Pseudomonas bijieensis Strain XL17 within the P. corrugata Subgroup Producing 2,4-Diacetylphloroglucinol and Lipopeptides Controls Bacterial Canker and Gray Mold Pathogens of Kiwifruit
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbial Strains
2.2. Assays of Antimicrobial Activities for Strain XL17
2.3. Scanning and Transmission Electron Microscopy on the Structure of Psa and B. cinerea
2.4. Leaf Assay of Control Efficacy on Psa
2.5. Fruit Assay of Control Efficacy on B. cinerea
2.6. Analysis of 16S rRNA Gene Sequences
2.7. Genome Sequencing and Assembly
2.8. Genome Relatedness Analysis
2.9. Genomic Analyses
2.10. Phylogenomic Analysis of the Pseudomonas Corrugata Subgroup
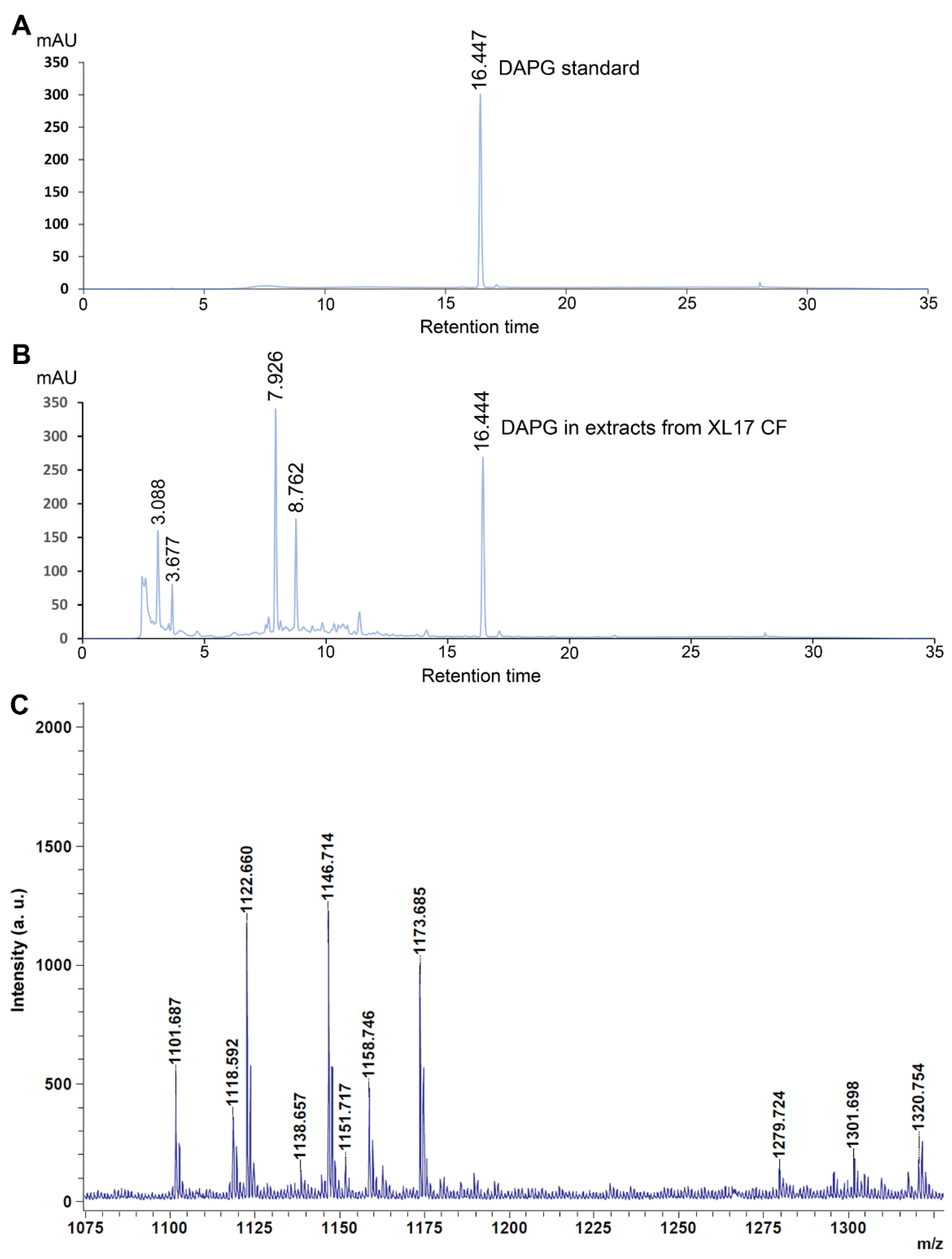
2.11. Determination of 2,4-Diacetylphloroglucinol (DAPG) Produced from XL17
2.12. Detection of Lipopeptides Produced from XL17 Cells
2.13. Detection of Fluorescent Pyoverdine Siderophore
2.14. Detection of Hydrolytic Enzyme Activities
2.15. Assay of Indole Acetic Acid (IAA) Production
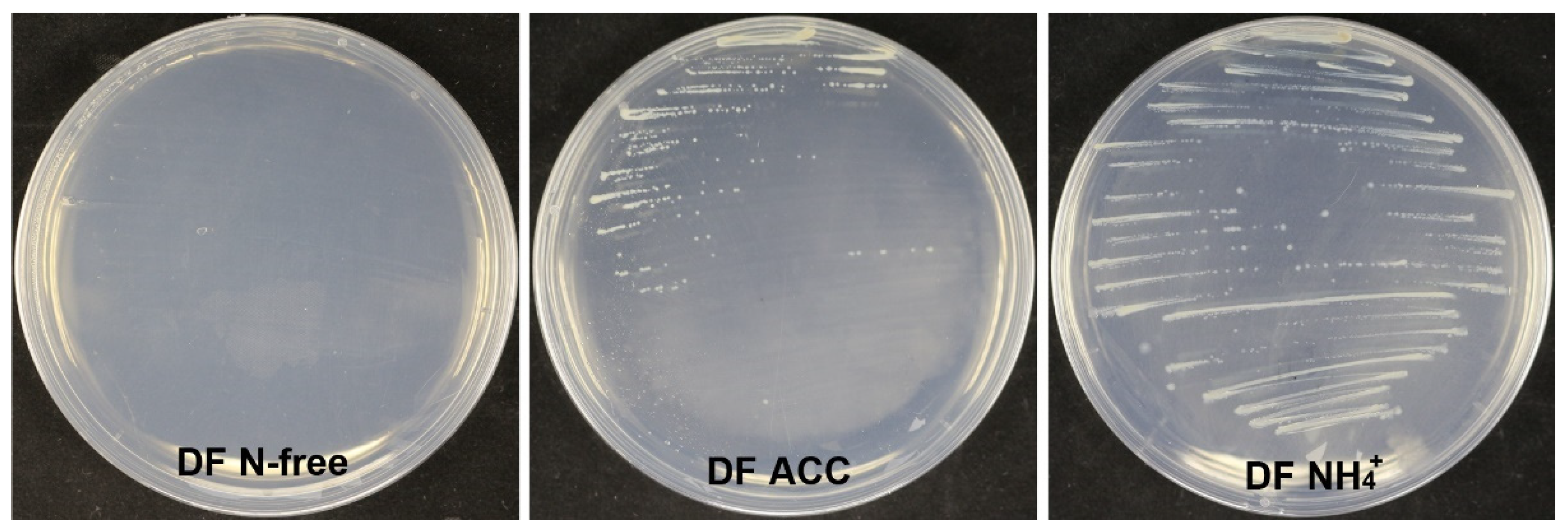
2.16. Detection of 1-Aminocyclopropane-1-Carboxylate Deaminase Activity
2.17. Assays of Toxic or Beneficial Potentials of Strain XL17 on Plants
2.18. Statistical Analysis
3. Results
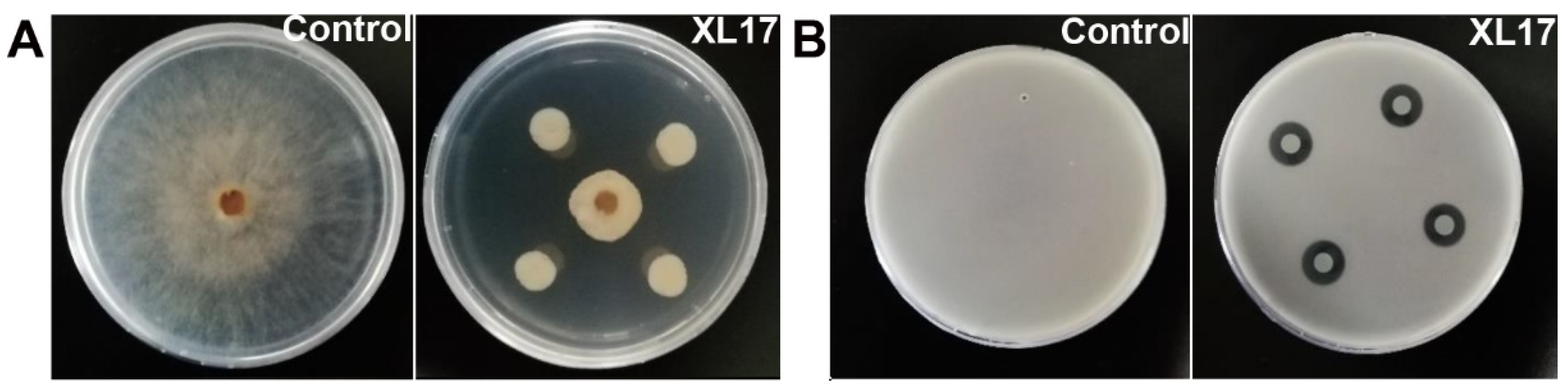
3.1. Strain XL17 Showed High Antimicrobial Activities against B. cinerea and Psa
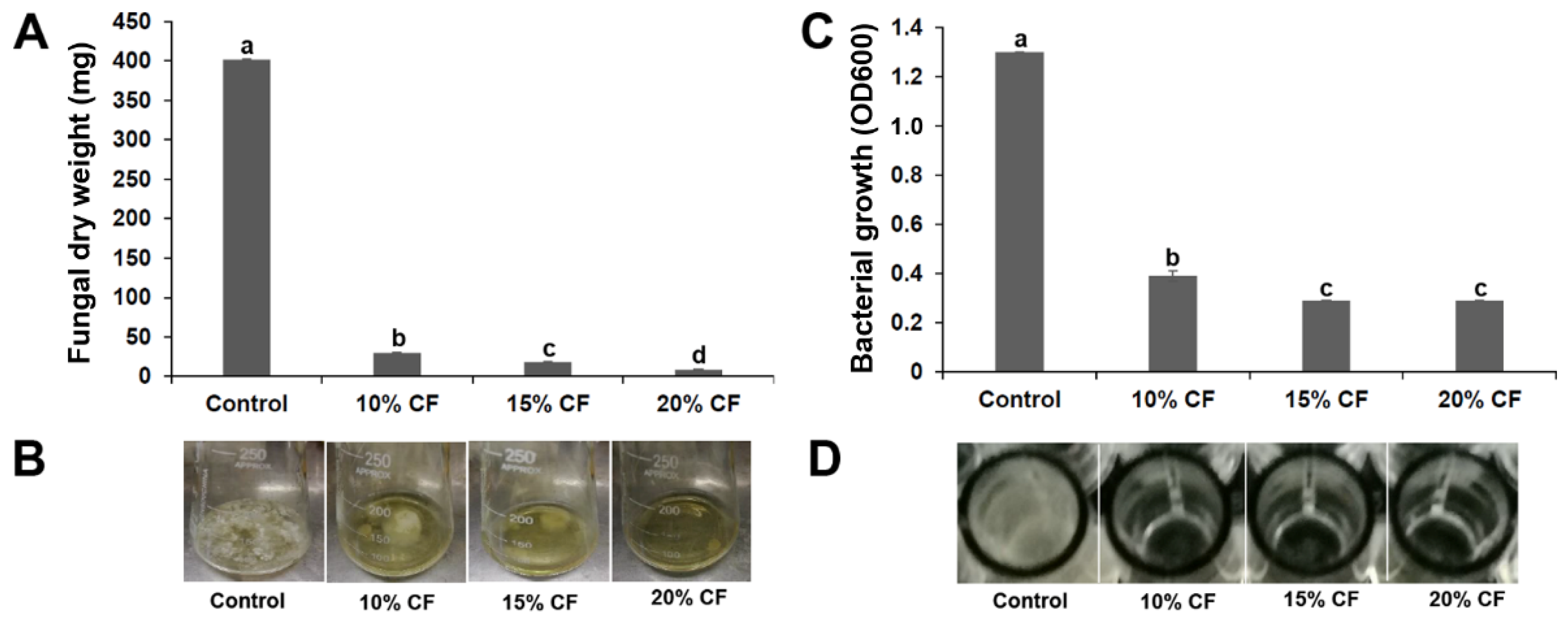
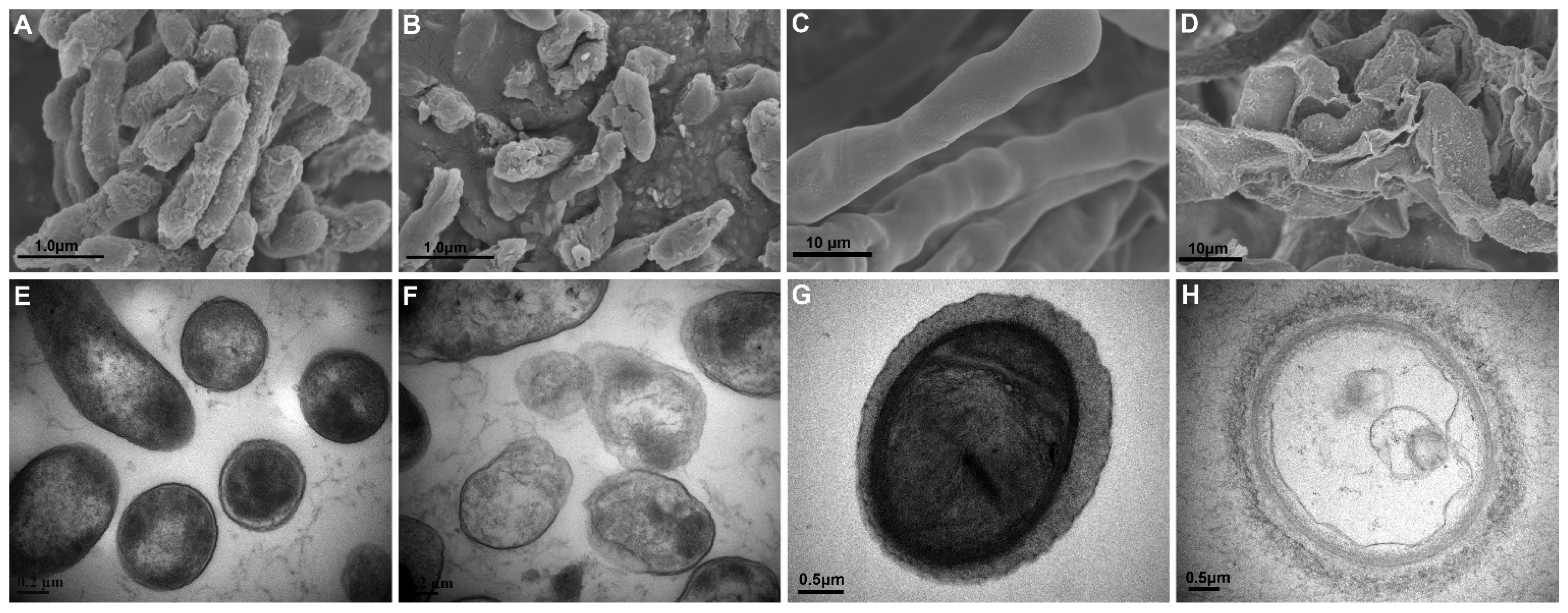
3.2. XL17 CF Damaged Psa Cells and B. cinerea Hyphae
3.3. Strain XL17 and XL17 CF Reduced Leaf Necrosis Caused by Psa and Gray Mold Lesions in Kiwifruits
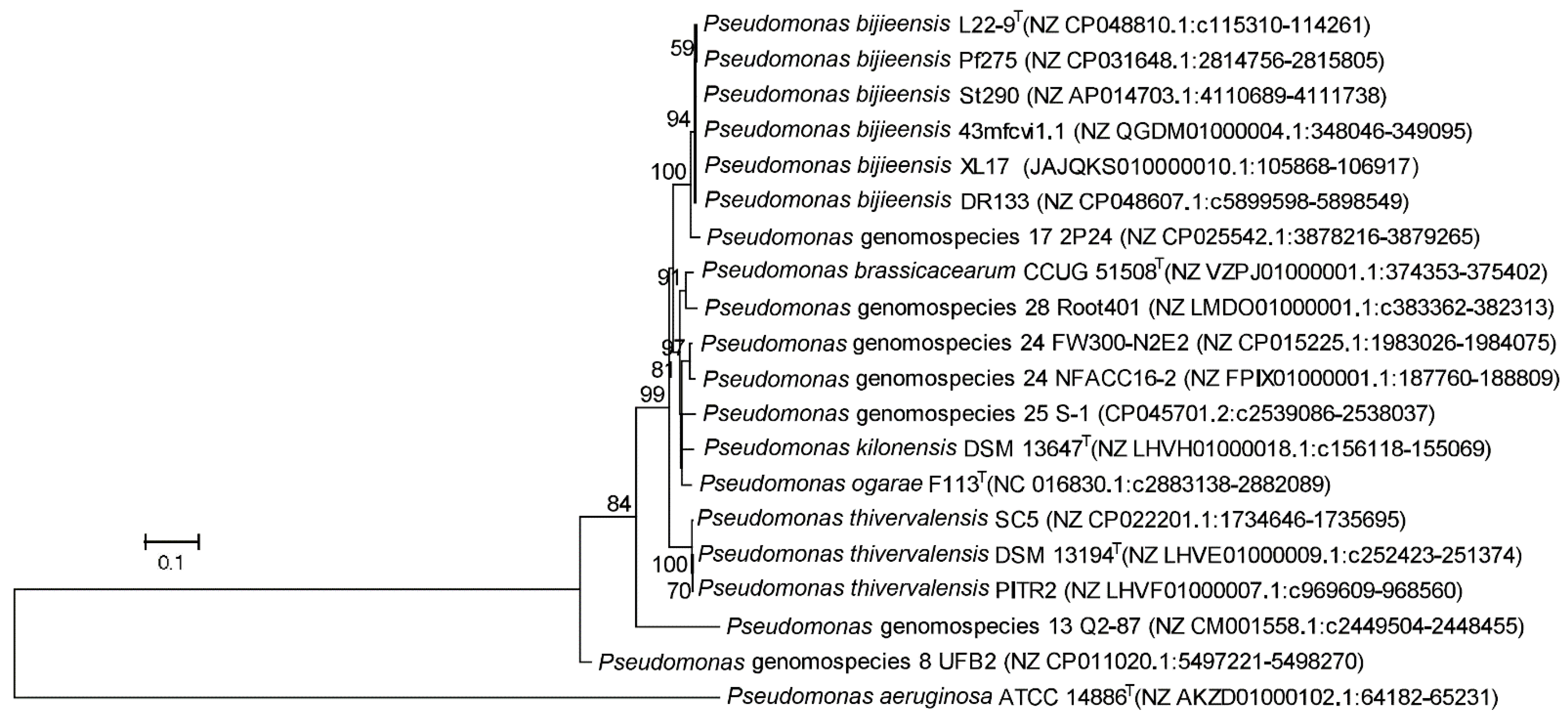
3.4. Strain XL17 Belongs to Pseudomonas bijieensis within the Pseudomonas corrugata Subgroup
3.5. Phylogeny and Genetic Markers Associated with Plant-Interaction Life Styles in the Pseudomonas corrugata Subgroup
3.6. Genes Associated with Plant-Interaction Life Style in Pseudomonas bijieensis and Strain 2P24
3.7. Strain XL17 Produced DAPG and Cyclic Lipopeptides of the Viscosin Family and Orfamide Family
3.8. Strain XL17 Produced Pyoverdine, β-1, 3-Glucanase, and Protease
3.9. Strain XL17 Produced IAA and ACC Deaminase
3.10. Strain XL17 and XL17 CF Were Not Toxic to Rice Seeds and Seedlings
