Chickpea Roots Undergoing Colonisation by Phytophthora medicaginis Exhibit Opposing Jasmonic Acid and Salicylic Acid Accumulation and Signalling Profiles to Leaf Hemibiotrophic Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Phytophthora Inoculations
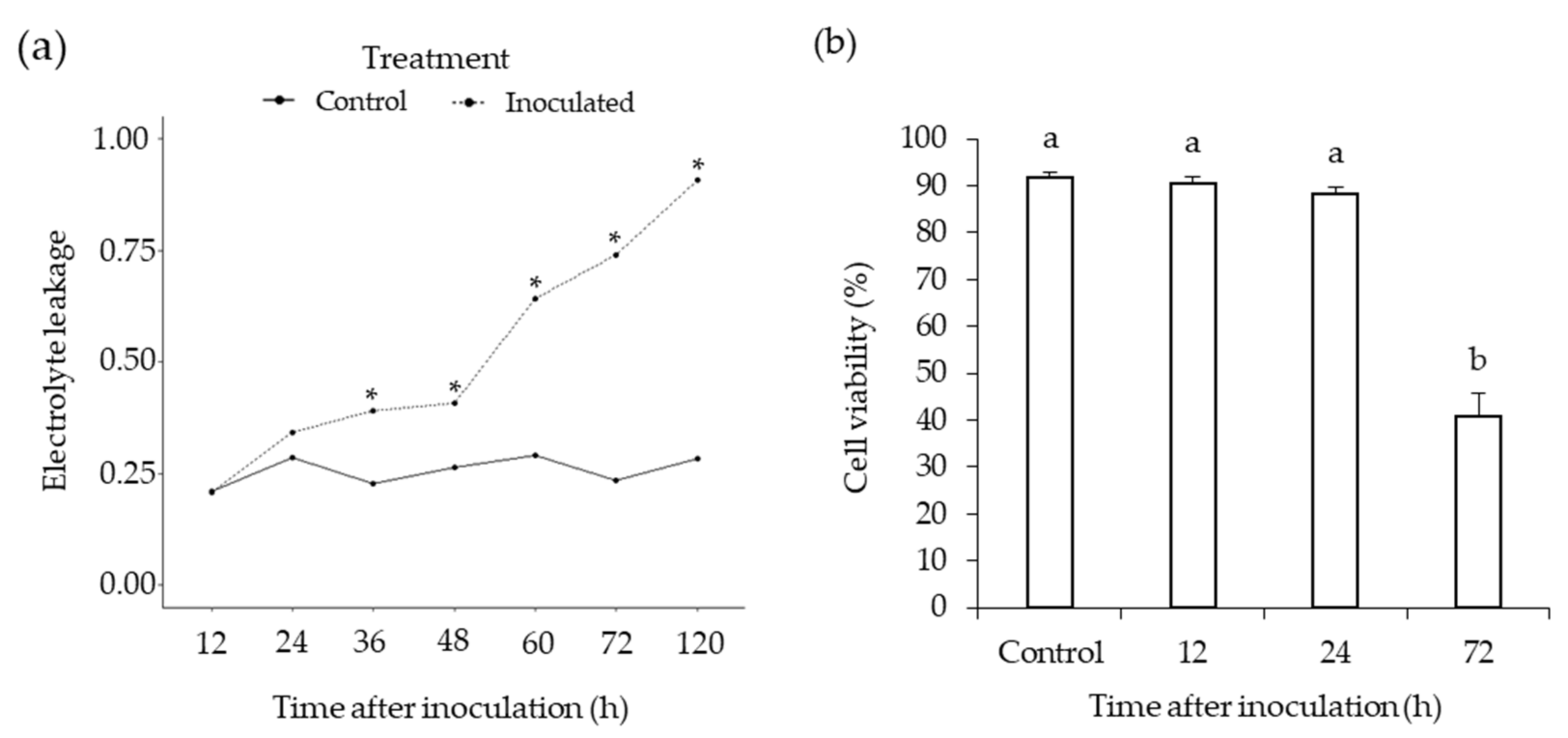
2.2. Electrolyte Leakage Root Cell Death Assay
2.3. Macroscopic Assessment of Symptoms
2.4. Root Cell Viability Analysis
2.5. Propidium Iodide/Wheat Germ Agglutinin Staining and Confocal Microscopy
2.6. RNA-Sequencing and Data Analysis
2.7. Quantitative PCR Expression Validation of RNA Sequencing
2.8. Gene Ontology (GO) Enrichment Analysis of the Unique Gene Sets at Each Phase
2.9. Expression Analysis of Chickpea Hormone Pathway Related Genes
2.10. In Vivo Root Salicylic and Jasmonic Acid Measurements
3. Results
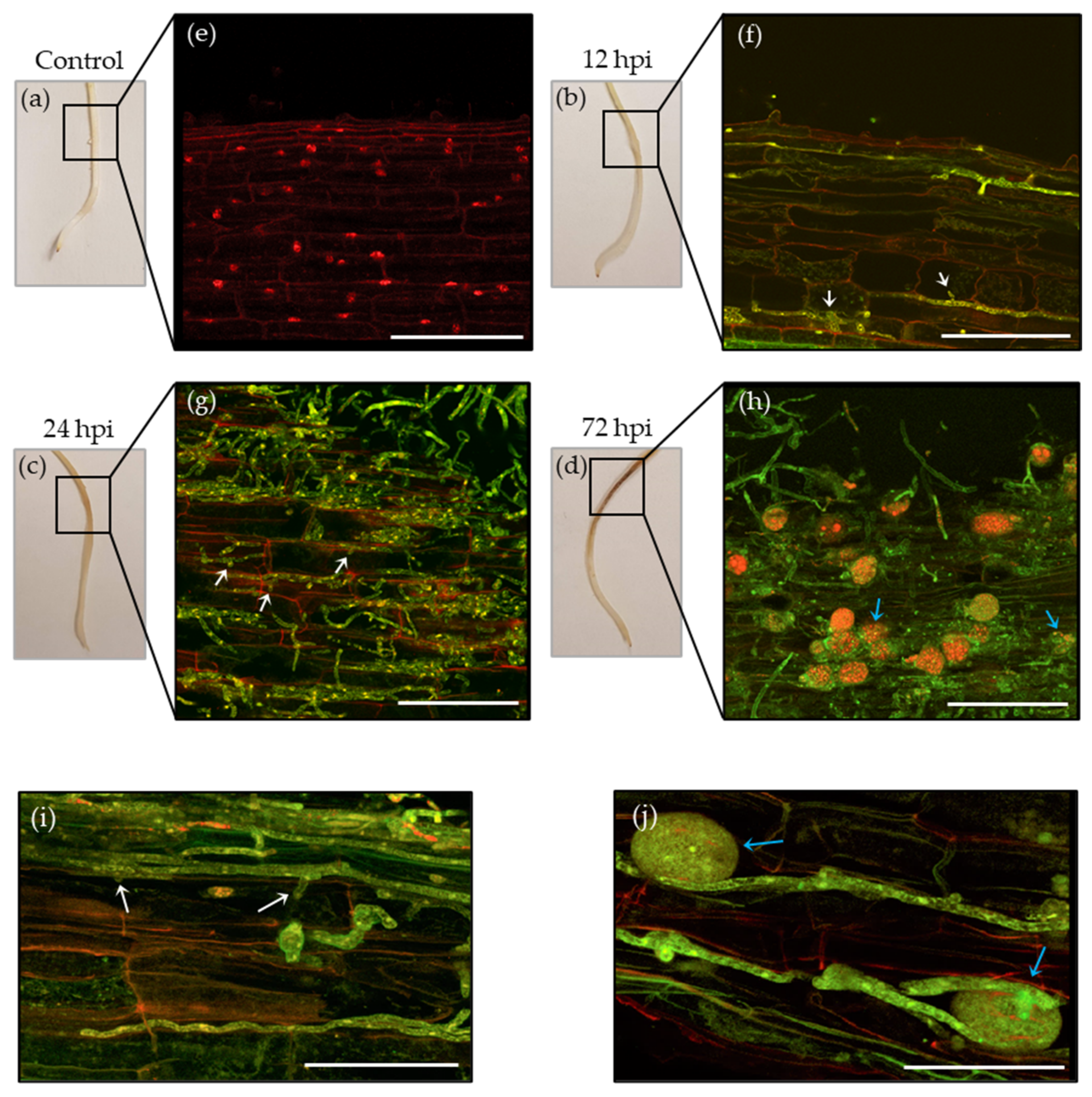
3.1. Phytophthora Medicaginis Infection of Chickpea Roots Features an Early Switch from Biotrophy to Necrotrophy
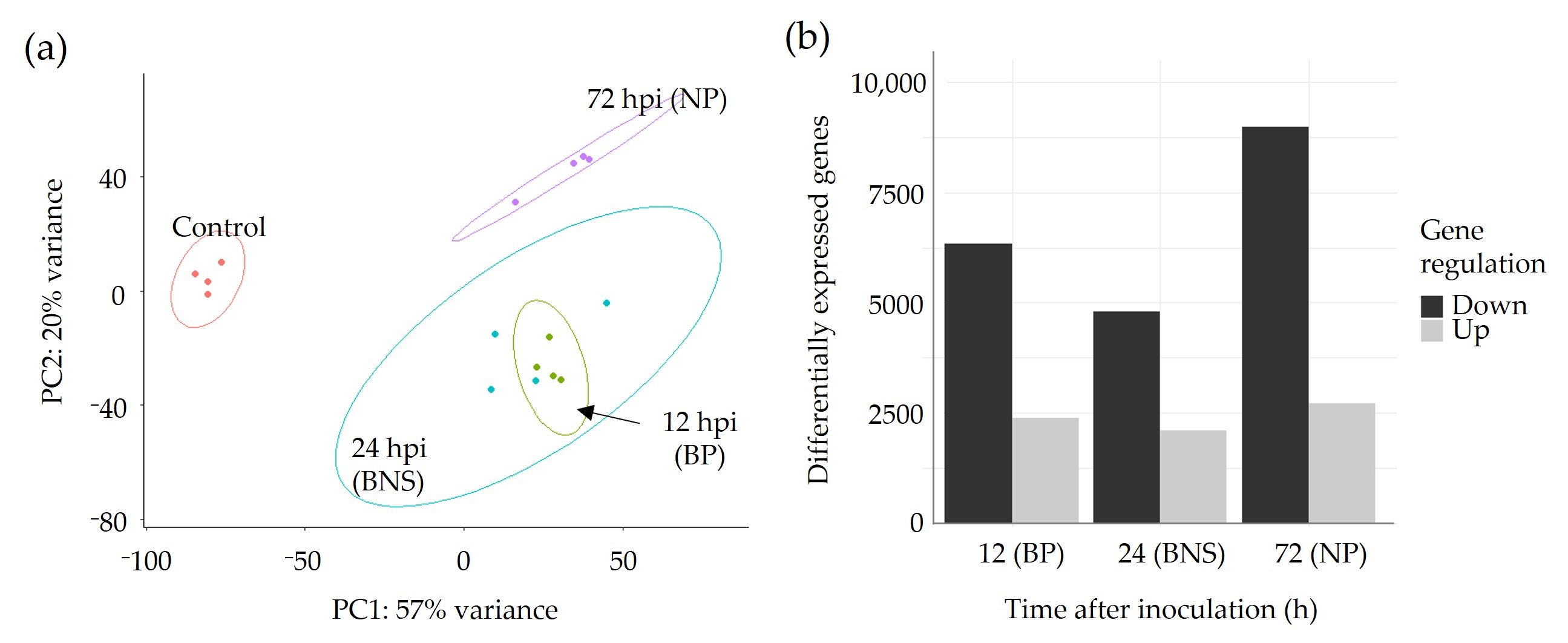
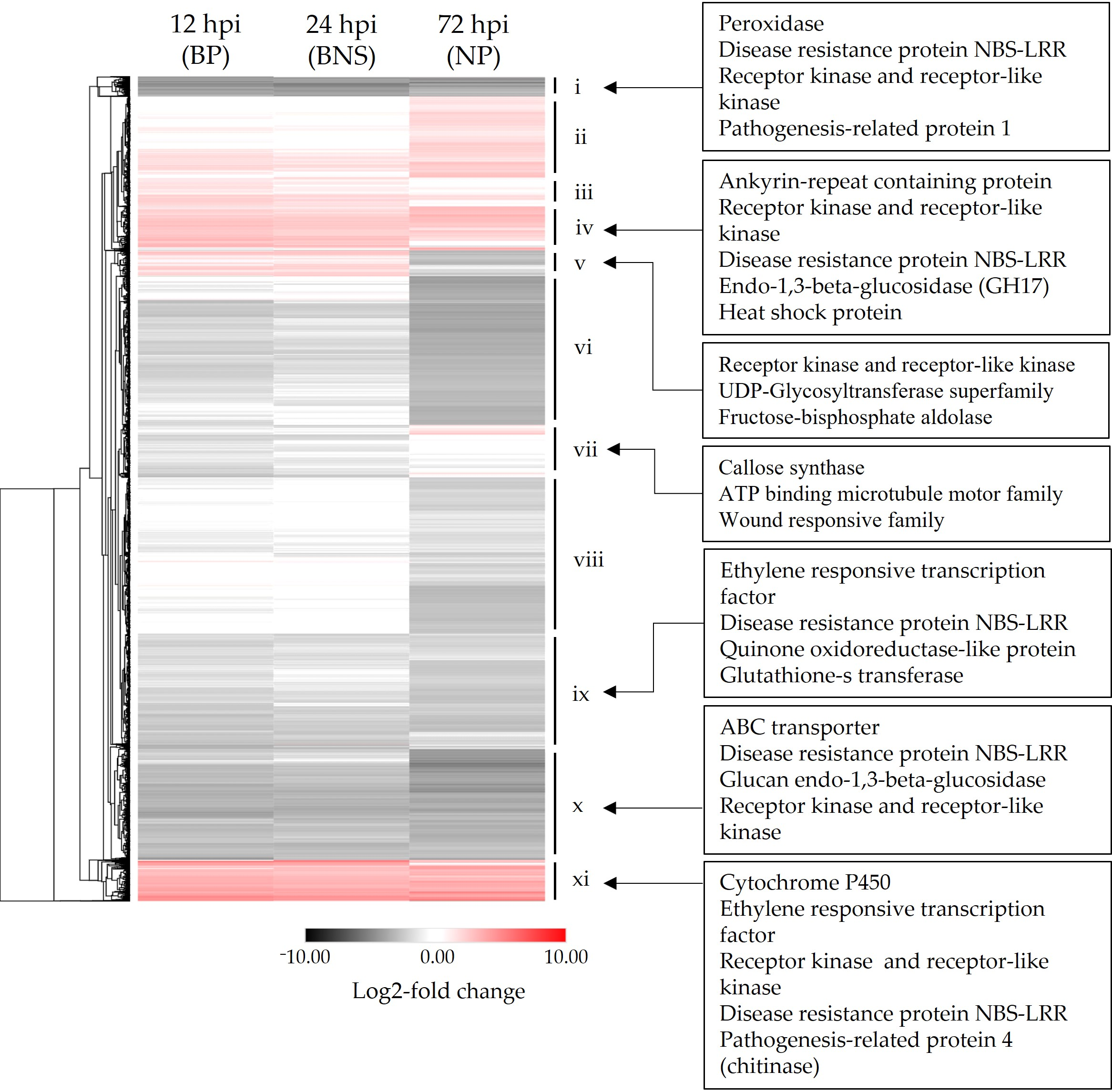
3.2. A Large Percentage of the Chickpea Genome Is Responsive to Phytophthora Infection
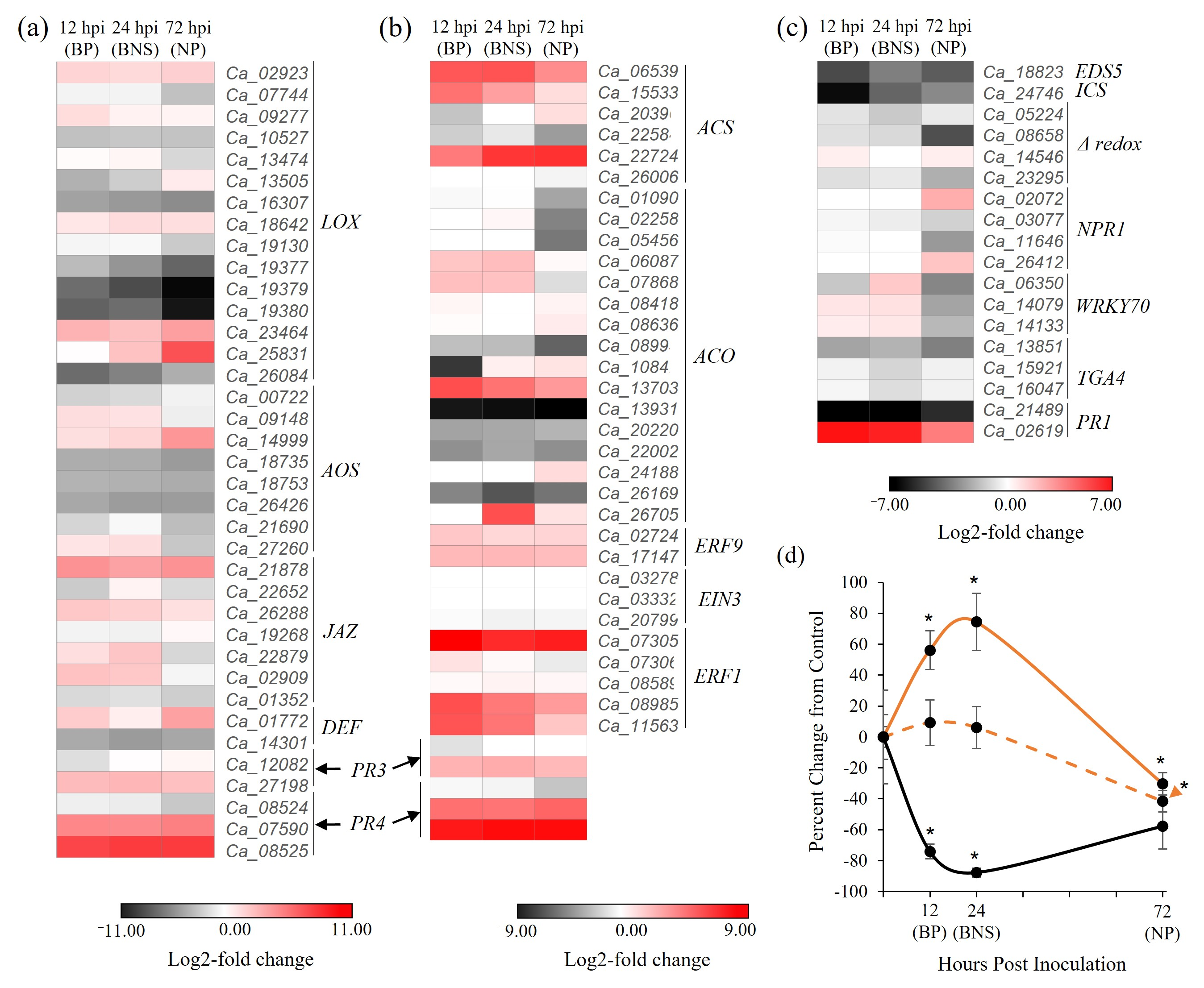
3.3. Chickpea Root Phytohormone Responses Do Not Conform to the Classical Leaf Model for Hemibiotrophic Pathogenesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schwessinger, B.; Sperschneider, J.; Cuddy, W.S.; Garnica, D.P.; Miller, M.E.; Taylor, J.M.; Dodds, P.N.; Figueroa, M.; Park, R.F.; Rathjen, J.P. A Near-Complete Haplotype-Phased Genome of the Dikaryotic Wheat Stripe Rust Fungus Puccinia striiformis f. sp. tritici Reveals High Interhaplotype Diversity. mBio 2018, 9, e02275-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spanu, P.D.; Panstruga, R. Editorial: Biotrophic Plant-Microbe Interactions. Front. Plant Sci. 2017, 8, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Kan, J.A.L.; Shaw, M.W.; Grant-Downton, R.T. Botrytis species: Relentless necrotrophic thugs or endophytes gone rogue? Mol. Plant Pathol. 