Detection and Molecular Identification of Eight Candida Species in Clinical Samples by Simplex PCR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Candida spp. Reference Strains and Clinical Isolates
2.2. Clinical Samples
2.3. DNA Extraction from Yeasts
2.4. DNA Extraction from Clinical Samples
2.5. Oligonucleotide Design
2.6. Specificity Evaluation and Detection Limit of the PCR Assay
2.7. Evaluation of the Diagnostic Utility of the PCR Assay
3. Results
3.1. Oligonucleotides Design
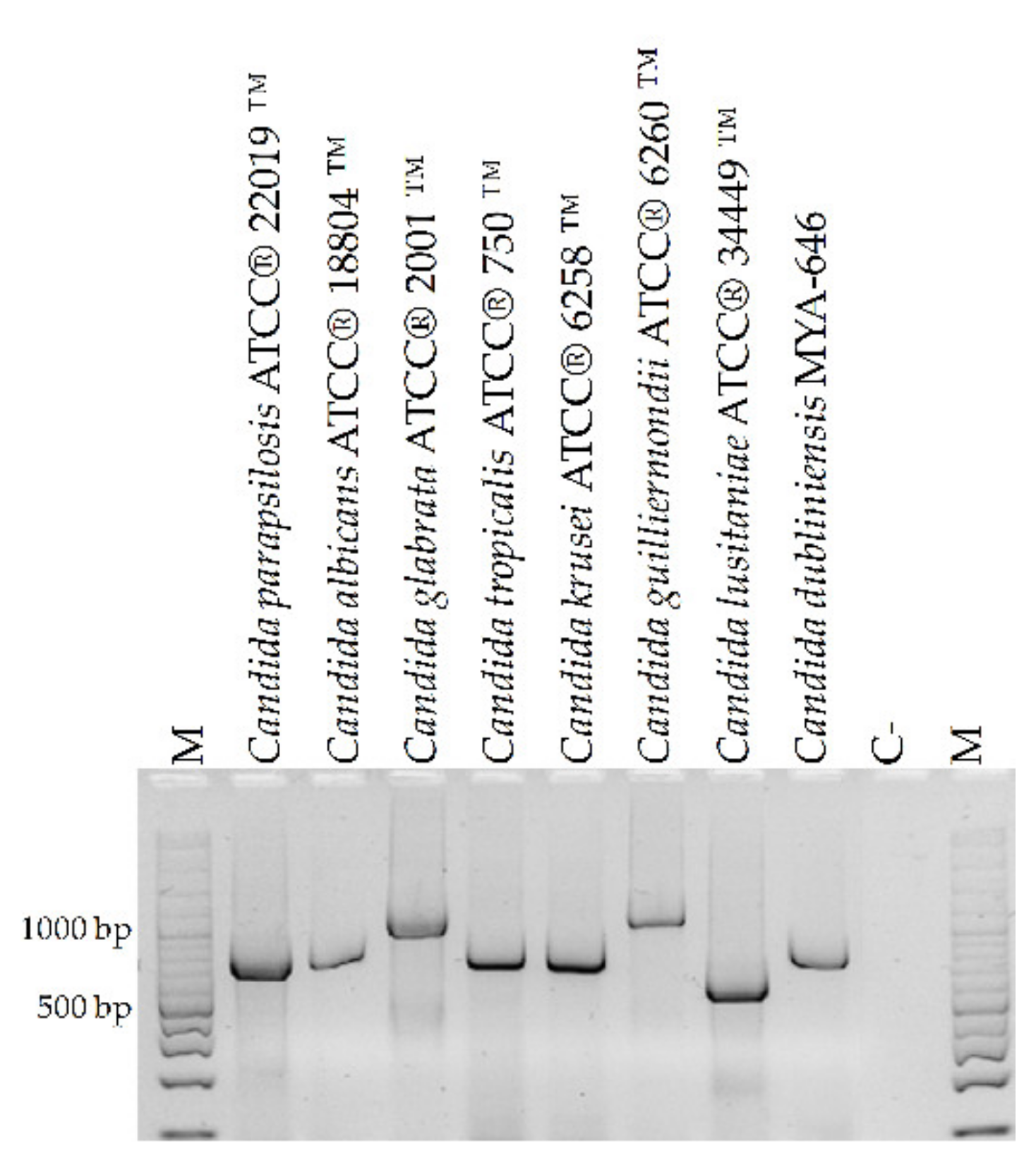
3.2. Specificity Evaluation and Detection Limit of the PCR Assay
3.3. Evaluation of the Diagnostic Utility of the PCR Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Gnat, S.; Łagowski, D.; Nowakiewicz, A.; Dyląg, M. A global view on fungal infections in humans and animals: Opportunistic infections and microsporidioses. J. Appl. Microbiol. 2021, 131, 2095–2113. [Google Scholar] [CrossRef] [PubMed]
- Garbee, D.D.; Pierce, S.S.; Manning, J. Opportunistic Fungal infections in critical care units. Crit. Care Nurs. Clin. N. Am. 2017, 29, 67–79. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Santos, G.C.; Vasconcelos, C.C.; Lopes, A.J.O.; de Sousa Cartágenes, M.D.S.; Filho, A.K.D.B.; do Nascimento, F.R.F.; Ramos, R.M.; Pires, E.R.R.B.; de Andrade, M.S.; Rocha, F.M.G.; et al. Candida infections and therapeutic strategies: Mechanisms of action for traditional and alternative agents. Front. Microbiol. 2018, 9, 1351. [Google Scholar] [CrossRef]
- Jenks, J.D.; Cornely, O.A.; Chen, S.C.; Thompson, G.R., III; Hoenigl, M. Breakthrough invasive fungal infections: Who is at risk? Mycoses 2020, 63, 1021–1032. [Google Scholar] [CrossRef]
- Pal, M.; Gebrezgabher, W.; Samajpati, N.; Manna, A.K. Growing role of non-Candida albicans species in clinical disorders of human and animals. J. Mycopathol. Res. 2015, 53, 41–48. [Google Scholar]
- Pappas, P.; Lionakis, M.; Arendrup, M.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Primers 2018, 4, 18026. [Google Scholar] [CrossRef]
- Bassetti, M.; Righi, E.; Costa, A.; Fasce, R.; Molinari, M.P.; Rosso, R.; Pallavicini, F.B.; Viscoli, C. Epidemiological trends in nosocomial candidemia in intensive care. BMC Infect. Dis. 2006, 6, 21. [Google Scholar] [CrossRef]
- Bassetti, M.; Giacobbe, D.R.; Vena, A.; Wolff, M. Diagnosis and treatment of candidemia in the intensive care unit. Semin. Respir. Crit. Care Med. 2019, 40, 524–539. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Jones, R.N.; Castanheira, M. Regional data analysis of Candida non-albicans strains collected in United States medical sites over a 6-year period, 2006–2011. Mycoses 2014, 57, 602–611. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Turnidge, J.D.; Castanheira, M.; Jones, R.N. Twenty Years of the SENTRY Antifungal Surveillance Program: Results for Candida Species From 1997–2016. Open Forum Infect. Dis. 2019, 6 (Suppl. S1), S79–S94. [Google Scholar] [CrossRef] [Green Version]
