Shiga Toxin-Producing Escherichia coli and Milk Fat Globules
Abstract
:1. Introduction
2. Raw Milk Sector and STEC
2.1. Importance of Raw Milk Cheeses
2.2. STEC
2.3. Milk Fat Globules
3. STEC in Raw Milk Cheeses
3.1. Prevalence and Behavior of STEC in Raw Milk Cheeses
3.2. Impact of Cheese-Making Parameters on STEC and MFGs
3.3. Location of STEC in Raw Milk Cheeses
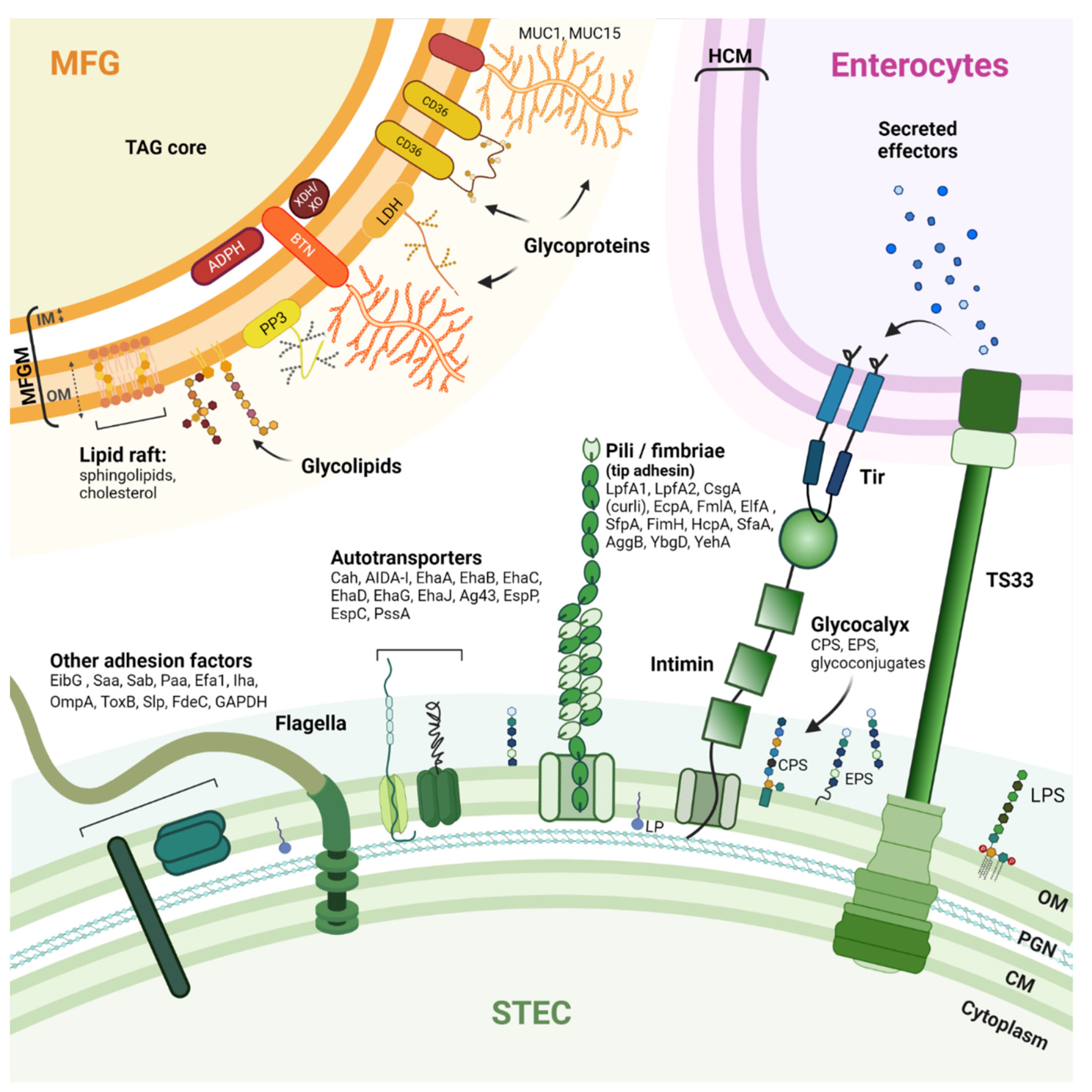
4. The Mechanism of STEC-MFG Association: What Do We Know?
4.1. General Information on Bacterial Adhesion
4.2. Physicochemical Interactions
4.2.1. Cell Surface Hydrophobicity
4.2.2. Electrostatic Forces
4.2.3. Van der Waals Forces
4.2.4. Lewis Acid/Base Interactions
4.3. Specific Molecular Interactions
4.3.1. MFGM as a Decoy Receptor for STEC
4.3.2. MFGM Proteins and Glycoproteins Potentially Targeted by STEC
5. Consequences of the STEC–MFG Association
5.1. Difficulties in Detecting STEC in Raw Milk Products

5.2. Impact of Creaming on the Presence of STEC in Milk
5.3. Anti-Adhesive Strategies
6. Conclusion and Future Directions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CNIEL Laits Liquides. Available online: https://www.filiere-laitiere.fr/fr/laits-liquides (accessed on 5 January 2022).
- International Dairy Federation. IDF Annual Report; International Dairy Federation: Schaerbeek, Belgium, 2021. [Google Scholar]
- Jost, R. Milk and Dairy Products. Ullmann’s Encycl. Ind. Chem. 2007, 23, 315–375. [Google Scholar]
- CNIEL Fromages. Available online: https://www.filiere-laitiere.fr/fr/fromages (accessed on 12 January 2022).
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The Complex Microbiota of Raw Milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skeie, S.B.; Håland, M.; Thorsen, I.M.; Narvhus, J.; Porcellato, D. Bulk Tank Raw Milk Microbiota Differs within and between Farms: A Moving Goalpost Challenging Quality Control. J. Dairy Sci. 2019, 102, 1959–1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado, S.G.; Baglinière, F.; Marchand, S.; Van Coillie, E.; Vanetti, M.C.D.; De Block, J.; Heyndrickx, M. The Biodiversity of the Microbiota Producing Heat-Resistant Enzymes Responsible for Spoilage in Processed Bovine Milk and Dairy Products. Front. Microbiol. 2017, 8, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucey, J.A. Raw Milk Consumption: Risks and Benefits. Nutr. Today 2015, 50, 189–193. [Google Scholar] [CrossRef] [Green Version]
- Crippa, G.; Zabzuni, D.; Bravi, E.; Piva, G.; De Noni, I.; Bighi, E.; Rossi, F. Randomized, Double Blind Placebo-Controlled Pilot Study of the Antihypertensive Effects of Grana Padano D.O.P. Cheese Consumption in Mild—Moderate Hypertensive Subjects. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7573–7581. [Google Scholar] [CrossRef]
- Loss, G.; Apprich, S.; Waser, M.; Kneifel, W.; Genuneit, J.; Büchele, G.; Weber, J.; Sozanska, B.; Danielewicz, H.; Horak, E.; et al. The Protective Effect of Farm Milk Consumption on Childhood Asthma and Atopy: The GABRIELA Study. J. Allergy Clin. Immunol. 2011, 128, 766–773.e4. [Google Scholar] [CrossRef]
- PASTURE Project Protection against Allergy: Study in Rural Environments. Available online: https://cordis.europa.eu/project/id/QLK4-CT-2001-00250/fr (accessed on 5 January 2022).
- French Ministry of Agriculture and Food Consumption of Cheeses Made from Raw Milk: Reminder of the Precautions to Take. Available online: https://agriculture.gouv.fr/consommation-de-fromages-base-de-lait-cru-rappel-des-precautions-prendre (accessed on 18 October 2021).