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Girard, L.; Lood, C.; Höfte, M.; Vandamme, P.; Rokni-Zadeh, H.; van Noort, V.; Lavigne, R.; De Mot, R. The ever-expanding Pseudomonas genus: Description of 43 new species and partition of the Pseudomonas putida group. Microorganisms 2021, 9, 1766. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Sanz, D.; Redondo-Nieto, M.; Martin, M.; Rivilla, R. Comparative genomics of the Pseudomonas corrugata subgroup reveals high species diversity and allows the description of Pseudomonas ogarae sp. nov. Microb. Genom. 2021, 7, 000593. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, I. Fruits of the Actinidia genus. Adv. Food Nutr. Res. 2007, 52, 293–324. [Google Scholar]
- Richardson, D.P.; Ansell, J.; Drummond, L.N. The nutritional and health attributes of kiwifruit: A review. Eur. J. Nutr. 2018, 57, 2659–2676. [Google Scholar] [CrossRef] [Green Version]
- Scortichini, M.; Marcelletti, S.; Ferrante, P.; Petriccione, M.; Firrao, G. Pseudomonas syringae pv. actinidiae: A re-emerging, multi-faceted, pandemic pathogen. Mol. Plant. Pathol. 2012, 13, 631–640. [Google Scholar] [CrossRef]
- Vanneste, J.L. The Scientific, Economic, and Social Impacts of the New Zealand outbreak of bacterial canker of Kiwifruit (Pseudomonas syringae pv. actinidiae). Annu. Rev. Phytopathol. 2017, 55, 377–399. [Google Scholar] [CrossRef]
- Dai, Y.; Wang, Z.; Leng, J.; Sui, Y.; Jiang, M.; Wisniewski, M.; Liu, J.; Wang, Q. Eco-friendly management of postharvest fungal decays in kiwifruit. Crit. Rev. Food Sci. Nutri. 2021, in press. [Google Scholar] [CrossRef]
- Michailides, T.J.; Elmer, P.A.G. Botrytis gray mold of kiwifruit caused by Botrytis cinerea in the United States and New Zealand. Plant Dis. 2000, 84, 208–223. [Google Scholar] [CrossRef] [Green Version]
- Cameron, A.; Sarojini, V. Pseudomonas syringae pv. actinidiae: Chemical control, resistance mechanisms and possible alternatives. Plant Pathol. 2014, 63, 1–11. [Google Scholar] [CrossRef]
- Vanneste, J.L.; Cornish, D.A.; Yu, J.; Boyd, R.J.; Morris, C.E. Isolation of copper and streptomycin resistant phytopathogenic Pseudomonas syringae from lakes and rivers in the central North Island of New Zealand. N. Z. Plant Prot. 2008, 61, 80–85. [Google Scholar] [CrossRef] [Green Version]
- Fan, F.; Hamada, M.S.; Li, N.; Li, G.Q.; Luo, C.X. Multiple fungicide resistance in Botrytis cinerea from greenhouse Strawberries in Hubei province, China. Plant Dis. 2017, 101, 601–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rupp, S.; Weber, R.W.; Rieger, D.; Detzel, P.; Hahn, M. Spread of Botrytis cinerea strains with multiple fungicide resistance in German horticulture. Front. Microbiol. 2016, 7, 2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbey, J.A.; Percival, D.; Abbey, L.; Asiedu, S.K.; Prithiviraj, B.; Schilder, A. Biofungicides as alternative to synthetic fungicide control of grey mould (Botrytis cinerea)—Prospects and challenges. Biocontrol. Sci. Technol. 2019, 29, 207–228. [Google Scholar] [CrossRef]