- Reyes-Montes, M.D.R.; Duarte-Escalante, E.; Martínez-Herrera, E.; Acosta-Altamirano, G.; Frías-De León, M.G. Current status of the etiology of candidiasis in Mexico. Rev. Iberoam. Micol. 2017, 34, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Lamoth, F.; Lockhart, S.R.; Berkow, E.L.; Calandra, T. Changes in the epidemiological landscape of invasive candidiasis. J. Antimicrob. Chemother. 2018, 73 (Suppl. S1), i4–i13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivero-Menendez, O.; Navarro-Rodriguez, P.; Bernal-Martinez, L.; Martin-Cano, G.; Lopez-Perez, L.; Sanchez-Romero, I.; Perez-Ayala, A.; Capilla, J.; Zaragoza, O.; Alastruey-Izquierdo, A. Clinical and laboratory development of echinocandin resistance in Candida glabrata: Molecular characterization. Front. Microbiol. 2019, 10, 1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pristov, K.E.; Ghannoum, M.A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef]
- Tosun, I.; Akyuz, Z.; Guler, N.C.; Gulmez, D.; Bayramoglu, G.; Kaklikkaya, N.; Arikan-Akdagli, S.; Aydin, F. Distribution, virulence attributes and antifungal susceptibility patterns of Candida parapsilosis complex strains isolated from clinical samples. Med. Mycol. 2013, 51, 483–492. [Google Scholar] [CrossRef] [Green Version]
- Neji, S.; Hadrich, I.; Trabelsi, H.; Abbes, S.; Cheikhrouhou, F.; Sellami, H.; Makni, F.; Ayadi, A. Virulence factors, antifungal susceptibility and molecular mechanisms of azole resistance among Candida parapsilosis complex isolates recovered from clinical specimens. J. Biomed. Sci. 2017, 24, 67. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.W.; Yu, S.Y.; Xiao, M.; Wang, H.; Kudinha, T.; Kong, F.; Xu, Y.C. Identification and Antifungal Susceptibility Profile of Candida guilliermondii and Candida fermentati from a Multicenter Study in China. J. Clin. Microbiol. 2016, 54, 2187–2189. [Google Scholar] [CrossRef] [Green Version]
- Hou, X.; Xiao, M.; Chen, S.C.A.; Kong, F.; Wang, H.; Chu, Y.Z.; Kang, M.; Sun, Z.Y.; Hu, Z.D.; Li, R.Y.; et al. Molecular Epidemiology and Antifungal Susceptibility of Candida glabrata in China (August 2009 to July 2014): A Multi-Center Study. Front. Microbiol. 2017, 8, 880. [Google Scholar] [CrossRef]
- Kim, H.J.; Brehm-Stecher, B.F. Design and evaluation of peptide nucleic acid probes for specific identification of Candida albicans. J. Clin. Microbiol. 2015, 53, 511–521. [Google Scholar] [CrossRef] [Green Version]
- Turhan, O.; Ozhak-Baysan, B.; Zaragoza, O.; Er, H.; Eres Sarıtas, Z.; Ongut, G.; Ogunc, D.; Colak, D.; Cuenca-Estrella, M. Evaluation of MALDI-TOF-MS for the identification of yeast isolates causing bloodstream infection. Clin. Lab. 2017, 63, 699–703. [Google Scholar] [CrossRef]
- Fidler, G.; Leiter, E.; Kocsube, S.; Biro, S.; Paholcsek, M. Validation of a simplex PCR assay enabling reliable identification of clinically relevant Candida species. BMC Infect. Dis. 2018, 18, 393. [Google Scholar] [CrossRef]