- Baylis, C.L. Raw Milk and Raw Milk Cheeses as Vehicles for Infection by Verotoxin-Producing Escherichia Coli. Int. J. Dairy Technol. 2009, 62, 293–307. [Google Scholar] [CrossRef]
- Currie, A. Outbreak of Escherichia Coli O157:H7 Infections Linked to Aged Raw Milk Gouda Cheese. J. Food Prot. 2018, 81, 325–331. [Google Scholar] [CrossRef]
- Espié, E.; Mariani-Kurkdjian, P.; Grimont, F.; Pihier, N.; Vaillant, V.; Francart, S.; Capek, I.; De Valk, H.; Vernozy-Rozand, C. Shiga-Toxin Producing Escherichia Coli O26 Infection and Unpasteurised Cows Cheese, France, 2005. In Proceedings of the 6th International Symposium on STEC, Melbourne, Australia, 30 October 2006. [Google Scholar]
- FAO; WHO. Attributing Illness Caused by Shiga Toxin-Producing Escherichia Coli (STEC) to Specific Foods; FAO: Rome, Italy, 2019. [Google Scholar]
- Honish, L.; Predy, G.; Hislop, N.; Chui, L.; Kowalewska-Grochowska, K.; Trottier, L.; Kreplin, C.; Zazulak, I. An Outbreak of E. Coli O157:H7 Hemorrhagic Colitis Associated with Unpasteurized Gouda Cheese. Can. J. Public Health 2005, 96, 182–184. [Google Scholar] [CrossRef]
- Mungai, E.A.; Behravesh, C.; Gould, L. Increased Outbreaks Associated with Nonpasteurized Milk, United States, 2007–2012. Emerg. Infect. Dis. 2015, 21, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Perrin, F.; Tenenhaus-Aziza, F.; Michel, V.; Miszczycha, S.; Bel, N.; Sanaa, M. Quantitative Risk Assessment of Haemolytic and Uremic Syndrome Linked to O157:H7 and Non-O157:H7 Shiga-Toxin Producing Escherichia Coli Strains in Raw Milk Soft Cheeses. Risk Anal. 2015, 35, 109–128. [Google Scholar] [CrossRef] [PubMed]
- Treacy, J. Outbreak of Shiga Toxin-Producing Escherichia Coli O157:H7 Linked to Raw Drinking Milk Resolved by Rapid Application of Advanced Pathogen Characterization Methods. Eurosurveillance 2019, 24, 1800191. [Google Scholar] [CrossRef] [PubMed]
- Etcheverría, A.I.; Padola, N.L. Shiga Toxin-Producing Escherichia Coli. Virulence 2013, 4, 366–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AFFSA. Bilan des Connaissances Relatives Aux Escherichia Coli Producteurs de Shiga-Toxines (STEC); French Food Safety Agency: Maisons-Alfort, France, 2003; p. 220. [Google Scholar]
- EU Directorate-General for Health and Food Safety RASFF. Available online: https://webgate.ec.europa.eu/rasff-window/screen/search (accessed on 2 February 2022).
- EFSA. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, e05926. [Google Scholar] [CrossRef] [Green Version]
- EFSA. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- EFSA. The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, e06971. [Google Scholar] [CrossRef]
- Douëllou, T.; Delannoy, S.; Ganet, S.; Mariani-Kurkdjian, P.; Fach, P.; Loukiadis, E.; Montel, M.; Thevenot-Sergentet, D. Shiga Toxin-Producing Escherichia Coli Strains Isolated from Dairy Products—Genetic Diversity and Virulence Gene Profiles. Int. J. Food Microbiol. 2016, 232, 52–62. [Google Scholar] [CrossRef]
- Claeys, W.L.; Cardoen, S.; Daube, G.; De Block, J.; Dewettinck, K.; Dierick, K.; De Zutter, L.; Huyghebaert, A.; Imberechts, H.; Thiange, P.; et al. Raw or Heated Cow Milk Consumption: Review of Risks and Benefits. Food Control 2013, 31, 251–262. [Google Scholar] [CrossRef]
- Douëllou, T.; Montel, M.C.; Thevenot Sergentet, D. Invited Review: Anti-Adhesive Properties of Bovine Oligosaccharides and Bovine Milk Fat Globule Membrane-Associated Glycoconjugates against Bacterial Food Enteropathogens. J. Dairy Sci. 2017, 100, 3348–3359. [Google Scholar] [CrossRef]
- Ofek, I.; Hasty, D.L.; Sharon, N. Anti-Adhesion Therapy of Bacterial Diseases: Prospects and Problems. FEMS Immunol. Med. Microbiol. 2003, 38, 181–191. [Google Scholar] [CrossRef]
- Yoon, Y.; Lee, S.; Choi, K.-H. Microbial Benefits and Risks of Raw Milk Cheese. Food Control 2016, 63, 201–215. [Google Scholar] [CrossRef]
- Kosmerl, E.; Rocha-Mendoza, D.; Ortega-Anaya, J.; Jiménez-Flores, R.; García-Cano, I. Improving Human Health with Milk Fat Globule Membrane, Lactic Acid Bacteria, and Bifidobacteria. Microorganisms 2021, 22, 341. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, T.A.; Lippolis, J.D. Bovine Milk Fat Globule Membrane Proteome. J. Dairy Res. 2006, 73, 406–416. [Google Scholar] [CrossRef] [Green Version]
- Spitsberg, V.L. Invited Review: Bovine Milk Fat Globule Membrane as a Potential Nutraceutical. J. Dairy Sci. 2005, 88, 2289–2294. [Google Scholar] [CrossRef]
- CNIEL. L’économie Laitière en Chiffre-Edition 2021; CNIEL: Paris, France, 2021; p. 204. [Google Scholar]
- Insée Principales Caractéristiques Des Entreprises En 2017−Caractéristiques Comptables, Financières et d’emploi Des Entreprises En 2017|Insee. Available online: https://www.insee.fr/fr/statistiques/4226019?sommaire=4226092#consulter-sommaire (accessed on 10 January 2022).
- CNIEL Centre National Interprofessionnel de l’Economie Laitière. Available online: https://www.filiere-laitiere.fr/fr/ (accessed on 12 January 2022).