- Mauri, S.; Cellini, A.; Buriani, G.; Donati, I.; Costa, G.; Spinelli, F. Optimization of cultural practices to reduce the development of Pseudomonas syringae pv. actinidiae, causal agent of the bacterial canker of kiwifruit. J. Berry Res. 2016, 6, 355–371. [Google Scholar]
- Stefani, E.; Giovanardi, D. Dissemination of Pseudomonas syringae pv. actinidiae through pollen and its epiphytic life on leaves and fruits. Phytopathol. Mediterr. 2011, 50, 489–496. [Google Scholar]
- Vanneste, J.L.; Yu, J.; Cornish, D.A.; Max, S.; Clark, G. Presence of Pseudomonas syringae pv. actinidiae, the causal agent of bacterial canker of kiwifruit, on symptomatic and asymptomatic tissues of kiwifruit. N. Z. Plant Prot. 2011, 64, 241–245. [Google Scholar] [CrossRef]
- Donati, I.; Cellini, A.; Sangiorgio, D.; Vanneste, J.L.; Scortichini, M.; Balestra, G.M.; Spinelli, F. Pseudomonas syringae pv. actinidiae: Ecology, infection dynamics and disease epidemiology. Microb. Ecol. 2020, 80, 81–102. [Google Scholar] [CrossRef]
- Figueira, D.; Garcia, E.; Ares, A.; Tiago, I.; Veríssimo, A.; Costa, J. Genetic diversity of Pseudomonas syringae pv. actinidiae: Seasonal and spatial population dynamics. Microorganisms 2020, 8, 931. [Google Scholar] [CrossRef]
- Gao, X.; Huang, Q.; Zhao, Z.; Han, Q.; Ke, X.; Qin, H.; Huang, L. Studies on the infection, colonization, and movement of Pseudomonas syringae pv. actinidiae in kiwifruit tissues using a GFPuv-labeled strain. PLoS ONE 2016, 11, e0151169. [Google Scholar] [CrossRef]
- Spinelli, F.; Donati, I.; Vanneste, J.L.; Costa, M.; Costa, G. Real time monitoring of the interactions between Pseudomonas syringae pv. actinidiae and Actinidia species. Acta Hortic. 2011, 913, 461–466. [Google Scholar] [CrossRef]
- Elad, Y.; Pertot, I.; Prado, A.M.C.; Stewart, A. Plant hosts of Botrytis spp. In Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems; Elad, Y., Fillinger, S., Eds.; Springer: Cham, Switzerland, 2016; pp. 413–486. [Google Scholar]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, M.A.; Lou, Y.; Hafeez, R.; Li, X.; Hossain, A.; Xie, T.; Lin, L.; Li, B.; Yin, Y.; Yan, J.; et al. Functional analysis and genome mining reveal high potential of biocontrol and plant growth promotion in nodule-inhabiting bacteria within Paenibacillus polymyxa complex. Front. Microbiol. 2021, 11, 618601. [Google Scholar] [CrossRef] [PubMed]