- Talmaci, R.; Traeger-Synodinos, J.; Kanavakis, E.; Coriu, D.; Colita, D.; Gavrila, L. Scanning of beta-globin gene for identification of beta-thalassemia mutation in Romanian population. J. Cell Mol. Med. 2004, 8, 232–240. [Google Scholar] [CrossRef]
- NCBI. An Online Data Base Currently Coordinated and Supported by National Center for Biothecnology. Available online: http://ncbi.nlm.nih.gov (accessed on 22 February 2018).
- Clustal Ω. Multiple Sequence Alignment. Available online: http://www.ebi.ac.uk/Tools/msa/clustalo/ (accessed on 17 April 2018).
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Nicholas, K.B.; Nicholas, H.B., Jr.; Deerfield, D.W., II. GeneDoc: Analysis and Visualization of Genetic Variation. EMBnet.news 1997, 4, 14. [Google Scholar]
- Michael, D.G.; Simon, T.M.; Andrew, N.D.; Elizabeth, A. Primaclade a flexible tool to find conserved PCR primers across multiple species. Bioinformatics 2005, 21, 1263–1264. [Google Scholar]
- Schabereiter-Gurtner, C.; Selitsch, B.; Rotter, M.L.; Hirschl, A.M.; Willinger, B. Development of novel real-time PCR assays for detection and differentiation of eleven medically important Aspergillus and Candida species in clinical specimens. J. Clin. Microbiol. 2007, 45, 906–914. [Google Scholar] [CrossRef] [Green Version]
- Taira, C.L.; Okay, T.S.; Delgado, A.F.; Rivero Ceccon, M.E.J.; Gottardo de Almeida, M.G.; Barbaro Del Negro, G.M. A multiplex nested PCR for the detection and identification of Candida species in blood samples of critically ill paediatric patients. BMC Infect. Dis. 2014, 14, 406. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi, R.; Abdi, S. Molecular identification of Candida species isolated from gastro-oesophageal candidiasis in Tehran, Iran. Gastroenterol. Hepatol. Bed Bench 2015, 8, 288–293. [Google Scholar]
- Chang, H.; Leaw, S.; Huang, A.; Wu, T.; Chang, T. Rapid identification of yeasts in positive blood cultures by a Multiplex PCR method. J. Clin. Microbiol. 2001, 39, 3466–3471. [Google Scholar] [CrossRef] [Green Version]
- Estrada-Barraza, D.; Dávalos, M.A.; Flores, P.L.; Mendoza, D.R.; Sánchez, V.L. Comparación entre métodos convencionales, ChromAgar Candida y el método de la PCR para la identificación de especies de Candida en aislamientos clínicos. Rev. Iberoam. Micol. 2011, 28, 36–42. [Google Scholar] [CrossRef]
- Merseguel, K.; Nishikaku, A.; Rodrigues, A.; Padovan, A.; e Ferreira, R.; Salles de Azevedo Melo, A.; Ribeiro da Silva Briones, M.; Lopes Colombo, A. Genetic diversity of medically important and emerging Candida species causing invasive infection. BMC Infect. Dis. 2015, 15, 57. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef]
- Bonfietti, L.X.; Dos Anjos Martins, M.; Walderez Szeszs, M.; Stolf Pukiskas, S.B.; Ueda Purisco, S.; Cortez Pimentel, F.; Hanna Pereira, G.; Silva, D.C.; Oliveira, L.; de Souza Carvalho Melhem, M. Prevalence, distribution and antifungal susceptibility profiles of Candida parapsilosis, Candida orthopsilosis and Candida metapsilosis bloodstream isolates. J. Med. Microbiol. 2012, 61, 1003–1008. [Google Scholar] [CrossRef] [Green Version]