- Karmali, M.A.; Gannon, V.; Sargeant, J.M. Verocytotoxin-Producing Escherichia Coli (VTEC). Vet. Microbiol. 2010, 140, 360–370. [Google Scholar] [CrossRef] [Green Version]
- Salaheen, S.; Kim, S.W.; Cao, H.; Wolfgang, D.R.; Hovingh, E.; Karns, J.S.; Haley, B.J.; Van Kessel, J.A.S. Antimicrobial Resistance Among Escherichia Coli Isolated from Veal Calf Operations in Pennsylvania. Foodborne Pathog. Dis. 2019, 16, 74–80. [Google Scholar] [CrossRef]
- Brown, C.A.; Harmon, B.G.; Zhao, T.; Doyle, M.P. Experimental Escherichia Coli O157:H7 Carriage in Calves. Appl. Environ. Microbiol. 1997, 63, 27–32. [Google Scholar] [CrossRef] [Green Version]
- Chapman, P.A.; Cerdán Malo, A.T.; Ellin, M.; Ashton, R.; Harkin, M.A. Escherichia Coli O157 in Cattle and Sheep at Slaughter, on Beef and Lamb Carcasses and in Raw Beef and Lamb Products in South Yorkshire, UK. Int. J. Food Microbiol. 2001, 64, 139–150. [Google Scholar] [CrossRef]
- Sarimehmetoglu, B.; Aksoy, M.H.; Ayaz, N.D.; Ayaz, Y.; Kuplulu, O.; Kaplan, Y.Z. Detection of Escherichia Coli O157:H7 in Ground Beef Using Immunomagnetic Separation and Multiplex PCR. Food Control 2009, 20, 357–361. [Google Scholar] [CrossRef]
- Ruegg, P.L. Practical Food Safety Interventions for Dairy Production. J. Dairy Sci. 2003, 68, E1–E9. [Google Scholar] [CrossRef] [Green Version]
- WHO; FAO. Shiga Toxin-Producing Escherichia Coli (STEC) and Food: Attribution, Characterization, and Monitoring: Report; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Bai, X.; Fu, S.; Zhang, J.; Fan, R.; Xu, Y.; Sun, H.; He, X.; Xu, J.; Xiong, Y. Identification and Pathogenomic Analysis of an Escherichia Coli Strain Producing a Novel Shiga Toxin 2 Subtype. Sci. Rep. 2018, 8, 6756. [Google Scholar] [CrossRef]
- Hughes, A.C.; Zhang, Y.; Bai, X.; Xiong, Y.; Wang, Y.; Yang, X.; Xu, Q.; He, X. Structural and Functional Characterization of Stx2k, a New Subtype of Shiga Toxin 2. Microorganisms 2019, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Patfield, S.; Rasooly, R.; Mavrici, D. Novel Monoclonal Antibodies against Stx1d and 1e and Their Use for Improving Immunoassays. J. Immunol. Methods 2017, 447, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bover-Cid, S.; Chemaly, M.; Davies, R.; Cesare, A.D.; Herman, L.; Hilbert, F.; Lindqvist, R.; et al. Pathogenicity Assessment of Shiga Toxin-Producing Escherichia Coli (STEC) and the Public Health Risk Posed by Contamination of Food with STEC. EFSA J. 2020, 18, e05967. [Google Scholar] [CrossRef]
- FAO/WHO STEC EXPERT GROUP Hazard Identification and Characterization: Criteria for Categorizing Shiga Toxin-Producing Escherichia Coli on a Risk Basis. J. Food Prot. 2019, 82, 7–21. [CrossRef] [PubMed] [Green Version]
- Newton, H.J.; Sloan, J.; Bulach, D.M.; Seemann, T.; Allison, C.C.; Tauschek, M.; Robins-Browne, R.M.; Paton, J.C.; Whittam, T.S.; Paton, A.W.; et al. Shiga Toxin–Producing Escherichia Coli Strains Negative for Locus of Enterocyte Effacement. Emerg. Infect. Dis. 2009, 15, 372–380. [Google Scholar] [CrossRef]
- Colello, R.; Krüger, A.; Velez, M.V.; Del Canto, F.; Etcheverría, A.I.; Vidal, R.; Padola, N.L. Identification and Detection of Iha Subtypes in LEE-Negative Shiga Toxin-Producing Escherichia Coli (STEC) Strains Isolated from Humans, Cattle and Food. Heliyon 2019, 5, e03015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frankel, G.; Lider, O.; Hershkoviz, R.; Mould, A.P.; Kachalsky, S.G.; Candy, D.C.A.; Cahalon, L.; Humphries, M.J.; Dougan, G. The Cell-Binding Domain of Intimin from Enteropathogenic Escherichia Coli Binds to Β1 Integrins. J. Biol. Chem. 1996, 271, 20359–20364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, J.F.; O’Brien, A.D. Cell Surface-Localized Nucleolin Is a Eukaryotic Receptor for the Adhesin Intimin-γ of Enterohemorrhagic Escherichia Coli O157:H7. J. Biol. Chem. 2002, 277, 2876–2885. [Google Scholar] [CrossRef] [Green Version]
- Sinclair, J.F.; O’Brien, A.D. Intimin Types α, β, and γ Bind to Nucleolin with Equivalent Affinity but Lower Avidity than to the Translocated Intimin Receptor. J. Biol. Chem. 2004, 279, 33751–33758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farfan, M.J.; Torres, A.G. Molecular Mechanisms That Mediate Colonization of Shiga Toxin-Producing Escherichia Coli Strains. Infect. Immun. 2012, 80, 903–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farfan, M.J.; Cantero, L.; Vidal, R.; Botkin, D.J.; Torres, A.G. Long Polar Fimbriae of Enterohemorrhagic Escherichia Coli O157:H7 Bind to Extracellular Matrix Proteins. Infect. Immun. 2011, 79, 3744–3750. [Google Scholar] [CrossRef] [Green Version]
- Herold, S.; Paton, J.C.; Paton, A.W. Sab, a Novel Autotransporter of Locus of Enterocyte Effacement-Negative Shiga-Toxigenic Escherichia Coli O113:H21, Contributes to Adherence and Biofilm Formation. Infect. Immun. 2009, 77, 3234–3243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia Coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- McWilliams, B.D.; Torres, A.G. EHEC Adhesins. Microbiol. Spectr. 2014, 2, EHEC-0003-2013. [Google Scholar] [CrossRef] [Green Version]
- Erdem, A.L.; Avelino, F.; Xicohtencatl-Cortes, J.; Girón, J.A. Host Protein Binding and Adhesive Properties of H6 and H7 Flagella of Attaching and Effacing Escherichia Coli. J. Bacteriol. 2007, 189, 7426–7435. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Iyoda, S.; Satou, H.; Satou, H.; Itoh, K.; Saitoh, T.; Watanabe, H. A New Immunoglobulin-Binding Protein, EibG, Is Responsible for the Chain-Like Adhesion Phenotype of Locus of Enterocyte Effacement-Negative, Shiga Toxin-Producing Escherichia Coli. Infect. Immun. 2006, 74, 5747–5755. [Google Scholar] [CrossRef] [Green Version]
- Rubin, D.; Zhang, W.; Karch, H.; Kuczius, T. Distinct Expression of Immunoglobulin-Binding Proteins in Shiga Toxin-Producing Escherichia Coli Implicates High Protein Stability and a Characteristic Phenotype. Toxins 2017, 9, 153. [Google Scholar] [CrossRef] [Green Version]
- Jaglic, Z.; Desvaux, M.; Weiss, A.; Nesse, L.L.; Meyer, R.L.; Demnerova, K.; Schmidt, H.; Giaouris, E.; Sipailiene, A.; Teixeira, P.; et al. Surface Adhesins and Exopolymers of Selected Foodborne Pathogens. Microbiology 2014, 160, 2561–2582. [Google Scholar] [CrossRef] [Green Version]
- Meynier, A.; Genot, C. Molecular and Structural Organization of Lipids in Foods: Their Fate during Digestion and Impact in Nutrition. OCL 2017, 24, D202. [Google Scholar] [CrossRef]
- Lopez, C.; Briard-Bion, V.; Ménard, O.; Beaucher, E.; Rousseau, F.; Fauquant, J.; Leconte, N.; Robert, B. Fat Globules Selected from Whole Milk According to Their Size: Different Compositions and Structure of the Biomembrane, Revealing Sphingomyelin-Rich Domains. Food Chem. 2011, 125, 355–368. [Google Scholar] [CrossRef]