- Faretra, F.; Antonacci, E. Production of apothecia of Botryotinia fuckeliana (de Bary) Whetz. under controlled environmental conditions. Phytopathol. Medit. 1987, 26, 29–35. [Google Scholar]
- Hossain, A.; Masum, M.M.I.; Wu, X.; Abdallah, Y.; Ogunyemi, S.O.; Wang, Y.; Sun, G.; Li, B.; An, Q. Screening of Bacillus strains in biocontrol of pathogen Dickeya dadantii causing stem and root rot disease of sweet potato. Biocontrol Sci. Technol. 2020, 30, 1180–1198. [Google Scholar] [CrossRef]
- Lee, H.J.; Choi, G.J.; Cho, K.Y. Correlation of lipid peroxidation in Botrytis cinerea caused by Dicarboximide fungicides with their fungicidal activity. J. Agric. Food Chem. 1998, 46, 737–741. [Google Scholar] [CrossRef]
- Koh, Y.J.; Lee, D.H.; Shin, J.S.; Hur, J.S. Chemical and cultural control of bacterial blossom blight of kiwifruit caused by Pseudomonas syringae in Korea. N. Z. J. Crop Hortic. Sci. 2001, 29, 29–34. [Google Scholar] [CrossRef]
- Ali, M.A.; Ren, H.; Ahmed, T.; Luo, J.; An, Q.; Qi, X.; Li, B. Antifungal effects of rhizospheric Bacillus species against Bayberry twig blight pathogen Pestalotiopsis versicolor. Agronomy 2020, 10, 1811. [Google Scholar] [CrossRef]
- Liu, J.; Sui, Y.; Chen, H.; Liu, Y.; Liu, Y. Proteomic analysis of kiwifruit in response to the postharvest pathogen, Botrytis cinerea. Front. Plant. Sci. 2018, 9, 158. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Imran, M.; Xiao, L.; Hu, Z.; Li, G.; Zhang, F.; Liu, X. Difenoconazole resistance shift in Botrytis cinerea from Tomato in China associated with inducible expression of CYP51. Plant Dis. 2021, 105, 400–407. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, H.J.; Lee, E.H.; Yoon, Y.; Chua, B.; Son, A. Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J. Appl. Microbiol. 2016, 120, 379–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.H.; Liao, Y.C. CISA: Contig integrator for sequence assembly of bacterial genomes. PLoS ONE 2013, 8, e60843. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, J.; Yang, J.; Sun, S.; Xiao, J.; Yu, J. PGAP: Pan-genomes analysis pipeline. Bioinformatics 2012, 28, 416–418. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Kuma, K.I.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Weller, D.M.; Thomashow, L.S. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl. Environ. Microbiol. 1997, 63, 881–887. [Google Scholar] [CrossRef] [Green Version]