2014, 15, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.S. After the famine: Plant pathology, Phytophthora infestans, and the late blight of potatoes, 1845–1960. Hist. Stud. Phys. Biol. Sci. 2005, 35, 341–370. [Google Scholar] [CrossRef]
- Rodenburg, S.Y.A.; Seidl, M.F.; Judelson, H.S.; Vu, A.L.; Govers, F.; de Ridder, D. Metabolic Model of the Phytophthora infestans-Tomato Interaction Reveals Metabolic Switches during Host Colonization. mBio 2019, 10, e00454-19. [Google Scholar] [CrossRef] [Green Version]
- Fawke, S.; Doumane, M.; Schornack, S. Oomycete interactions with plants: Infection strategies and resistance principles. Microbiol. Mol. Biol. Rev. 2015, 79, 263–280. [Google Scholar] [CrossRef] [Green Version]
- Stotz, H.U.; Mitrousia, G.K.; de Wit, P.J.G.M.; Fitt, B.D.L. Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 2014, 19, 491–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Lee, S.-J.; Rose, J.K.C. Mediation of the transition from biotrophy to necrotrophy in hemibiotrophic plant pathogens by secreted effector proteins. Plant Signal. Behav. 2010, 5, 769–772. [Google Scholar] [CrossRef]
- Qutob, D.; Kamoun, S.; Gijzen, M. Expression of a Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. TPJ 2002, 32, 361–373. [Google Scholar] [CrossRef] [Green Version]
- Haddadi, P.; Ma, L.; Wang, H.; Borhan, M.H. Genome-wide transcriptomic analyses provide insights into the lifestyle transition and effector repertoire ofLeptosphaeria maculansduring the colonization ofBrassica napusseedlings. Mol. Plant Pathol. 2016, 17, 1196–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glazebrook, J. Contrasting Mechanisms of Defense Against Biotrophic and Necrotrophic Pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.-J. Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: Do we understand what they are whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; Zhang, X.; Fernando, W.G.D. Directing trophic divergence in plant-pathogen interactions: Antagonistic phytohormones with NO doubt? Front. Plant Sci. 2020, 11, 600063. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Mir, Z.A.; Tyagi, A.; Bhat, J.A.; Chandrashekar, N.; Papolu, P.K.; Rawat, S.; Grover, A. Identification and comparative analysis of Brassica juncea pathogenesis-related genes in response to hormonal, biotic and abiotic stresses. Acta Physiol. Plant. 2017, 39, 268. [Google Scholar] [CrossRef]
- Cao, H.; Bowling, S.A.; Gordon, A.S.; Dong, X. Characterization of an Arabidopsis Mutant That Is Nonresponsive to Inducers of Systemic Acquired Resistance. Plant Cell 1994, 6, 1583–1592. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 2018, 173, 1454–1467. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212, 29–37. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, L.; Duan, J.; Miki, B.; Wu, K. HISTONE DEACETYLASE19 Is Involved in Jasmonic Acid and Ethylene Signaling of Pathogen Response in Arabidopsis. Plant Cell 2005, 17, 1196–1204. [Google Scholar] [CrossRef] [Green Version]
- Onkokesung, N.; Gális, I.; von Dahl, C.C.; Matsuoka, K.; Saluz, H.-P.; Baldwin, I.T. Jasmonic Acid and Ethylene Modulate Local Responses to Wounding and Simulated Herbivory in Nicotiana attenuata leaves. Plant Physiol. 2010, 153, 785–798. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, S.; Basu, A.; Kundu, S. Biotrophy-necrotrophy switch in pathogen evoke differential response in resistant and susceptible sesame involving multiple signaling pathways at different phases. Sci. Rep. 2017, 7, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Zuluaga, A.P.; Vega-Arreguin, J.; Fei, Z.; Matas, A.J.; Patev, S.; Fry, W.E.; Rose, J.K.C. Analysis of the tomato leaf transcriptome during successive hemibiotrophic stages of a compatible interaction with the oomycete pathogen Phytophthora infestans. Mol. Plant Pathol. 2015, 17, 42–54. [Google Scholar] [CrossRef]