- Raynal-Ljutovac, K.; Bouvier, J.; Gayet, C.; Simon, N.; Joffre, F.; Fine, F.; Vendeuvre, J.-L.; Lopez, C.; Chardigny, J.-M.; Michalski, M.-C.; et al. Organisation structurale et moléculaire des lipides dans les aliments: Impacts possibles sur leur digestion et leur assimilation par l’Homme. OCL 2011, 18, 324–351. [Google Scholar] [CrossRef] [Green Version]
- Evers, J.M.; Haverkamp, R.G.; Holroyd, S.E.; Jameson, G.B.; Mackenzie, D.D.S.; McCarthy, O.J. Heterogeneity of Milk Fat Globule Membrane Structure and Composition as Observed Using Fluorescence Microscopy Techniques. Int. Dairy J. 2008, 18, 1081–1089. [Google Scholar] [CrossRef]
- Keenan, T.; Mather, I. Intracellular Origin of Milk Fat Globules and the Nature of the Milk Fat Globule Membrane. In Advanced Dairy Chemistry Volume 2 Lipids; Springer: Berlin/Heidelberg, Germany, 2006; pp. 137–171. [Google Scholar]
- Lopez, C. Intracellular Origin of Milk Fat Globules, Composition and Structure of the Milk Fat Globule Membrane Highlighting the Specific Role of Sphingomyelin. In Advanced Dairy Chemistry, Volume 2: Lipids; McSweeney, P.L.H., Fox, P.F., O’Mahony, J.A., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 107–131. ISBN 978-3-030-48686-0. [Google Scholar]
- El-Zeini, H.M. Microstructure, Rheological and Geometrical Properties of Fat Globules of Milk from Different Animal Species. Pol. J. Food Nutr. Sci. 2006, 56, 147–154. [Google Scholar]
- Mather, I.H. A Review and Proposed Nomenclature for Major Proteins of the Milk-Fat Globule Membrane1,2. J. Dairy Sci. 2000, 83, 203–247. [Google Scholar] [CrossRef]
- Abd El-Salam, M.H.; El-Shibiny, S. Milk Fat Globule Membrane: An Overview with Particular Emphasis on Its Nutritional and Health Benefits. Int. J. Dairy Technol. 2020, 73, 639–655. [Google Scholar] [CrossRef]
- Jiménez-Flores, R.; Brisson, G. The Milk Fat Globule Membrane as an Ingredient: Why, How, When? Dairy Sci. Technol. 2008, 88, 5–18. [Google Scholar] [CrossRef] [Green Version]
- Farrokh, C.; Jordan, K.; Auvray, F.; Glass, K.; Oppegaard, H.; Raynaud, S.; Thevenot, D.; Condron, R.; De Reu, K.; Govaris, A.; et al. Review of Shiga-Toxin-Producing Escherichia Coli (STEC) and Their Significance in Dairy Production. Int. J. Food Microbiol. 2013, 162, 190–212. [Google Scholar] [CrossRef]
- Rivero, M.A.; Passucci, J.A.; Rodriguez, E.M.; Parma, A.E. Role and Clinical Course of Verotoxigenic Escherichia Coli Infections in Childhood Acute Diarrhoea in Argentina. J. Med. Microbiol. 2010, 59, 345–352. [Google Scholar] [CrossRef] [Green Version]
- Miszczycha, S.D.; Perrin, F.; Ganet, S.; Jamet, E.; Tenenhaus-Aziza, F.; Montel, M.-C.; Thevenot-Sergentet, D. Behavior of Different Shiga Toxin-Producing Escherichia Coli Serotypes in Various Experimentally Contaminated Raw-Milk Cheeses. Appl. Environ. Microbiol. 2013, 79, 150–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miszczycha, S.D.; Bel, N.; Gay-Perret, P.; Michel, V.; Montel, M.C.; Sergentet-Thevenot, D. Short Communication: Behavior of Different Shiga Toxin-Producing Escherichia Coli Serotypes (O26:H11, O103:H2, O145:H28, O157:H7) during the Manufacture, Ripening, and Storage of a White Mold Cheese. J. Dairy Sci. 2016, 99, 5224–5229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donnelly, C. Review of Controls for Pathogen Risks in Scottish Artisan Cheeses Made from Unpasteurised Milk; Food Standards Scotland: Aberdeen, Scotland, 2018. [Google Scholar]
- Chon, J.-W.; Kim, J.-W.; Song, K.-Y.; Lim, J.-S.; Bae, D.; Kim, H.; Seo, K.-H. Fate and Survival of Listeria Monocytogenes and Escherichia Coli O157:H7 during Ripening of Cheddar Cheeses Manufactured from Unpasteurized Raw Milk. LWT 2020, 133, 109944. [Google Scholar] [CrossRef]
- Gill, A.; Oudit, D. Enumeration of Escherichia Coli O157 in Outbreak-Associated Gouda Cheese Made with Raw Milk. J. Food Prot. 2015, 78, 1733–1737. [Google Scholar] [CrossRef]
- Bonanno, L.; Delubac, B.; Michel, V.; Auvray, F. Influence of Stress Factors Related to Cheese-Making Process and to STEC Detection Procedure on the Induction of Stx Phages from STEC O26:H11. Front. Microbiol. 2017, 8, 296. [Google Scholar] [CrossRef] [Green Version]
- Lopez, C. Focus on the Supramolecular Structure of Milk Fat in Dairy Products. Reprod. Nutr. Dev. 2005, 45, 497–511. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.H.; Jimenez-Flores, R. Heat-Induced Interactions between the Proteins of Milk Fat Globule Membrane and Skim Milk. J. Dairy Sci. 1995, 78, 24–35. [Google Scholar] [CrossRef]
- Sharma, S.K.; Dalgleish, D.G. Interactions between Milk Serum Proteins and Synthetic Fat Globule Membrane during Heating of Homogenized Whole Milk. J. Agric. Food Chem. 1993, 41, 1407–1412. [Google Scholar] [CrossRef]
- Lopez, C.; Cauty, C.; Guyomarc’h, F. Organization of Lipids in Milks, Infant Milk Formulas and Various Dairy Products: Role of Technological Processes and Potential Impacts. Dairy Sci. Technol. 2015, 95, 863–893. [Google Scholar] [CrossRef]
- Pieters, R.J. Carbohydrate Mediated Bacterial Adhesion. In Bacterial Adhesion: Chemistry, Biology and Physics; Linke, D., Goldman, A., Eds.; Advances in Experimental Medicine and Biology; Springer: Dordrecht, The Netherlands, 2011; pp. 227–240. ISBN 978-94-007-0940-9. [Google Scholar]
- Keenan, T.W.; Dylewski, D.P.; Woodford, T.A.; Ford, R.H. Origin of Milk Fat Globules and the Nature of the Milk Fat Globule Membrane. In Developments in Dairy Chemistry—2; Springer: Dordrecht, The Netherlands, 1983; Volume 2, pp. 83–118. [Google Scholar]
- Douëllou, T.; Galia, W.; Kerangart, S.; Marchal, T.; Milhau, N.; Bastien, R.; Bouvier, M.; Buff, S.; Montel, M.-C.; Sergentet-Thevenot, D. Milk Fat Globules Hamper Adhesion of Enterohemorrhagic Escherichia Coli to Enterocytes: In Vitro and In Vivo Evidence. Front. Microbiol. 2018, 9, 947. [Google Scholar] [CrossRef]
- Brewster, J.D.; Paul, M. Short Communication: Improved Method for Centrifugal Recovery of Bacteria from Raw Milk Applied to Sensitive Real-Time Quantitative PCR Detection of Salmonella spp. J. Dairy Sci. 2016, 99, 3375–3379. [Google Scholar] [CrossRef] [PubMed]
- D’Incecco, P.; Faoro, F.; Silvetti, T.; Schrader, K.; Pellegrino, L. Mechanisms of Clostridium Tyrobutyricum Removal through Natural Creaming of Milk: A Microscopy Study. J. Dairy Sci. 2015, 98, 5164–5172. [Google Scholar] [CrossRef] [PubMed]