- Dutta, S.; Yu, S.M.; Jeong, S.C.; Lee, Y.H. High throughput analysis of genes involved in biocontrol performance of Pseudomonas fluorescens NBC275 against gray mold. J. Appl. Microbiol. 2019, 128, 265–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarwar, M.; Kremer, R.J. Determination of bacterially derived auxins using a microplate method. Lett. Appl. Microbiol. 1995, 20, 282–285. [Google Scholar] [CrossRef]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef] [Green Version]
- OECD. Terrestrial plants, growth test. In OECD Guideline for Testing of Chemicals No. 208; OECD: Paris, France, 1984; pp. 1–21. [Google Scholar]
- Melnyk, R.A.; Hossain, S.S.; Haney, C.H. Convergent gain and loss of genomic islands drive lifestyle changes in plant-associated Pseudomonas. ISME J. 2019, 13, 1575–1588. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083T the type strain (U5/41T) of Escherichia coli and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef] [Green Version]
- Licciardello, G.; Bertani, I.; Steindler, L.; Bella, P.; Venturi, V. Pseudomonas corrugata contains a conserved N-acyl homoserine lactone quorum sensing system; its role in tomato pathogenicity and tobacco hypersensitivity response. FEMS Microbiol. Ecol. 2007, 61, 222–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Wei, Z.; Zhang, L.Q.; Liu, X.; Wei, H.L. Supramolecular structure and functional analysis of the type III secretion system in Pseudomonas fluorescens 2P24. Front. Plant Sci. 2016, 6, 1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Luo, Y.; Gu, Y.; Wei, H.L. Characterization of the SPI-1 Type III secretion system in Pseudomonas fluorescens 2P24. Front. Microbiol. 2021, 12, 749037. [Google Scholar] [CrossRef] [PubMed]
- Gerard, J.; Lloyd, R.; Barsby, T.; Haden, P.; Kelly, M.T.; Andersen, R.J. Massetolides A-H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. J. Nat. Prod. 1997, 60, 223–229. [Google Scholar] [CrossRef]
- Nielsen, T.H.; Christophersen, C.; Anthoni, U.; Sørensen, J. Viscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54. J. Appl. Microbiol. 1999, 87, 80–90. [Google Scholar] [CrossRef]
- De Bruijn, I.; De Kock, M.J.; Yang, M.; De Waard, P.; Van Beek, T.A.; Raaijmakers, J.M. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol. Microbiol. 2007, 63, 417–428. [Google Scholar] [CrossRef]