- Spoel, S.H.; Koornneef, A.; Claessens, S.M.C.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Buchala, A.J.; Métraux, J.-P.; Brown, R.; Kazan, K.; et al. (NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 2003, 15, 760–770. [Google Scholar] [CrossRef] [Green Version]
- Bhadauria, V.; Bett, K.E.; Zhou, T.; Vandenberg, A.; Wei, Y.; Banniza, S. Identification of Lens culinaris defense genes responsive to the anthracnose pathogen Colletotrichum truncatum. BMC Genet. 2013, 14, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jupe, J.; Stam, R.; Howden, A.J.M.; Morris, J.A.; Zhang, R.; Hedley, P.E.; Huitema, E. Phytophthora capsici-tomato interaction features dramatic shifts in gene expression associated with a hemi-biotrophic lifestyle. Genome Biol. 2013, 14, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudd, J.J.; Kanyuka, K.; Hassani-Pak, K.; Derbyshire, M.; Andongabo, A.; Devonshire, J.; Lysenko, A.; Saqi, M.; Desai, N.M.; Powers, S.J.; et al. Transcriptome and Metabolite Profiling of the Infection Cycle of Zymoseptoria tritici on Wheat Reveals a Biphasic Interaction with Plant Immunity Involving Differential Pathogen Chromosomal Contributions and a Variation on the Hemibiotrophic Lifestyle Definition. Plant Physiol. 2015, 167, 1158–1185. [Google Scholar] [CrossRef] [Green Version]
- Vargas, W.A.; Martín, J.M.S.; Rech, G.E.; Rivera, L.P.; Benito, E.P.; Díaz-Mínguez, J.M.; Thon, M.R.; Sukno, S.A. Plant Defense Mechanisms Are Activated during Biotrophic and Necrotrophic Development of Colletotricum graminicola in Maize. Plant Physiol. 