- Laloy, E.; Vuillemard, J.-C.; El Soda, M.; Simard, R.E. Influence of the Fat Content of Cheddar Cheese on Retention and Localization of Starters. Int. Dairy J. 1996, 6, 729–740. [Google Scholar] [CrossRef]
- Oberg, C.J.; Wr, M.; Dj, M. Microstructure of Mozzarella Cheese during Manufacture. Food Struct. 1993, 12, 251–258. [Google Scholar]
- Pitino, I.; Randazzo, C.L.; Cross, K.L.; Parker, M.L.; Bisignano, C.; Wickham, M.S.J.; Mandalari, G.; Caggia, C. Survival of Lactobacillus Rhamnosus Strains Inoculated in Cheese Matrix during Simulated Human Digestion. Food Microbiol. 2012, 31, 57–63. [Google Scholar] [CrossRef]
- Brisson, G.; Payken, H.F.; Sharpe, J.P.; Jiménez-Flores, R. Characterization of Lactobacillus Reuteri Interaction with Milk Fat Globule Membrane Components in Dairy Products. J. Agric. Food Chem. 2010, 58, 5612–5619. [Google Scholar] [CrossRef]
- Lopez, C.; Maillard, M.-B.; Briard-Bion, V.; Camier, B.; Hannon, J.A. Lipolysis during Ripening of Emmental Cheese Considering Organization of Fat and Preferential Localization of Bacteria. J. Agric. Food Chem. 2006, 54, 5855–5867. [Google Scholar] [CrossRef]
- Sun, L.; Dicksved, J.; Priyashantha, H.; Lundh, Å.; Johansson, M. Distribution of Bacteria between Different Milk Fractions, Investigated Using Culture-Dependent Methods and Molecular-Based and Fluorescent Microscopy Approaches. J. Appl. Microbiol. 2019, 127, 1028–1037. [Google Scholar] [CrossRef]
- Burdikova, Z.; Svindrych, Z.; Hickey, C.; Wilkinson, M.G.; Auty, M.A.E.; Samek, O.; Bernatova, S.; Krzyzanek, V.; Periasamy, A.; Sheehan, J.J. Application of Advanced Light Microscopic Techniques to Gain Deeper Insights into Cheese Matrix Physico-Chemistry. Dairy Sci. Technol. 2015, 95, 687–700. [Google Scholar] [CrossRef] [Green Version]
- Hickey, C.D.; Sheehan, J.J.; Wilkinson, M.G.; Auty, M.A.E. Growth and Location of Bacterial Colonies within Dairy Foods Using Microscopy Techniques: A Review. Front. Microbiol. 2015, 6, 99. [Google Scholar] [CrossRef]
- Klančnik, A.; Šimunović, K.; Sterniša, M.; Ramić, D.; Smole Možina, S.; Bucar, F. Anti-Adhesion Activity of Phytochemicals to Prevent Campylobacter Jejuni Biofilm Formation on Abiotic Surfaces. Phytochem. Rev. 2020, 20, 55–84. [Google Scholar] [CrossRef] [Green Version]
- Asadi, A.; Razavi, S.; Talebi, M.; Gholami, M. A Review on Anti-Adhesion Therapies of Bacterial Diseases. Infection 2019, 47, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Ofek, I.; Bayer, E.A.; Abraham, S.N. Bacterial Adhesion. In The Prokaryotes: Human Microbiology; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 107–123. ISBN 978-3-642-30144-5. [Google Scholar]
- Boks, N.P.; Kaper, H.J.; Norde, W.; Busscher, H.J.; van der Mei, H.C. Residence Time Dependent Desorption of Staphylococcus Epidermidis from Hydrophobic and Hydrophilic Substrata. Colloids Surf. B Biointerfaces 2008, 67, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Hermansson, M. The DLVO Theory in Microbial Adhesion. Colloids Surf. B Biointerfaces 1999, 14, 105–119. [Google Scholar] [CrossRef]
- Bayoudh, S.; Othmane, A.; Mora, L.; Ben Ouada, H. Assessing Bacterial Adhesion Using DLVO and XDLVO Theories and the Jet Impingement Technique. Colloids Surf. B Biointerfaces 2009, 73, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Achinas, S.; Charalampogiannis, N.; Euverink, G.J.W. A Brief Recap of Microbial Adhesion and Biofilms. Appl. Sci. 2019, 9, 2801. [Google Scholar] [CrossRef] [Green Version]
- Beloin, C.; Houry, A.; Froment, M.; Ghigo, J.-M.; Henry, N. A Short–Time Scale Colloidal System Reveals Early Bacterial Adhesion Dynamics. PLoS Biol. 2008, 6, e167. [Google Scholar] [CrossRef]
- Katsikogianni, M.; Missirlis, Y. Concise Review of Mechanisms of Bacterial Adhesion to Biomaterials and of Techniques Used in Estimating Bacteria-Material Interactions. eCM 2004, 8, 37–57. [Google Scholar] [CrossRef]
- Dunne, W.M. Bacterial Adhesion: Seen Any Good Biofilms Lately? Clin. Microbiol. Rev. 2002, 15, 155–166. [Google Scholar] [CrossRef] [Green Version]
- van Loosdrecht, M.C.; Lyklema, J.; Norde, W.; Schraa, G.; Zehnder, A.J. The Role of Bacterial Cell Wall Hydrophobicity in Adhesion. Appl. Environ. Microbiol. 1987, 53, 1893–1897. [Google Scholar] [CrossRef] [Green Version]
- Zita, A.; Hermansson, M. Determination of Bacterial Cell Surface Hydrophobicity of Single Cells in Cultures and in Wastewater in Situ. FEMS Microbiol. Lett. 1997, 152, 299–306. [Google Scholar] [CrossRef]
- Berne, C.; Ellison, C.K.; Ducret, A.; Brun, Y.V. Bacterial Adhesion at the Single-Cell Level. Nat. Rev. Microbiol. 2018, 16, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Krasowska, A.; Sigler, K. How Microorganisms Use Hydrophobicity and What Does This Mean for Human Needs? Front. Cell Infect. Microbiol. 2014, 4, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Law, K.-Y. Water–Surface Interactions and Definitions for Hydrophilicity, Hydrophobicity and Superhydrophobicity. Pure Appl. Chem. 2015, 87, 759–765. [Google Scholar] [CrossRef]
- Chevalier, F.; Sommerer, N. Analytical Methods | Mass Spectrometric Methods. In Encyclopedia of Dairy Sciences; Elsevier: Amsterdam, The Netherlands, 2011; pp. 198–205. ISBN 978-0-12-374407-4. [Google Scholar]
- Corredig, M.; Dalgleish, D.G. Effect of Heating of Cream on the Properties of Milk Fat Globule Membrane Isolates. J. Agric. Food Chem. 1998, 46, 2533–2540. [Google Scholar] [CrossRef]
- Hong, Y.; Brown, D.G. Electrostatic Behavior of the Charge-Regulated Bacterial Cell Surface. Langmuir 2008, 24, 5003–5009. [Google Scholar] [CrossRef] [PubMed]
- Kłodzińska, E.; Szumski, M.; Dziubakiewicz, E.; Hrynkiewicz, K.; Skwarek, E.; Janusz, W.; Buszewski, B. Effect of Zeta Potential Value on Bacterial Behavior during Electrophoretic Separation. Electrophoresis 2010, 31, 1590–1596. [Google Scholar] [CrossRef]
- Ng, W. Zeta Potential of Escherichia Coli DH5α Grown in Different Growth Media; PeerJ Inc.: Corte Madera, CA, USA, 2018. [Google Scholar]
- Lytle, D.A.; Rice, E.W.; Johnson, C.H.; Fox, K.R. Electrophoretic Mobilities of Escherichia Coli O157:H7 and Wild-Type Escherichia Coli Strains. Appl. Environ. Microbiol. 1999, 65, 3222–3225. [Google Scholar] [CrossRef] [Green Version]
- Ukuku, D.O.; Fett, W.F. Relationship of Cell Surface Charge and Hydrophobicity to Strength of Attachment of Bacteria to Cantaloupe Rind†. J. Food Prot. 2002, 65, 1093–1099. [Google Scholar] [CrossRef]
- Obeid, S.; Guyomarc’h, F.; Tanguy, G.; Leconte, N.; Rousseau, F.; Dolivet, A.; Leduc, A.; Wu, X.; Cauty, C.; Jan, G.; et al. The Adhesion of Homogenized Fat Globules to Proteins Is Increased by Milk Heat Treatment and Acidic PH: Quantitative Insights Provided by AFM Force Spectroscopy. Food Res. Int. 2019, 129, 108847. [Google Scholar] [CrossRef]