- D’aes, J.; Kieu, N.P.; Léclère, V.; Tokarski, C.; Olorunleke, F.E.; De Maeyer, K.; Jacques, P.; Höfte, M.; Ongena, M. To settle or to move? The interplay between two classes of cyclic lipopeptides in the biocontrol strain Pseudomonas CMR12a. Environ. Microbiol. 2014, 16, 2282–2300. [Google Scholar] [CrossRef]
- Gross, H.; Stockwell, V.O.; Henkels, M.D.; Nowak-Thompson, B.; Loper, J.E.; Gerwick, W.H. The genomisotopic approach: A systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol. 2007, 14, 53–63. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Geudens, N.; Kieu, N.P.; Sinnaeve, D.; Ongena, M.; Martins, J.C.; Höfte, M. Biosynthesis, chemical structure, and structure-activity relationship of orfamide lipopeptides produced by Pseudomonas protegens and related species. Front. Microbiol. 2016, 7, 382. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Meier-Kolthoff, J.P.; Göker, M.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS ONE 2016, 11, e0153733. [Google Scholar] [CrossRef] [Green Version]
- Lalucat, J.; Mulet, M.; Gomila, M.; García-Valdés, E. Genomics in bacterial taxonomy: Impact on the genus Pseudomonas. Genes (Basel) 2020, 11, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catara, V. Pseudomonas corrugata: Plant pathogen and/or biological resource? Mol. Plant Pathol. 2007, 8, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.L.; Zhou, H.Y.; Zhang, L.Q.; Wang, Y.; Tang, W.H. Experimental evidence on the functional agent of 2,4-diacetylphloroglucinol in biocontrol activity of Pseudomonas fluorescens 2P24. Acta Microbiol. Sin. 2004, 44, 663–666. [Google Scholar]
- Zhang, W.; Zhao, Z.; Zhang, B.; Wu, X.G.; Ren, Z.G.; Zhang, L.Q. Posttranscriptional regulation of 2,4-Diacetylphloroglucinol production by GidA and TrmE in Pseudomonas fluorescens 2P24. Appl. Environ. Microbiol. 2014, 80, 3972–3981. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; Wang, S.; Yiming, A.; Fu, L.; Ahmad, I.; Chen, G.; Zhu, B. Pseudomonas bijieensis sp. nov., isolated from cornfield soil. Int. J. Syst. Evol. Microbiol. 2019, 71, 004676. [Google Scholar] [CrossRef]
- Dutta, S.; Yu, S.M.; Nagendran, R.; Jeong, S.C.; Lee, Y.H. Complete genome sequencing of Pseudomonas fluorescens NBC275, a biocontrol agent against fungal pathogens of plants and insects. Korean J. Microbiol. 2019, 55, 157–159. [Google Scholar]
