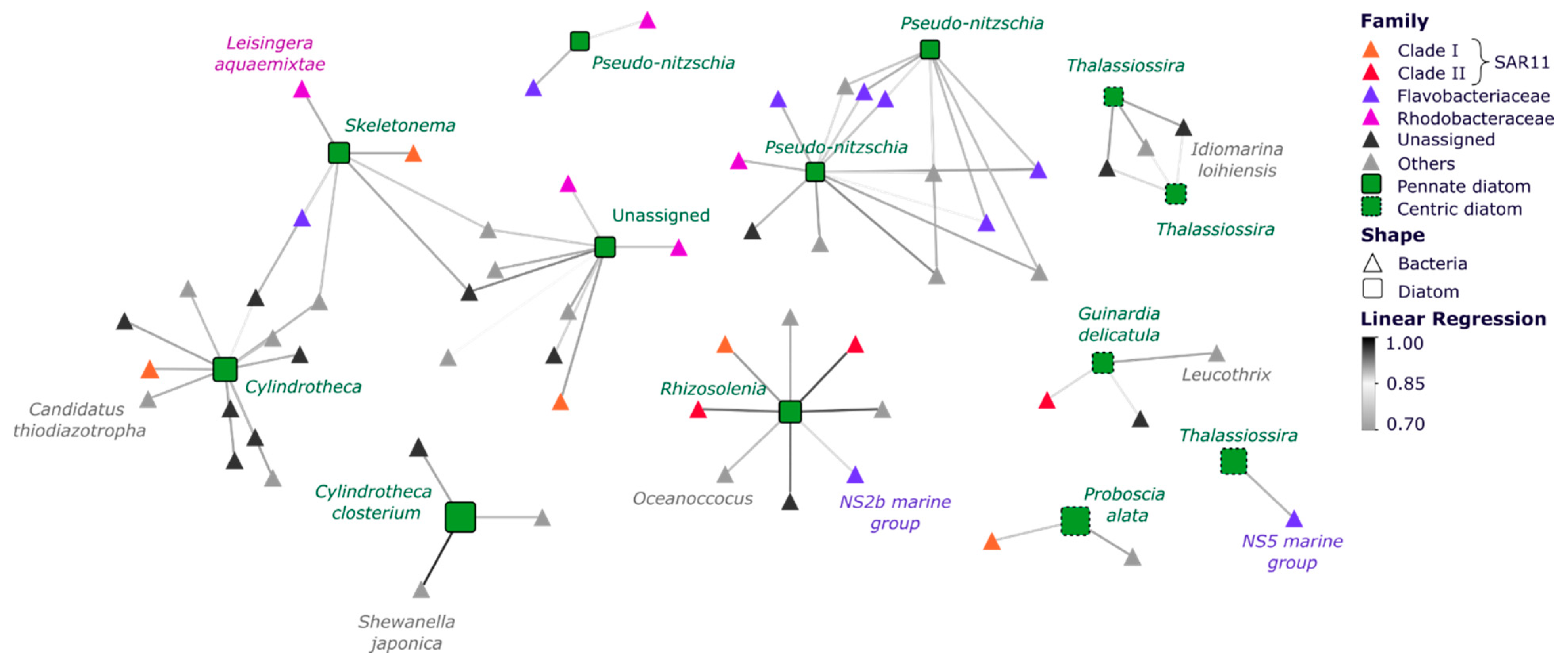
3.3. Specific Bacterial Taxa Are Strongly Correlated with Diatoms across Space and Time
4. Discussion
4.1. Environmental and Bacterial Drivers of Continental-Wide Diatom Spatial Distribution
4.2. Bacteria Contribute to Seasonal Variability of Diatom Communities
4.3. Biological Factors Influence Diatom Community Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saade, A.; Bowler, C. Molecular tools for discovering the secrets of diatoms. Bioscience 2009, 59, 757–765. [Google Scholar] [CrossRef] [Green Version]
- Falkowski, P.G.; Barber, R.T.; Smetacek, V. Biogeochemical controls and feedbacks on ocean primary production. Science 1998, 281, 200–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Field, C.B.; Behrenfeld, M.J.; Randerson, J.T. Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauly, D.; Christensen, V. Primary production required to sustain global fisheries. Nature 1995, 374, 255–257. [Google Scholar] [CrossRef]
- Armbrust, E.V. The life of diatoms in the world’s oceans. Nature 2009, 459, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Malviya, S.; Scalco, E.; Audic, S.; Vincent, F.; Veluchamy, A.; Poulain, J.; Wincker, P.; Iudicone, D.; De Vargas, C.; Bittner, L.; et al. Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. USA 2016, 113, E1516–E1525. [Google Scholar] [CrossRef] [Green Version]
- Pierella Karlusich, J.J.; Ibarbalz, F.M.; Bowler, C. Phytoplankton in the Tara Ocean. Ann. Rev. Mar. Sci. 2020, 12, 233–265. [Google Scholar] [CrossRef] [Green Version]
- Tréguer, P.; Bowler, C.; Moriceau, B.; Dutkiewicz, S.; Gehlen, M.; Aumont, O.; Bittner, L.; Dugdale, R.; Finkel, Z.; Iudicone, D.; et al. Influence of diatom diversity on the ocean biological carbon pump. Nat. Geosci. 2018, 11, 27–37. [Google Scholar] [CrossRef]
- Henson, S.A.; Robinson, I.; Allen, J.T.; Waniek, J.J.; Thomas, A.C. Effect of meteorological conditions on interannual variability in timing and magnitude of the spring bloom in the Irminger Basin, North Atlantic. Deep Sea Res. Part I Oceanogr. Res. Pap. 2006, 53, 1601–1615. [Google Scholar] [CrossRef]
- Henson, S.A.; Thomas, A.C. Interannual variability in timing of bloom initiation in the California Current System. J. Geophys. Res. Ocean. 2007, 112, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Edwards, K.F.; Thomas, M.K.; Klausmeier, C.A.; Litchman, E. Phytoplankton growth and the interaction of light and temperature: A synthesis at the species and community level. Limnol. Oceanogr. 2016, 61, 1232–1244. [Google Scholar] [CrossRef] [Green Version]
- Ajani, P.A.; Hallegraeff, G.M.; Allen, D.; Coughlan, A.; Richardson, A.J.; Armand, L.K.; Ingleton, T.; Murray, S.A. Establishing baselines: Eighty years of phytoplankton diversity and biomass in south-eastern Australia. Oceanogr. Mar. Biol. Annu. Rev. 2016, 54, 387–412. [Google Scholar] [CrossRef] [Green Version]