- Michalski, M.-C.; Michel, F.; Sainmont, D.; Briard, V. Apparent ζ-Potential as a Tool to Assess Mechanical Damages to the Milk Fat Globule Membrane. Colloids Surf. B Biointerfaces 2002, 23, 23–30. [Google Scholar] [CrossRef]
- Michalski, M.-C.; Camier, B.; Briard, V.; Leconte, N.; Gassi, J.-Y.; Goudédranche, H.; Michel, F.; Fauquant, J. The Size of Native Milk Fat Globules Affects Physico-Chemical and Functional Properties of Emmental Cheese. Le Lait 2004, 84, 343–358. [Google Scholar] [CrossRef]
- Verma, A.; Ghosh, T.; Bhushan, B.; Packirisamy, G.; Navani, N.K.; Sarangi, P.P.; Ambatipudi, K. Characterization of Difference in Structure and Function of Fresh and Mastitic Bovine Milk Fat Globules. PLoS ONE 2019, 14, e0221830. [Google Scholar] [CrossRef]
- Malik, P.; Danthine, S.; Paul, A.; Blecker, C. Physical-Chemical Properties of Milk Fat Globule Membrane at Different Stages of Isolation. Sci. Bull. Ser. F Biotechnol. 2015, 19, 154–159. [Google Scholar]
- Azghani, A.O.; Clark, C.A. Bacterial Infection Process: An Overview. In Regulation of the Inflammatory Response in Health and Disease; Research Signpost: Thiruvananthapuram, India, 2009; pp. 37–55. ISBN 978-81-308-0372-2. [Google Scholar]
- Kendall, K.; Roberts, A.D. Van Der Waals Forces Influencing Adhesion of Cells. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140078. [Google Scholar] [CrossRef] [Green Version]
- Burgain, J.; Scher, J.; Francius, G.; Borges, F.; Corgneau, M.; Revol-Junelles, A.M.; Cailliez-Grimal, C.; Gaiani, C. Lactic Acid Bacteria in Dairy Food: Surface Characterization and Interactions with Food Matrix Components. Adv. Colloid Interface Sci. 2014, 213, 21–35. [Google Scholar] [CrossRef]
- Kiely, L.J.; Olson, N.F. The Physicochemical Surface Characteristics of Lactobacillus Casei. Food Microbiol. 2000, 17, 277–291. [Google Scholar] [CrossRef]
- Kiely, L.J.; Olson, N.F. Short Communication: Estimate of Non Electrostatic Interaction Free Energy Parameters for Milk Fat Globules. J. Dairy Sci. 2003, 86, 3110–3112. [Google Scholar] [CrossRef]
- Shoaf-Sweeney, K.D.; Hutkins, R.W. Chapter 2 Adherence, Anti-Adherence, and Oligosaccharides. In Advances in Food and Nutrition Research; Elsevier: Amsterdam, The Netherlands, 2008; Volume 55, pp. 101–161. ISBN 978-0-12-374120-2. [Google Scholar]
- Cho, S.-H.; Lee, K.M.; Kim, C.-H.; Kim, S.S. Construction of a Lectin–Glycan Interaction Network from Enterohemorrhagic Escherichia Coli Strains by Multi-Omics Analysis. IJMS 2020, 21, 2681. [Google Scholar] [CrossRef]
- Ielasi, F.S.; Alioscha-Perez, M.; Donohue, D.; Claes, S.; Sahli, H.; Schols, D.; Willaert, R.G. Lectin-Glycan Interaction Network-Based Identification of Host Receptors of Microbial Pathogenic Adhesins. mBio 2016, 7, e00584-16. [Google Scholar] [CrossRef] [Green Version]
- Sharon, N.; Ofek, I. Microbial Lectins. In Comprehensive Glycoscience-From Chemistry to Systems Biology; Elsevier: Amsterdam, The Netherlands, 2007; pp. 623–659. [Google Scholar]
- Nie, S.; Cui, S.W.; Xie, M. Bioactive Polysaccharides; Academic Press: London, UK; Elsevier: San Diego, CA, USA, 2018; ISBN 978-0-12-809418-1. [Google Scholar]
- Chevalier, L.; Selim, J.; Genty, D.; Baste, J.M.; Piton, N.; Boukhalfa, I.; Hamzaoui, M.; Pareige, P.; Richard, V. Electron Microscopy Approach for the Visualization of the Epithelial and Endothelial Glycocalyx. Morphologie 2017, 101, 55–63. [Google Scholar] [CrossRef]
- Varki, A. Biological Roles of Glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef] [Green Version]
- Monteiro, R.; Ageorges, V.; Rojas-Lopez, M.; Schmidt, H.; Weiss, A.; Bertin, Y.; Forano, E.; Jubelin, G.; Henderson, I.R.; Livrelli, V.; et al. A Secretome View of Colonisation Factors in Shiga Toxin-Encoding Escherichia Coli (STEC): From Enterohaemorrhagic E. Coli (EHEC) to Related Enteropathotypes. FEMS Microbiol. Lett. 2016, 363, fnw179. [Google Scholar] [CrossRef] [Green Version]
- Dhakal, B.K.; Bower, J.M.; Mulvey, M.A.; Yang, X.H. Pili, Fimbriae☆. In Encyclopedia of Microbiology, 4th ed.; Schmidt, T.M., Ed.; Academic Press: Oxford, UK, 2019; pp. 595–613. ISBN 978-0-12-811737-8. [Google Scholar]
- Ross, S.A.; Lane, J.A.; Kilcoyne, M.; Joshi, L.; Hickey, R.M. Defatted Bovine Milk Fat Globule Membrane Inhibits Association of Enterohaemorrhagic Escherichia Coli O157:H7 with Human HT-29 Cells. Int. Dairy J. 2016, 59, 36–43. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Juanes, F.; Alonso, J.M.; Zancada, L.; Hueso, P. Glycosphingolipids from Bovine Milk and Milk Fat Globule Membranes: A Comparative Study. Adhesion to Enterotoxigenic Escherichia Coli Strains. Biol. Chem. 2009, 390, 31–40. [Google Scholar] [CrossRef]
- Novakovic, P.; Huang, Y.Y.; Lockerbie, B.; Shahriar, F.; Kelly, J.; Gordon, J.R.; Middleton, D.M.; Loewen, M.E.; Kidney, B.A.; Simko, E. Identification of Escherichia Coli F4ac-Binding Proteins in Porcine Milk Fat Globule Membrane. Can. J. Vet. Res. 2015, 79, 120–128. [Google Scholar]
- Bergström, J.H.; Birchenough, G.M.H.; Katona, G.; Schroeder, B.O.; Schütte, A.; Ermund, A.; Johansson, M.E.V.; Hansson, G.C. Gram-Positive Bacteria Are Held at a Distance in the Colon Mucus by the Lectin-like Protein ZG16. Proc. Natl. Acad. Sci. USA 2016, 113, 13833–13838. [Google Scholar] [CrossRef] [Green Version]
- Barboza, M.; Pinzon, J.; Wickramasinghe, S.; Froehlich, J.W.; Moeller, I.; Smilowitz, J.T.; Ruhaak, L.R.; Huang, J.; Lönnerdal, B.; German, J.B.; et al. Glycosylation of Human Milk Lactoferrin Exhibits Dynamic Changes During Early Lactation Enhancing Its Role in Pathogenic Bacteria-Host Interactions. Mol. Cell Proteom. 2012, 11, M111.015248. [Google Scholar] [CrossRef] [Green Version]
- Ye, A.; Singh, H.; Taylor, M.; Anema, S. Interactions of Whey Proteins with Milk Fat Globule Membrane Proteins during Heat Treatment of Whole Milk. Le Lait 2004, 84, 269–283. [Google Scholar] [CrossRef] [Green Version]
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin Glycan Foraging in the Human Gut Microbiome. Front. Genet. 2015, 6, 81. [Google Scholar] [CrossRef] [Green Version]
- Josenhans, C.; Müthing, J.; Elling, L.; Bartfeld, S.; Schmidt, H. How Bacterial Pathogens of the Gastrointestinal Tract Use the Mucosal Glyco-Code to Harness Mucus and Microbiota: New Ways to Study an Ancient Bag of Tricks. Int. J. Med. Microbiol. 2020, 310, 151392. [Google Scholar] [CrossRef]
- Harvey, K.L.; Jarocki, V.M.; Charles, I.G.; Djordjevic, S.P. The Diverse Functional Roles of Elongation Factor Tu (EF-Tu) in Microbial Pathogenesis. Front. Microbiol. 2019, 10, 2351. [Google Scholar] [CrossRef]