- Rezzonico, F.; Zala, M.; Keel, C.; Duffy, B.; Moënne-Loccoz, Y.; Défago, G. Is the ability of biocontrol fluorescent pseudomonads to produce the antifungal metabolite 2,4-diacetylphloroglucinol really synonymous with higher plant protection? New Phytol. 2007, 17, 861–872. [Google Scholar] [CrossRef]
- Almario, J.; Bruto, M.; Vacheron, J.; Prigent-Combaret, C.; Moënne-Loccoz, Y.; Muller, D. Distribution of 2,4-Diacetylphloroglucinol biosynthetic genes among the Pseudomonas spp. reveals unexpected polyphyletism. Front. Microbiol. 2017, 8, 1218. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-García, K.; Neira-González, A.; Pérez-Gutiérrez, R.M.; Granados-Ramírez, G.; Zarraga, R.; Wrobel, K.; Barona-Gómez, F.; Flores-Cotera, L.B. Phylogenomics of 2,4-Diacetylphloroglucinol-producing Pseudomonas and novel antiglycation endophytes from Piper auritum. J. Nat. Prod. 2017, 80, 1955–1963. [Google Scholar] [CrossRef]
- Dutta, S.; Yu, S.M.; Lee, Y.H. Assessment of the contribution of antagonistic secondary metabolites to the antifungal and biocontrol activities of Pseudomonas fluorescens NBC275. Plant Pathol. J. 2020, 36, 491–496. [Google Scholar] [CrossRef]
- Geudens, N.; Martins, J.C. Cyclic lipodepsipeptides from Pseudomonas spp.–biological Swiss-army knives. Front. Microbiol. 2018, 9, 1867. [Google Scholar] [CrossRef] [PubMed]
- Girard, L.; Höfte, M.; De Mot, R. Lipopeptide families at the interface between pathogenic and beneficial Pseudomonas-plant interactions. Crit. Rev. Microbiol. 2020, 46, 397–419. [Google Scholar] [CrossRef] [PubMed]
- Julian, W.T.; Vasilchenko, A.V.; Shpindyuk, D.D.; Poshvina, D.V.; Vasilchenko, A.S. Bacterial-derived plant protection metabolite 2,4-diacetylphloroglucinol: Effects on bacterial cells at inhibitory and subinhibitory concentrations. Biomolecules 2020, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- De Souza, J.T.; Arnould, C.; Deulvot, C.; Lemanceau, P.; Gianinazzi-Pearson, V.; Raaijmakers, J.M. Effect of 2, 4-diacetylphloroglucinol on Pythium: Cellular responses and variation in sensitivity among propagules and species. Phytopathology 2003, 93, 966–975. [Google Scholar] [CrossRef] [Green Version]
- Schouten, A.; Maksimova, O.; Cuesta-Arenas, Y.; Van Den Berg, G.; Raaijmakers, J.M. Involvement of the ABC transporter BcAtrB and the laccase BcLCC2 in defence of Botrytis cinerea against the broad-spectrum antibiotic 2,4-diacetylphloroglucinol. Environ. Microbiol. 2008, 10, 1145–1157. [Google Scholar] [CrossRef]