- Marcos-Zambrano, L.J.; Puig-Asensio, M.; Pérez-García, F.; Escribano, P.; Sánchez-Carrillo, C.; Zaragoza, O.; Padilla, B.; Cuenca-Estrella, M.; Almirante, B.; Martín-Gómez, M.T.; et al. Candida guilliermondii Complex Is Characterized by High Antifungal Resistance but Low Mortality in 22 Cases of Candidemia. Antimicrob. Agents Chemother. 2017, 61, e00099-17. [Google Scholar] [CrossRef] [Green Version]
- Małek, M.; Mrowiec, P.; Klesiewicz, K.; Skiba-Kurek, I.; Szczepański, A.; Białecka, J.; Żak, I.; Bogusz, B.; Kędzierska, J.; Budak, A.; et al. Prevalence of human pathogens of the clade Nakaseomyces in a culture collection—Tthe first report on Candida bracarensis in Poland. Folia Microbiol. 2019, 64, 307–312. [Google Scholar] [CrossRef] [Green Version]




| Isolation | Species |
|---|---|
| 828 | Candida albicans |
| 423 | Candida albicans |
| 71 | Candida albicans |
| 7 | Candida albicans |
| 119 | Candida albicans |
| 80 | Candida albicans |
| 423 | Candida albicans |
| 108 | Candida albicans |
| 316 | Candida albicans |
| 209 | Candida albicans |
| 44 | Candida albicans |
| 742 | Candida albicans |
| 227 | Candida glabrata |
| 149 | Candida glabrata |
| 22 | Candida glabrata |
| 52 | Candida glabrata |
| 28 | Candida glabrata |
| 120 | Candida glabrata |
| 15 | Candida glabrata |
| 18 | Candida glabrata |
| 109 | Candida glabrata |
| 133 | Candida glabrata |
| 4 | Candida glabrata |
| 216 | Candida parapsilosis |
| 70 | Candida parapsilosis |
| 11 | Candida parapsilosis |
| 754 | Candida parapsilosis |
| 751 | Candida parapsilosis |
| 756 | Candida parapsilosis |
| 147 | Candida parapsilosis |
| 518 | Candida parapsilosis |
| 625 | Candida parapsilosis |
| 626 | Candida parapsilosis |
| 557 | Candida parapsilosis |
| 609 | Candida parapsilosis |
| 832 | Candida tropicalis |
| 266 | Candida tropicalis |
| 70 | Candida tropicalis |
| 854 | Candida tropicalis |
| 73 | Candida tropicalis |
| 752 | Candida krusei/Pichia kudriazevii |
| 75 | Candida krusei/Pichia kudriazevii |
| Species | GenBank Access Number |
|---|---|
| C. albicans | KF241849.1, KF241847.1, MH545917.1, KC905077.1, KC905075.1, KC905076.1 |
| C. glabrata sensu stricto | KP675655.1, KP068740.1, KP675693.1, MH545922.1, KC253980.1, JN391276.1 |
| C. nivariensis | KP068747.1, KP068746.1, KP131740.1 |
| C. bracarensis | JN882338.1, KP674833.1, JN882340.1, GU199439.1 |
| C. tropicalis | JF709971.1, MH545915.1, JQ008834.1, EU589204.1 |
| C. parapsilosis sensu stricto | EU552502.1, EU564205.1, EU552500.1, EU564204.1 |
| C. metapsilosis | EU484055.1, KM014585.1, EU564207.1 |
| C. orthopsilosis | EU557370.1, EU557371.1, EU552495.1, EU557373.1, EU557372.1 |
| C. krusei/P. kudriazevii | KC886644.1, MK394162.1, KF959839.1, KF959838.1 |
| C. guilliermondii/M. guilliermondii sensu stricto | U45709.1, MH545918.1 |
| C. fermentati | AY187283.1 |
| C. carpophila/C. xestobii | U45707.1 |
| C. smithsonii | AY518525.1 |
| C. athensensis | AY518525.1 |