- Bao, J.; Pan, G.; Poncz, M.; Wei, J.; Ran, M.; Zhou, Z. Serpin Functions in Host-Pathogen Interactions. PeerJ 2018, 6, e4557. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Zhang, W.; Chang, Q.; Su, Z.; Gong, D.; Zhou, Y.; Xiao, J.; Drelich, A.; Liu, Y.; Popov, V.; et al. A New Role for Host Annexin A2 in Establishing Bacterial Adhesion to Vascular Endothelial Cells: Lines of Evidence from Atomic Force Microscopy and an in Vivo Study. Lab. Investig. 2019, 99, 1650–1660. [Google Scholar] [CrossRef]
- Li, X.; Pei, G.; Zhang, L.; Cao, Y.; Wang, J.; Yu, L.; Dianjun, W.; Gao, S.; Zhang, Z.-S.; Yao, Z.; et al. Compounds Targeting YadC of Uropathogenic Escherichia Coli and Its Host Receptor Annexin A2 Decrease Bacterial Colonization in Bladder. EBioMedicine 2019, 50, 23–33. [Google Scholar] [CrossRef] [Green Version]
- Rand, J.H.; Wu, X.-X.; Lin, E.Y.; Griffel, A.; Gialanella, P.; McKitrick, J.C. Annexin A5 Binds to Lipopolysaccharide and Reduces Its Endotoxin Activity. mBio 2012, 3, e00292-11. [Google Scholar] [CrossRef] [Green Version]
- Hari-Dass, R.; Shah, C.; Meyer, D.J.; Raynes, J.G. Serum Amyloid A Protein Binds to Outer Membrane Protein A of Gram-Negative Bacteria. J. Biol. Chem. 2005, 280, 18562–18567. [Google Scholar] [CrossRef] [Green Version]
- Beck, W.H.J.; Adams, C.P.; Biglang-awa, I.M.; Patel, A.B.; Vincent, H.; Haas-Stapleton, E.J.; Weers, P.M.M. Apolipoprotein A-I Binding to Anionic Vesicles and Lipopolysaccharides: Role for Lysine Residues in Antimicrobial Properties. Biochim. Biophys. Acta 2013, 1828, 1503–1510. [Google Scholar] [CrossRef] [Green Version]
- Han, Q.; Han, Y.; Wen, H.; Pang, Y.; Li, Q. Molecular Evolution of Apolipoprotein Multigene Family and the Original Functional Properties of Serum Apolipoprotein (LAL2) in Lampetra Japonica. Front. Immunol. 2020, 11, 1751. [Google Scholar] [CrossRef]
- Huang, Y.; Hui, K.; Jin, M.; Yin, S.; Wang, W.; Ren, Q. Two Endoplasmic Reticulum Proteins (Calnexin and Calreticulin) Are Involved in Innate Immunity in Chinese Mitten Crab (Eriocheir Sinensis). Sci. Rep. 2016, 6, 27578. [Google Scholar] [CrossRef]
- van Harten, R.M.; van Woudenbergh, E.; van Dijk, A.; Haagsman, H.P. Cathelicidins: Immunomodulatory Antimicrobials. Vaccines 2018, 6, 63. [Google Scholar] [CrossRef] [Green Version]
- Baranova, I.N.; Kurlander, R.; Bocharov, A.V.; Vishnyakova, T.G.; Chen, Z.; Remaley, A.T.; Csako, G.; Patterson, A.P.; Eggerman, T.L. Role of Human CD36 in Bacterial Recognition, Phagocytosis and Pathogen-Induced C-Jun N-Terminal Kinase (JNK)-Mediated Signaling. J. Immunol. 2008, 181, 7147–7156. [Google Scholar] [CrossRef] [PubMed]
- Martinez, V.G.; Escoda-Ferran, C.; Tadeu Simões, I.; Arai, S.; Orta Mascaró, M.; Carreras, E.; Martínez-Florensa, M.; Yelamos, J.; Miyazaki, T.; Lozano, F. The Macrophage Soluble Receptor AIM/Api6/CD5L Displays a Broad Pathogen Recognition Spectrum and Is Involved in Early Response to Microbial Aggression. Cell Mol. Immunol. 2014, 11, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.; van Pijkeren, J.-P.; Svensson, L.; Claesson, M.J.; Sturme, M.; Li, Y.; Cooney, J.C.; van Sinderen, D.; Walker, A.W.; Parkhill, J.; et al. Fibrinogen-Binding and Platelet-Aggregation Activities of a Lactobacillus Salivarius Septicaemia Isolate Are Mediated by a Novel Fibrinogen-Binding Protein. Mol. Microbiol. 2012, 85, 862–877. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J. The MSCRAMM Family of Cell-Wall-Anchored Surface Proteins of Gram-Positive Cocci. Trends Microbiol. 2019, 27, 927–941. [Google Scholar] [CrossRef] [PubMed]
- Hymes, J.P.; Klaenhammer, T.R. Stuck in the Middle: Fibronectin-Binding Proteins in Gram-Positive Bacteria. Front. Microbiol. 2016, 7, 1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, Y.-P.; Flick, M.J. Fibrinogen Is at the Interface of Host Defense and Pathogen Virulence in Staphylococcus Aureus Infection. Semin. Thromb Hemost 2016, 42, 408–421. [Google Scholar] [CrossRef] [Green Version]
- Oh, Y.J.; Hubauer-Brenner, M.; Gruber, H.J.; Cui, Y.; Traxler, L.; Siligan, C.; Park, S.; Hinterdorfer, P. Curli Mediate Bacterial Adhesion to Fibronectin via Tensile Multiple Bonds. Sci. Rep. 2016, 6, 33909. [Google Scholar] [CrossRef] [PubMed]
- Pizarro-Cerdá, J.; Cossart, P. Bacterial Adhesion and Entry into Host Cells. Cell 2006, 124, 715–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasta, G.R. Roles of Galectins in Infection. Nat. Rev. Microbiol. 2009, 7, 424–438. [Google Scholar] [CrossRef] [Green Version]
- Bucki, R.; Byfield, F.J.; Kulakowska, A.; McCormick, M.E.; Drozdowski, W.; Namiot, Z.; Hartung, T.; Janmey, P.A. Extracellular Gelsolin Binds Lipoteichoic Acid and Modulates Cellular Response to Proinflammatory Bacterial Wall Components. J. Immunol. 2008, 181, 4936–4944. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, C.; Ohlsen, K.; Hauck, C.R. Integrin-Mediated Uptake of Fibronectin-Binding Bacteria. Eur. J. Cell Biol. 2011, 90, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Krukonis, E.S.; Isberg, R.R. Microbial Pathogens and Integrin Interactions. In Integrin-Ligand Interaction; Springer: Boston, MA, USA, 1997; pp. 175–197. ISBN 978-1-4757-4066-0. [Google Scholar]
- Scibelli, A.; Roperto, S.; Manna, L.; Pavone, L.M.; Tafuri, S.; Morte, R.D.; Staiano, N. Engagement of Integrins as a Cellular Route of Invasion by Bacterial Pathogens. Vet. J. 2007, 173, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Ulanova, M.; Gravelle, S.; Barnes, R. The Role of Epithelial Integrin Receptors in Recognition of Pulmonary Pathogens. J. Innate Immun. 2008, 1, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Shahriar, F.; Ngeleka, M.; Gordon, J.R.; Simko, E. Identification by Mass Spectroscopy of F4ac-Fimbrial-Binding Proteins in Porcine Milk and Characterization of Lactadherin as an Inhibitor of F4ac-Positive Escherichia Coli Attachment to Intestinal Villi in Vitro. Dev. Comp. Immunol. 2006, 30, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Juarez, R.C.; Romero, M.J.; Ascencio, F. Adhesive Properties of a LamB-like Outer-Membrane Protein and Its Contribution to Aeromonas Veronii Adhesion. J. Appl. Microbiol. 2004, 96, 700–708. [Google Scholar] [CrossRef]
- Peiser, L.; Gough, P.J.; Kodama, T.; Gordon, S. Macrophage Class A Scavenger Receptor-Mediated Phagocytosis of Escherichia Coli: Role of Cell Heterogeneity, Microbial Strain, and Culture Conditions In Vitro. Infect. Immun. 2000, 68, 1953–1963. [Google Scholar] [CrossRef] [Green Version]
- Naughton, J.; Duggan, G.; Bourke, B.; Clyne, M. Interaction of Microbes with Mucus and Mucins. Gut Microbes 2014, 5, 48–52. [Google Scholar] [CrossRef] [Green Version]