- Chae, D.H.; Kim, D.R.; Cho, G.; Moon, S.; Kwak, Y.S. Genome-wide investigation of 2,4-Diacetylphloroglucinol protection genes in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2020, 33, 1072–1079. [Google Scholar] [CrossRef]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]




| Treatment | Germination (%) | Root Length (mm) | Shoot Height (mm) | Dry Weight (mg) |
|---|---|---|---|---|
| Control | 100 ± 0.0 a* | 45.6 ± 0.0 ab | 48.2 ± 0.0 b | 6.8 ± 0.1 b |
| XL17 | 100 ± 0.0 a | 45.9 ± 0.0 a | 50.8 ± 0.0 a | 7.4 ± 0.1 a |
| Streptomycin | 98.3 ± 1.7 a | 38.9 ± 0.0 d | 42.0 ± 0.1 c | 3.9 ± 0.1 d |
| Difenoconazole | 98.3 ± 1.7 a | 42.1 ± 0.1 c | 49.5 ± 0.0 ab | 6.3 ± 0.2 c |
| 10% CF | 100 ± 0.0 a | 44.5 ± 0.0 ab | 49.9 ± 0.1 a | 7.2 ± 0.1 a |
| 15% CF | 100 ± 0.0 a | 44.3 ± 0.0 b | 50.1 ± 0.1 a | 7.3 ± 0.2 a |
| 20% CF | 100 ± 0.0 a | 44.4 ± 0.1 b | 50.5 ± 0.0 a | 7.2 ± 0.0 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, M.A.; Luo, J.; Ahmed, T.; Zhang, J.; Xie, T.; Dai, D.; Jiang, J.; Zhu, J.; Hassan, S.; Alorabi, J.A.; et al. Pseudomonas bijieensis Strain XL17 within the P. corrugata Subgroup Producing 2,4-Diacetylphloroglucinol and Lipopeptides Controls Bacterial Canker and Gray Mold Pathogens of Kiwifruit. Microorganisms 2022, 10, 425. https://doi.org/10.3390/microorganisms10020425
Ali MA, Luo J, Ahmed T, Zhang J, Xie T, Dai D, Jiang J, Zhu J, Hassan S, Alorabi JA, et al. Pseudomonas bijieensis Strain XL17 within the P. corrugata Subgroup Producing 2,4-Diacetylphloroglucinol and Lipopeptides Controls Bacterial Canker and Gray Mold Pathogens of Kiwifruit. Microorganisms. 2022; 10(2):425. https://doi.org/10.3390/microorganisms10020425
Chicago/Turabian StyleAli, Md. Arshad, Jinyan Luo, Temoor Ahmed, Jiannan Zhang, Ting Xie, Dejiang Dai, Jingyong Jiang, Jie Zhu, Sabry Hassan, Jamal A. Alorabi, and et al. 2022. "Pseudomonas bijieensis Strain XL17 within the P. corrugata Subgroup Producing 2,4-Diacetylphloroglucinol and Lipopeptides Controls Bacterial Canker and Gray Mold Pathogens of Kiwifruit" Microorganisms 10, no. 2: 425. https://doi.org/10.3390/microorganisms10020425
APA StyleAli, M. A., Luo, J., Ahmed, T., Zhang, J., Xie, T., Dai, D., Jiang, J., Zhu, J., Hassan, S., Alorabi, J. A., Li, B., & An, Q. (2022). Pseudomonas bijieensis Strain XL17 within the P. corrugata Subgroup Producing 2,4-Diacetylphloroglucinol and Lipopeptides Controls Bacterial Canker and Gray Mold Pathogens of Kiwifruit. Microorganisms, 10(2), 425. https://doi.org/10.3390/microorganisms10020425