- Behrenfeld, M.J.; Boss, E.S. Resurrecting the ecological underpinnings of ocean plankton blooms. Ann. Rev. Mar. Sci. 2014, 6, 167–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calbet, A.; Landry, M.R. Phytoplankton growth, microzooplankton grazing, and carbon cycling in marine systems. Limnol. Oceanogr. 2004, 49, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Smetacek, V.; Assmy, P.; Henjes, J. The role of grazing in structuring Southern Ocean pelagic ecosystems and biogeochemical cycles. Antarct. Sci. 2004, 16, 541–558. [Google Scholar] [CrossRef]
- Bonnet, S.; Berthelot, H.; Turk-Kubo, K.; Cornet-Barthaux, V.; Fawcett, S.; Berman-Frank, I.; Barani, A.; Grégori, G.; Dekaezemacker, J.; Benavides, M.; et al. Diazotroph derived nitrogen supports diatom growth in the South West Pacific: A quantitative study using nanoSIMS. Limnol. Oceanogr. 2016, 61, 1549–1562. [Google Scholar] [CrossRef] [Green Version]
- Buchan, A.; LeCleir, G.R.; Gulvik, C.A.; González, J.M. Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 2014, 12, 686–698. [Google Scholar] [CrossRef]
- Durham, B.P.; Van Mooy, B.A.S.; Moran, M.A.; Amin, S.A.; Luo, H.; Armbrust, E.V.; Campagna, S.R.; Dearth, S.P.; Kujawinski, E.B.; Sharma, S.; et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl. Acad. Sci. USA 2014, 112, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Shibl, A.A.; Isaac, A.; Ochsenkühn, M.A.; Cárdenas, A.; Fei, C.; Behringer, G.; Arnoux, M.; Drou, N.; Santos, M.P.; Gunsalus, K.C.; et al. Diatom modulation of select bacteria through use of two unique secondary metabolites. Proc. Natl. Acad. Sci. USA 2020, 202012088. [Google Scholar] [CrossRef]
- Amin, S.A.; Hmelo, L.R.; Van Tol, H.M.; Durham, B.P.; Carlson, L.T.; Heal, K.R.; Morales, R.L.; Berthiaume, C.T.; Parker, M.S.; Djunaedi, B.; et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 2015, 522, 98–101. [Google Scholar] [CrossRef]
- Vuong, T.T.; Kwon, B.R.; Eom, J.I.; Shin, B.K.; Kim, S.M. Interaction between marine bacterium Stappia sp. K01 and diatom Phaeodactylum tricornutum through extracellular fatty acids. J. Appl. Phycol. 2020, 32, 71–82. [Google Scholar] [CrossRef]
- Hosie, G.W.; Fukuchi, M.; Kawaguchi, S. Development of the Southern Ocean Continuous Plankton Recorder survey. Prog. Oceanogr. 2003, 57, 263–283. [Google Scholar] [CrossRef]
- Pesant, S.; Not, F.; Picheral, M.; Kandels-Lewis, S.; Le Bescot, N.; Gorsky, G.; Iudicone, D.; Karsenti, E.; Speich, S.; Trouble, R.; et al. Open science resources for the discovery and analysis of Tara Oceans data. Sci. Data 2015, 2, 150023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunagawa, S.; Acinas, S.G.; Bork, P.; Bowler, C.; Acinas, S.G.; Babin, M.; Bork, P.; Boss, E.; Bowler, C.; Cochrane, G.; et al. Tara Oceans: Towards global ocean ecosystems biology. Nat. Rev. Microbiol. 2020, 18, 428–445. [Google Scholar] [CrossRef]