| C. elateridarum | AY518530.1 |
| C. lusitaniae/C. lusitaniae | KP131851.1, KY102563.1, AY493434.1, KP131846.1, KP131844.1, KP131839.1 |
| C. kefyr/Kluyveromyces marxianus | KC905771.1 |
| C. famata/D. hansenii | GQ376085.1 |
| C. rugosa | GU144663.1 |
| C. dubliniensis | KP131696.1, KP131697.1, MH545916.1, KC905080.1, KC905078.1, KC905079.1 |
| C. norvegensis | AB278166.1, AB278165.1, AB278169.1, AB278163.1, AB278162.1 |
| C. lipolytica | KP132909.1, KP132908.1, KP132907.1 |
| C. sake | FJ515167.1, KM384608.1, KM384082.1, KM384081.1 |
| C. pelliculosa | KP132885.1, KP132887.1, KP132884.1 |
| C. apícola | EU926481.1, EU926480.1, EU926479.1, EU926486.1 |
| C. zeylanoides | HE799676.1, HE799675.1, AB278160.1 |
| C. valida | KF057628.1, KF057621.1, KF057630.1, KF057617.1 |
| C. intermedia | KP131722.1 |
| C. pulcherrima | EF449525.1, AY301026.1 |
| C. haemulonii | JX459675.1, JX459674.1, KJ706229.1, JX459661.1 |
| C. utilis | KP132000.1, KP131999.1 |
| C. humícola | HM459599.1, KC118118.1, GU256753.1, JN882338.1, KP674833.1, JN882340.1, GU199439.1, J515176.1 |
| C. lambica | KF646205.1, KF646196.1, KP132501.1, KF646181.1 |
| C. ciferrii | KP132796.1, KP132795.1 |
| C. colliculosa | HE799671.1 |
| C. marina | KJ707187.1, KJ706412.1, KJ707232.1, KJ707196.1 |
| C. sphaerica | HE799667.1 |
| C. holmii | KM374151.1 |
| Oligonucleotide Name | Sequence (5′-3′) | Identified Candida Species | Amplicon Size (pb) |
|---|---|---|---|
| CandF | AGCTTGCGTTGATTACGTCCCTGCCC | C. albicans | 850 |
| C. glabrata | 1000 | ||
| C. tropicalis | 790 | ||
| C. parapsilosis | 731 | ||
| CandR | TTCACTCGCCGCTACTAAGGCAATCCC | C. krusei/P. kudriazevii | 800 |
| C. guilliermondii/M. guilliermondii | 1100 | ||
| C. lusitaniae/C. lusitaniae | 590 | ||
| C. dubliniensis | 810 |
| Sample Number | Type of Sample | PCR with Cand Primers | Culture | |||
|---|---|---|---|---|---|---|
| Positive (P)/Negative (N) | Amplicon Size (bp) | Species | Positive (P)/Negative (N) | Species * | ||
| 1 | blood | N | N | |||
| 2 | blood | N | P | C. albicans | ||
| 3 | blood | N | N | |||
| 4 | blood | P | 731 | C. parapsilosis | P | C. parapsilosis |
| 5 | blood | P | 731 | C. parapsilosis | P | C. parapsilosis |
| 6 | blood | P | 850 | C. albicans | N | |
| 7 | blood | N | N | |||
| 8 | blood | P | 850 | C. albicans | P | C. albicans |
| 9 | blood | N | N | |||
| 10 | blood | N | N | |||
| 11 | blood | N | N | |||
| 12 | blood | N | P | C. glabrata | ||
| 13 | blood | P | 850 | C. albicans | P | C. albicans |
| 14 | blood | P | 850 | C. albicans | P | C. albicans |
| 15 | blood | P | 731 | C. parapsilosis | P | C. parapsilosis |
| 16 | blood | P | 850 | C. albicans | P | C. albicans |
| 17 | blood | N | N | |||
| 18 | blood | P | 850 | C. albicans | P | C. albicans |
| 19 | blood | N | N | |||
| 20 | blood | N | N | |||
| 21 | blood | N | P | C. parapsilosis | ||
| 22 | blood | P | 731 | C. parapsilosis | P | C. parapsilosis |
| 23 | blood | N | N | |||