2012, 158, 1342–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; Li, W.; Jørgensen, H.J.L. Transcriptional Reprogramming of Wheat and the Hemibiotrophic Pathogen Septoria tritici during Two Phases of the Compatible Interaction. PLoS ONE 2013, 8, e81606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlink, K. Down-regulation of defense genes and resource allocation into infected roots as factors for compatibility between Fagus sylvatica and Phytophthora citricola. Funct. Integr. Genomics 2010, 10, 253–264. [Google Scholar] [CrossRef]
- Lyons, R.; Stiller, J.; Powell, J.; Rusu, A.; Manners, J.M.; Kazan, K. Fusarium oxysporum Triggers Tissue-Specific Transcriptional Reprogramming in Arabidopsis thaliana. PLoS ONE 2015, 10, e0121902. [Google Scholar] [CrossRef]
- Marcel, S.; Sawers, R.; Oakeley, E.; Angliker, H.; Paszkowski, U. Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell 2010, 22, 3177–3187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Berre, J.-Y.; Gourgues, M.; Samans, B.; Keller, H.; Panabières, F.; Attard, A. Transcriptome dynamic of Arabidopsis roots infected with Phytophthora parasitica identifies VQ29, a gene induced during the penetration and involved in the restriction of infection. PLoS ONE 2017, 12, e0190341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholz, S.S.; Schmidt-Heck, W.; Guthke, R.; Furch, A.C.U.; Reichelt, M.; Gershenzon, J.; Oelmüller, R. Verticillium dahliae-Arabidopsis Interaction Causes Changes in Gene Expression Profiles and Jasmonate Levels on Different Time Scales. Front. Microbiol. 2018, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Moy, P.; Qutob, D.; Chapman, B.P.; Atkinson, I.; Gijzen, M. Patterns of Gene Expression Upon Infection of Soybean Plants by Phytophthora sojae. Mol. Plant. Microbe Interact. 2004, 17, 1051–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Mideros, S.X.; Bao, L.; Hanlon, R.; Arredondo, F.D.; Tripathy, S.; Krampis, K.; Jerauld, A.; Evans, C.; St Martin, S.K.; et al. Infection and genotype remodel the entire soybean transcriptome. BMC Genom. 2009, 10, 49. [Google Scholar] [CrossRef] [Green Version]