- Ibarbalz, F.M.; Henry, N.; Brandão, M.C.; Martini, S.; Busseni, G.; Byrne, H.; Coelho, L.P.; Endo, H.; Gasol, J.M.; Gregory, A.C.; et al. Global Trends in Marine Plankton Diversity across Kingdoms of Life. Cell 2019, 179, 1084–1097. [Google Scholar] [CrossRef]
- Villar, E.; Farrant, G.K.; Follows, M.; Garczarek, L.; Speich, S.; Audic, S.; Bittner, L.; Blanke, B.; Brum, J.R.; Brunet, C.; et al. Environmental characteristics of Agulhas rings affect interocean plankton transport. Science 2015, 348. [Google Scholar] [CrossRef] [Green Version]
- Amin, S.A.; Parker, M.S.; Armbrust, E.V. Interactions between Diatoms and Bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 667–684. [Google Scholar] [CrossRef] [Green Version]
- Cirri, E.; Pohnert, G. Algae−bacteria interactions that balance the planktonic microbiome. New Phytol. 2019, 223, 100–106. [Google Scholar] [CrossRef] [Green Version]
- Martin-Platero, A.M.; Cleary, B.; Kauffman, K.; Preheim, S.P.; McGillicuddy, D.J.; Alm, E.J.; Polz, M.F. High resolution time series reveals cohesive but short-lived communities in coastal plankton. Nat. Commun. 2018, 9, 266. [Google Scholar] [CrossRef] [Green Version]
- Ajani, P.A.; Larsson, M.E.; Woodcock, S.; Rubio, A.; Farrell, H.; Brett, S.; Murray, S.A. Fifteen years of Pseudo-nitzschia in an Australian estuary, including the first potentially toxic P. delicatissima bloom in the southern hemisphere. Estuar. Coast. Shelf Sci. 2020, 236, 106651. [Google Scholar] [CrossRef]
- Delegrange, A.; Lefebvre, A.; Gohin, F.; Courcot, L.; Vincent, D. Pseudo-nitzschia sp. diversity and seasonality in the southern North Sea, domoic acid levels and associated phytoplankton communities. Estuar. Coast. Shelf Sci. 2018, 214, 194–206. [Google Scholar] [CrossRef] [Green Version]
- Muñiz, O.; Rodríguez, J.G.; Revilla, M.; Laza-Martínez, A.; Seoane, S.; Franco, J. Seasonal variations of phytoplankton community in relation to environmental factors in an oligotrophic area of the European Atlantic coast (southeastern Bay of Biscay). Reg. Stud. Mar. Sci. 2018, 17, 59–72. [Google Scholar] [CrossRef]
- Ajani, P.A.; Allen, A.P.; Ingleton, T.; Armand, L. A decadal decline in relative abundance and a shift in microphytoplankton composition at a long-term coastal station off southeast Australia. Limnol. Oceanogr. 2014, 59, 519–531, Erratum in Limnol. Oceanogr. 2014, 59, 2240–2242. [Google Scholar]
- Teeling, H.; Fuchs, B.M.; Bennke, C.M.; Krüger, K.; Chafee, M.; Kappelmann, L.; Reintjes, G.; Waldmann, J.; Quast, C.; Glöckner, F.O.; et al. Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. Elife 2016, 5, e11888. [Google Scholar] [CrossRef] [PubMed]
- Luria, C.M.; Amaral-Zettler, L.A.; Ducklow, H.W.; Repeta, D.J.; Rhyne, A.L.; Rich, J.J. Seasonal shifts in bacterial community responses to phytoplankton-derived dissolved organic matter in the Western Antarctic Peninsula. Front. Microbiol. 2017, 8, 2117. [Google Scholar] [CrossRef]