| 24 | blood | P | 790 | C. tropicalis | P | C. tropicalis |
| 25 | blood | N | N | |||
| 26 | blood | N | N | |||
| 27 | blood | N | N | |||
| 28 | blood | P | 850 | C. albicans | P | C. albicans |
| 29 | blood | N | N | |||
| 30 | blood | P | 850 | C. albicans | P | C. albicans |
| 31 | blood | N | N | |||
| 32 | blood | P | 850 | C. albicans | P | C. albicans |
| 33 | blood | N | P | C. famata/D. hansenii | ||
| 34 | blood | P | 850 | C. albicans | P | C. albicans |
| 35 | blood | P | 731 | C. parapsilosis | P | C. parapsilosis |
| 36 | blood | N | N | |||
| 37 | blood | N | P | C. albicans | ||
| 38 | blood | N | N | |||
| 39 | blood | P | 1000 | C. glabrata | P | C. glabrata |
| 40 | blood | N | N | |||
| 41 | BAL | N | N | |||
| 42 | BAL | N | N | |||
| 43 | BAL | P | 850 | C. albicans | P | C. albicans |
| 44 | BAL | N | N | |||
| 45 | BAL | N | N | |||
| 46 | BAL | N | N | |||
| 47 | BAL | N | N | |||
| 48 | BAL | N | P | C. albicans | ||
| 49 | BAL | N | N | |||
| 50 | BAL | N | N | |||
| Gold Standard (Culture) | ||||
| PCR with Cand primers | Present Disease | Absent Disease | Total | |
| Positive PCR | true positives: 17 | false positives: 1 | individuals with positive test: 18 | |
| Negative PCR | false negatives: 6 | true negatives: 26 | individuals with negative test: 32 | |
| Total | sick individuals: 23 | non-sick individuals: 27 | individuals included: 50 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Salazar, E.; Acosta-Altamirano, G.; Betancourt-Cisneros, P.; Reyes-Montes, M.d.R.; Rosas-De-Paz, E.; Duarte-Escalante, E.; Sánchez-Conejo, A.R.; Ocharan Hernández, E.; Frías-De-León, M.G. Detection and Molecular Identification of Eight Candida Species in Clinical Samples by Simplex PCR. Microorganisms 2022, 10, 374. https://doi.org/10.3390/microorganisms10020374
García-Salazar E, Acosta-Altamirano G, Betancourt-Cisneros P, Reyes-Montes MdR, Rosas-De-Paz E, Duarte-Escalante E, Sánchez-Conejo AR, Ocharan Hernández E, Frías-De-León MG. Detection and Molecular Identification of Eight Candida Species in Clinical Samples by Simplex PCR. Microorganisms. 2022; 10(2):374. https://doi.org/10.3390/microorganisms10020374
Chicago/Turabian StyleGarcía-Salazar, Eduardo, Gustavo Acosta-Altamirano, Paola Betancourt-Cisneros, María del Rocío Reyes-Montes, Emmanuel Rosas-De-Paz, Esperanza Duarte-Escalante, Alma Rosa Sánchez-Conejo, Esther Ocharan Hernández, and María Guadalupe Frías-De-León. 2022. "Detection and Molecular Identification of Eight Candida Species in Clinical Samples by Simplex PCR" Microorganisms 10, no. 2: 374. https://doi.org/10.3390/microorganisms10020374
APA StyleGarcía-Salazar, E., Acosta-Altamirano, G., Betancourt-Cisneros, P., Reyes-Montes, M. d. R., Rosas-De-Paz, E., Duarte-Escalante, E., Sánchez-Conejo, A. R., Ocharan Hernández, E., & Frías-De-León, M. G. (2022). Detection and Molecular Identification of Eight Candida Species in Clinical Samples by Simplex PCR. Microorganisms, 10(2), 374. https://doi.org/10.3390/microorganisms10020374