- Attard, A.; Gourgues, M.; Callemeyn-Torre, N.; Keller, H. The immediate activation of defense responses in Arabidopsis roots is not sufficient to prevent Phytophthora parasitica infection. New Phytol. 2010, 187, 449–460. [Google Scholar] [CrossRef]
- Plett, J.M.; Plett, K.L.; Bithell, S.L.; Mitchell, C.; Moore, K.; Powell, J.R.; Anderson, I.C. Improved Phytophthora resistance in commercial chickpea (Cicer arietinum) varieties negatively impacts symbiotic gene signalling and symbiotic potential in some varieties. Plant Cell Environ. 2016, 39, 1858–1869. [Google Scholar] [CrossRef]
- Amalraj, A.; Taylor, J.; Bithell, S.; Li, Y.; Moore, K.; Hobson, K.; Sutton, T. Mapping resistance to Phytophthora root rot identifies independent loci from cultivated (Cicer arietinum L.) and wild (Cicer echinospermum PH Davis) chickpea. Theor. Appl. Genet. 2019, 132, 1017–1033. [Google Scholar] [CrossRef]
- Merga, B.; Haji, J. Economic importance of chickpea: Production, value, and world trade. Cogent Food Agric. 2019, 5, 1615718. [Google Scholar] [CrossRef]
- Bithell, S.; Moore, K.; McKay, A.; Harden, S.; Simpfendorfer, S. Phytophthora root rot of chickpea: Inoculum concentration and seasonally dependent success for qPCR based predictions of disease and yield loss. Australas. Plant Pathol. 2021, 50, 91–103. [Google Scholar] [CrossRef]
- Miller, S.A.; Maxwell, D.P. Ultrastructure of susceptible, host resistant and nonhost resistant interactions of alfalfa with Phytophthora megasperma. Can. J. Bot. 1984, 62, 117–128. [Google Scholar] [CrossRef]
- Enkerli, K.; Mims, C.; Hahn, M. Ultrastructure of compatible and incompatible interactions of soybean roots infected with the plant pathogenic oomycete Phytophthora sojae. Can. J. Bot. 1997, 75, 1493–1508. [Google Scholar] [CrossRef]