- Ajani, P.; Lee, R.; Pritchard, T.; Krogh, M. Phytoplankton dynamics at a long-term coastal station off sydney, Australia. J. Coast. Res. 2001, 60–73. [Google Scholar]
- Biller, S.J.; Berube, P.M.; Dooley, K.; Williams, M.; Satinsky, B.M.; Hackl, T.; Hogle, S.L.; Coe, A.; Bergauer, K.; Bouman, H.A.; et al. Data descriptor: Marine microbial metagenomes sampled across space and time. Sci. Data 2018, 5, 1–7. [Google Scholar] [CrossRef]
- Karl, D.M.; Church, M.J. Microbial oceanography and the Hawaii Ocean Time-series programme. Nat. Rev. Microbiol. 2014, 12, 699–713. [Google Scholar] [CrossRef]
- Brown, M.V.; Van De Kamp, J.; Ostrowski, M.; Seymour, J.R.; Ingleton, T.; Messer, L.F.; Jeffries, T.; Siboni, N.; Laverock, B.; Bibiloni-Isaksson, J.; et al. Data Descriptor: Systematic, continental scale temporal monitoring of marine pelagic microbiota by the Australian Marine Microbial Biodiversity Initiative. Sci. Data 2018, 5, 180130. [Google Scholar] [CrossRef]
- Lynch, T.P.; Morello, E.B.; Evans, K.; Richardson, A.J.; Rochester, W.; Steinberg, C.R.; Roughan, M.; Thompson, P.; Middleton, J.F.; Feng, M.; et al. IMOS National Reference Stations: A continental-wide physical, chemical and biological coastal observing system. PLoS ONE 2014, 9, e113652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridgway, K.; Hill, K. The East Australian Current. In A Marine Climate Change Impacts and Adaptation Report Card for Australia; Poloczanska, E.S., Hobday, A.J., Richardson, A.J., Eds.; NCCARF: Southport, Australia, 2009; ISBN 978-1-921609-03-9. [Google Scholar]
- Cresswell, G. The Leeuwin Current near Rottnest Island, Western Australia. Mar. Freshw. Res. 1996, 47, 483–487. [Google Scholar] [CrossRef]
- Rees, C.; Pender, L.; Sherrin, K.; Schwanger, C.; Hughes, P.; Tibben, S.; Marouchos, A.; Rayner, M. Methods for reproducible shipboard SFA nutrient measurement using RMNS and automated data processing. Limnol. Oceanogr. Methods 2019, 17, 25–41. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; Wong, J.; Heiner, C.; Oh, S.; Theriot, C.M.; Gulati, S.; Mcgill, S.K.; Dougherty, M.K. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019, 47, e103. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R.; Al, W.E.T. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Ludwig, W.; Pruesse, E.; Quast, C.; Schweer, T.; Glo, F.O. The SILVA and “All-species Living Tree Project (LTP )” taxonomic frameworks. Nucleic Acids Res. 2014, 42, 643–648. [Google Scholar] [CrossRef] [Green Version]
- Mcmurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Package ‘vegan’. Community Ecology Package. 2020. version 2.5-7. pp. 1–295. Available online: https://CRAN.R-project.org/package=vegan (accessed on 13 December 2021).
- Lin, H.; Peddada, S. Das Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Needham, D.M.; Fuhrman, J.A. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat. Microbiol. 2016, 1, 16005. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Busseni, G.; Caputi, L.; Piredda, R.; Fremont, P.; Hay Mele, B.; Campese, L.; Scalco, E.; de Vargas, C.; Bowler, C.; d’Ovidio, F.; et al. Large scale patterns of marine diatom richness: Drivers and trends in a changing ocean. Glob. Ecol. Biogeogr. 2020, 29, 1915–1928. [Google Scholar] [CrossRef]
- Hallegraeff, G.M.; Reid, D.D. Phytoplankton species successions and their hydrological environment at a coastal station off sydney. Mar. Freshw. Res. 1986, 37, 377. [Google Scholar] [CrossRef]
- Garcia, M.; Odebrecht, C. Morphology and ecology of the planktonic diatom Palmerina hardmaniana (Greville) Hasle in southern Brazil. Biota Neotrop. 2008, 8, 85–90. [Google Scholar] [CrossRef]
- Liu, Y.; Debeljak, P.; Rembauville, M.; Blain, S.; Obernosterer, I. Diatoms shape the biogeography of heterotrophic prokaryotes in early spring in the Southern Ocean. Environ. Microbiol. 2019, 21, 1452–1465. [Google Scholar] [CrossRef]
- Armbrecht, L.H.; Schaeffer, A.; Roughan, M.; Armand, L.K. Interactions between seasonality and oceanic forcing drive the phytoplankton variability in the tropical-temperate transition zone (~30°S) of Eastern Australia. J. Mar. Syst. 2015, 144, 92–106. [Google Scholar] [CrossRef]
- Luria, C.M.; Amaral-Zettler, L.A.; Ducklow, H.W.; Rich, J.J. Seasonal succession of free-living bacterial communities in coastal waters of the western antarctic peninsula. Front. Microbiol. 2016, 7, 1731. [Google Scholar] [CrossRef] [Green Version]
- Teeling, H.; Fuchs, B.M.; Becher, D.; Klockow, C.; Gardebrecht, A.; Bennke, C.M.; Kassabgy, M.; Huang, S.; Mann, A.J.; Waldmann, J.; et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012, 336, 608–611. [Google Scholar] [CrossRef]
- Yang, C.; Li, Y.; Zhou, B.; Zhou, Y.; Zheng, W.; Tian, Y.; Van Nostrand, J.D.; Wu, L.; He, Z.; Zhou, J.; et al. Illumina sequencing-based analysis of free-living bacterial community dynamics during an Akashiwo sanguine bloom in Xiamen sea, China. Sci. Rep. 2015, 5, 8476. [Google Scholar] [CrossRef] [Green Version]
- Moore, L.R.; Huang, T.; Ostrowski, M.; Mazard, S.; Kumar, S.S.; Gamage, H.K.A.H.; Brown, M.V.; Messer, L.F.; Seymour, J.R.; Paulsen, I.T. Unicellular cyanobacteria are important components of phytoplankton communities in Australia’s northern oceanic ecoregions. Front. Microbiol. 2019, 10, 3356. [Google Scholar] [CrossRef] [Green Version]
- Krüger, K.; Chafee, M.; Ben Francis, T.; Glavina del Rio, T.; Becher, D.; Schweder, T.; Amann, R.I.; Teeling, H. In marine Bacteroidetes the bulk of glycan degradation during algae blooms is mediated by few clades using a restricted set of genes. ISME J. 2019, 13, 2800–2816. [Google Scholar] [CrossRef] [Green Version]
- Rajapitamahuni, S.; Bachani, P.; Sardar, R.K.; Mishra, S. Co-cultivation of siderophore-producing bacteria Idiomarina loihiensis RS14 with Chlorella variabilis ATCC 12198, evaluation of micro-algal growth, lipid, and protein content under iron starvation. J. Appl. Phycol. 2019, 31, 29–39. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Reun, N.; Bramucci, A.; O’Brien, J.; Ostrowski, M.; Brown, M.V.; Van de Kamp, J.; Bodrossy, L.; Raina, J.-B.; Ajani, P.; Seymour, J. Diatom Biogeography, Temporal Dynamics, and Links to Bacterioplankton across Seven Oceanographic Time-Series Sites Spanning the Australian Continent. Microorganisms 2022, 10, 338. https://doi.org/10.3390/microorganisms10020338
Le Reun N, Bramucci A, O’Brien J, Ostrowski M, Brown MV, Van de Kamp J, Bodrossy L, Raina J-B, Ajani P, Seymour J. Diatom Biogeography, Temporal Dynamics, and Links to Bacterioplankton across Seven Oceanographic Time-Series Sites Spanning the Australian Continent. Microorganisms. 2022; 10(2):338. https://doi.org/10.3390/microorganisms10020338
Chicago/Turabian StyleLe Reun, Nine, Anna Bramucci, James O’Brien, Martin Ostrowski, Mark V. Brown, Jodie Van de Kamp, Levente Bodrossy, Jean-Baptiste Raina, Penelope Ajani, and Justin Seymour. 2022. "Diatom Biogeography, Temporal Dynamics, and Links to Bacterioplankton across Seven Oceanographic Time-Series Sites Spanning the Australian Continent" Microorganisms 10, no. 2: 338. https://doi.org/10.3390/microorganisms10020338