- Vock, N.T.; Langdon, P.W.; Pegg, K.G. Root rot of chickpea caused by Phytophthora megasperma var. sojae in Queensland. Australas. Plant Pathol. 1980, 9, 117. [Google Scholar] [CrossRef]
- Beakes, G.W.; El-Hamalawi, Z.A.; Erwin, D.C. Ultrastructure of mature oospores of Phytophthora megasperma f.sp. medicaginis: Preparation protocols and effects of MTT vital staining and permanganate pre-treatment. Trans. Br. Mycol. Soc. 1986, 86, 195–206. [Google Scholar] [CrossRef]
- El-Hamalawi, Z.A.; Erwin, D. Components in alfalfa root extract and root exudate that increase oospore germination of Phytophthora megasperma f. sp. medicaginis. Phytopathology 1986, 76, 508–513. [Google Scholar] [CrossRef]
- Jain, M.; Misra, G.; Patel, R.K.; Priya, P.; Jhanwar, S.; Khan, A.W.; Shah, N.; Singh, V.K.; Garg, R.; Jeena, G.; et al. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). TPJ 2013, 74, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Amalraj, A. Molecular Genetics and Genomics Approaches to Improve Phytophthora Root Rot Resistance in Chickpea. Ph.D. Thesis, University of Adelaide, Adelaide, SA, Australia, 2019. [Google Scholar]
- Feng, G.; Yang, H.; Li, Y. Kinetics of relative electrical conductivity and correlation with gas composition in modified atmosphere packaged bayberries (Myrica rubra Siebold and Zuccarini). LWT-Food Sci. Technol. 2005, 38, 249–254. [Google Scholar] [CrossRef]
- GenStat Committee. The Guide to GenStat Release 12–Parts 1–3; VSN International: Oxford, UK, 2009. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 25 April 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 13 June 2021).
- Fox, J.; Friendly, M.; Weisberg, S. Hypothesis Tests for Multivariate Linear Models Using the car Package. R J. 2013, 5, 39. [Google Scholar] [CrossRef] [Green Version]
- Lenth R Emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.1. 2020. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 10 April 2021).
- Cheng, W.; Lin, M.; Qiu, M.; Kong, L.; Xu, Y.; Li, Y.; Wang, Y.; Ye, W.; Dong, S.; He, S.; et al. Chitin synthase is involved in vegetative growth, asexual reproduction and pathogenesis of Phytophthora capsici and Phytophthora sojae. Environ. Microbiol. 2019, 21, 4537–4547. [Google Scholar] [CrossRef]
- Jones, K.; Kim, D.W.; Park, J.S.; Khang, C.H. Live-cell fluorescence imaging to investigate the dynamics of plant cell death during infection by the rice blast fungus Magnaporthe oryzae. BMC Plant Biol. 2016, 16, 69. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexa, A.; Rahnenführer, J. Gene set enrichment analysis with topGO. Bioconductor. Improv. 2009, 27, 1–26. [Google Scholar]
- Zuluaga, A.P.; Vega-Arreguín, J.C.; Fei, Z.; Ponnala, L.; Lee, S.J.; Matas, A.J.; Patev, S.; Fry, W.E.; Rose, J.K.C. Transcriptional dynamics of Phytophthora infestansduring sequential stages of hemibiotrophic infection of tomato. Mol. Plant Pathol. 2015, 17, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Keon, J.; Antoniw, J.; Carzaniga, R.; Deller, S.; Ward, J.L.; Baker, J.M.; Beale, M.H.; Hammond-Kosack, K.; Rudd, J.J. Transcriptional Adaptation of Mycosphaerella graminicola to Programmed Cell Death (PCD) of Its Susceptible Wheat Host. Mol. Plant. Microbe Interact. 2007, 20, 178–193. [Google Scholar] [CrossRef] [Green Version]
- Diniz, I.; Figueiredo, A.; Loureiro, A.; Batista, D.; Azinheira, H.; Várzea, V.; Pereira, A.P.; Gichuru, E.; Moncada, P.; Guerra-Guimarães, L.; et al. A first insight into the involvement of phytohormones pathways in coffee resistance and susceptibility to Colletotrichum kahawae. PLoS ONE 2017, 12, e0178159. [Google Scholar] [CrossRef] [Green Version]
- Smart, C.D.; Myers, K.L.; Restrepo, S.; Martin, G.B.; Fry, W.E. Partial Resistance of Tomato to Phytophthora infestans is not dependent upon ethylene, jasmonic acid, or salicylic acid signaling pathways. Mol. Plant-Microbe Interact. 2003, 16, 141–148. [Google Scholar] [CrossRef] [Green Version]
- Mehta, A.P.; Abdelwahed, S.H.; Begley, T.P. Molybdopterin biosynthesis—Mechanistic studies on a novel MoaA catalyzed insertion of a purine carbon into the ribose of GTP. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2015, 1854, 1073–1077. [Google Scholar] [CrossRef] [Green Version]
- Cheval, C.; Aldon, D.; Galaud, J.-P.; Ranty, B. Calcium/calmodulin-mediated regulation of plant immunity. Biochim. Biophys. Acta 2013, 1833, 1766–1771. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.H.; Gill, W.M.; Pinkard, E.; Mohammed, C.L. Anatomical and histochemical defence responses induced in juvenile leaves of Eucalyptus globulus and Eucalyptus nitens by Mycosphaerella infection. For. Pathol. 2007, 37, 361–373. [Google Scholar] [CrossRef]
- Su, X.; Lu, G.; Guo, H.; Zhang, K.; Li, X.; Cheng, H. The dynamic transcriptome and metabolomics profiling in Verticillium dahliae inoculated Arabidopsis thaliana. Sci. Rep. 2018, 8, 15404. [Google Scholar] [CrossRef] [Green Version]
- Raman, H.; Raman, R.; Diffey, S.; Qiu, Y.; McVittie, B.; Barbulescu, D.M.; Salisbury, P.A.; Marcroft, S.; Delourme, R. Stable Quantitative Resistance Loci to Blackleg Disease in Canola (Brassica napus L.) Over Continents. Front. Plant Sci. 2018, 9, 1622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browse, J. Jasmonate passes muster: A Receptor and Targets For The Defense Hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef]
- Ren, C.; Zhu, Q.; Gao, B.-D.; Ke, S.-Y.; Yu, W.-C.; Xie, D.-X.; Peng, W. Transcription factor WRKY70 displays important but no indispensable roles in jasmonate and salicylic acid signaling. J. Integr. Plant Biol. 2008, 50, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Neu, E.; Domes, H.S.; Menz, I.; Kaufmann, H.; Linde, M.; Debener, T. Interaction of roses with a biotrophic and a hemibiotrophic leaf pathogen leads to differences in defense transcriptome activation. Plant Mol. Biol. 2019, 99, 299–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coles, D.W.; Bithell, S.L.; Mikhael, M.; Cuddy, W.S.; Plett, J.M. Chickpea Roots Undergoing Colonisation by Phytophthora medicaginis Exhibit Opposing Jasmonic Acid and Salicylic Acid Accumulation and Signalling Profiles to Leaf Hemibiotrophic Models. Microorganisms 2022, 10, 343. https://doi.org/10.3390/microorganisms10020343
Coles DW, Bithell SL, Mikhael M, Cuddy WS, Plett JM. Chickpea Roots Undergoing Colonisation by Phytophthora medicaginis Exhibit Opposing Jasmonic Acid and Salicylic Acid Accumulation and Signalling Profiles to Leaf Hemibiotrophic Models. Microorganisms. 2022; 10(2):343. https://doi.org/10.3390/microorganisms10020343
Chicago/Turabian StyleColes, Donovin W., Sean L. Bithell, Meena Mikhael, William S. Cuddy, and Jonathan M. Plett. 2022. "Chickpea Roots Undergoing Colonisation by Phytophthora medicaginis Exhibit Opposing Jasmonic Acid and Salicylic Acid Accumulation and Signalling Profiles to Leaf Hemibiotrophic Models" Microorganisms 10, no. 2: 343. https://doi.org/10.3390/microorganisms10020343