- Iovino, F.; Engelen-Lee, J.-Y.; Brouwer, M.; van de Beek, D.; van der Ende, A.; Valls Seron, M.; Mellroth, P.; Muschiol, S.; Bergstrand, J.; Widengren, J.; et al. PIgR and PECAM-1 Bind to Pneumococcal Adhesins RrgA and PspC Mediating Bacterial Brain Invasion. J. Exp. Med. 2017, 214, 1619–1630. [Google Scholar] [CrossRef]
- Johansen, F.-E.; Kaetzel, C.S. Regulation of the Polymeric Immunoglobulin Receptor and IgA Transport: New Advances in Environmental Factors That Stimulate PIgR Expression and Its Role in Mucosal Immunity. Mucosal Immunol. 2011, 4, 598–602. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.; Bowden, G.H.W.; Myal, Y. Identification of Mouse Submaxillary Gland Protein in Mouse Saliva and Its Binding to Mouse Oral Bacteria. Arch. Oral Biol. 2002, 47, 327–332. [Google Scholar] [CrossRef]
- Schenkels, L.C.P.M.; Walgreen-Weterings, E.; Oomen, L.C.J.M.; Bolscher, J.G.M.; Veerman, E.C.I.; Nieuw Amerongen, A.V. In Vivo Binding of the Salivary Glycoprotein EP-GP (Identical to GCDFP-15) to Oral and Non-Oral Bacteria Detection and Identification of EP-GP Binding Species. Biol. Chem. 1997, 378, 83–88. [Google Scholar] [CrossRef]
- Lu, X.; Wang, M.; Qi, J.; Wang, H.; Li, X.; Gupta, D.; Dziarski, R. Peptidoglycan Recognition Proteins Are a New Class of Human Bactericidal Proteins. J. Biol. Chem. 2006, 281, 5895–5907. [Google Scholar] [CrossRef] [Green Version]
- Green, R.S.; Naimi, W.A.; Oliver, L.D.; O’Bier, N.; Cho, J.; Conrad, D.H.; Martin, R.K.; Marconi, R.T.; Carlyon, J.A. Binding of Host Cell Surface Protein Disulfide Isomerase by Anaplasma Phagocytophilum Asp14 Enables Pathogen Infection. mBio 2020, 11, e03141-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawasaki, T.; Kawai, T. Toll-Like Receptor Signaling Pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mossman, K.L.; Mian, M.F.; Lauzon, N.M.; Gyles, C.L.; Lichty, B.; Mackenzie, R.; Gill, N.; Ashkar, A.A. Cutting Edge: FimH Adhesin of Type 1 Fimbriae Is a Novel TLR4 Ligand. J. Immunol. 2008, 181, 6702–6706. [Google Scholar] [CrossRef]
- Oliveira-Nascimento, L.; Massari, P.; Wetzler, L.M. The Role of TLR2 in Infection and Immunity. Front. Immunol. 2012, 3, 79. [Google Scholar] [CrossRef] [Green Version]
- Tükel, Ç.; Nishimori, J.H.; Wilson, R.P.; Winter, M.G.; Keestra, A.M.; van Putten, J.P.M.; Bäumler, A.J. Toll-like Receptors 1 and 2 Cooperatively Mediate Immune Responses to Curli, a Common Amyloid from Enterobacterial Biofilms. Cell Microbiol. 2010, 12, 1495–1505. [Google Scholar] [CrossRef] [Green Version]
- Pak, J.; Pu, Y.; Zhang, Z.T.; Hasty, D.L.; Wu, X.R. Tamm-Horsfall Protein Binds to Type 1 Fimbriated Escherichia Coli and Prevents E. Coli from Binding to Uroplakin Ia and Ib Receptors. J. Biol. Chem. 2001, 276, 9924–9930. [Google Scholar] [CrossRef] [Green Version]
- Yanagihara, S.; Kanaya, T.; Fukuda, S.; Nakato, G.; Hanazato, M.; Wu, X.-R.; Yamamoto, N.; Ohno, H. Uromodulin–SlpA Binding Dictates Lactobacillus Acidophilus Uptake by Intestinal Epithelial M Cells. Int. Immunol. 2017, 29, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Mak, T.N.; Brüggemann, H. Vimentin in Bacterial Infections. Cells 2016, 5, 18. [Google Scholar] [CrossRef]
- Singh, B.; Su, Y.-C.; Riesbeck, K. Vitronectin in Bacterial Pathogenesis: A Host Protein Used in Complement Escape and Cellular Invasion. Mol. Microbiol. 2010, 78, 545–560. [Google Scholar] [CrossRef]
- Baik, J.E.; Choe, H.-I.; Hong, S.W.; Kang, S.-S.; Ahn, K.B.; Cho, K.; Yun, C.-H.; Han, S.H. Human Salivary Proteins with Affinity to Lipoteichoic Acid of Enterococcus Faecalis. Mol. Immunol. 2016, 77, 52–59. [Google Scholar] [CrossRef]
- Gomand, F.; Borges, F.; Guerin, J.; El-Kirat-Chatel, S.; Francius, G.; Dumas, D.; Burgain, J.; Gaiani, C. Adhesive Interactions Between Lactic Acid Bacteria and β-Lactoglobulin: Specificity and Impact on Bacterial Location in Whey Protein Isolate. Front. Microbiol. 2019, 10, 1512. [Google Scholar] [CrossRef] [Green Version]
- Guerin, J.; Bacharouche, J.; Burgain, J.; Lebeer, S.; Francius, G.; Borges, F.; Scher, J.; Gaiani, C. Pili of Lactobacillus Rhamnosus GG Mediate Interaction with β-Lactoglobulin. Food Hydrocoll. 2016, 58, 35–41. [Google Scholar] [CrossRef]
- Mather, I.H. Milk Lipids | Milk Fat Globule Membrane. In Encyclopedia of Dairy Sciences; Elsevier: Amsterdam, The Netherlands, 2011; pp. 680–690. ISBN 978-0-12-374407-4. [Google Scholar]
- Yang, M.; Cong, M.; Peng, X.; Wu, J.; Wu, R.; Liu, B.; Ye, W.; Yue, X. Quantitative Proteomic Analysis of Milk Fat Globule Membrane (MFGM) Proteins in Human and Bovine Colostrum and Mature Milk Samples through ITRAQ Labeling. Food Funct. 2016, 7, 2438–2450. [Google Scholar] [CrossRef]
- Bachiero, D.; Uson, S.; JimÃ, R. Lipid Binding Characterization of Lactic Acid Bacteria in Dairy Products. J. Dairy Sci. 2007, 90 (Suppl. S1), 490. [Google Scholar]
- Zhang, L.; García-Cano, I.; Jiménez-Flores, R. Characterization of Adhesion between Limosilactobacillus Reuteri and Milk Phospholipids by Density Gradient and Gene Expression. JDS Commun. 2020, 1, 29–35. [Google Scholar] [CrossRef]
- Janganan, T.K.; Mullin, N.; Tzokov, S.B.; Stringer, S.; Fagan, R.P.; Hobbs, J.K.; Moir, A.; Bullough, P.A. Characterization of the Spore Surface and Exosporium Proteins of Clostridium Sporogenes; Implications for Clostridium Botulinum Group I Strains. Food Microbiol. 2016, 59, 205–212. [Google Scholar] [CrossRef] [Green Version]
- Shuster, B.; Khemmani, M.; Nakaya, Y.; Holland, G.; Iwamoto, K.; Abe, K.; Imamura, D.; Maryn, N.; Driks, A.; Sato, T.; et al. Expansion of the Spore Surface Polysaccharide Layer in Bacillus Subtilis by Deletion of Genes Encoding Glycosyltransferases and Glucose Modification Enzymes. J. Bacteriol. 2019, 201, e00321-19. [Google Scholar] [CrossRef] [Green Version]
- D’Incecco, P.; Ong, L.; Pellegrino, L.; Faoro, F.; Barbiroli, A.; Gras, S. Effect of Temperature on the Microstructure of Fat Globules and the Immunoglobulin-Mediated Interactions between Fat and Bacteria in Natural Raw Milk Creaming. J. Dairy Sci. 2018, 101, 2984–2997. [Google Scholar] [CrossRef] [Green Version]
- LeBouder, E.; Rey-Nores, J.E.; Raby, A.-C.; Affolter, M.; Vidal, K.; Thornton, C.A.; Labéta, M.O. Modulation of Neonatal Microbial Recognition: TLR-Mediated Innate Immune Responses Are Specifically and Differentially Modulated by Human Milk. J. Immunol. 2006, 176, 3742–3752. [Google Scholar] [CrossRef] [Green Version]
- Honkanen-Buzalski, T.; Sandholm, M. Association of Bovine Secretory Immunoglobulins with Milk Fat Globule Membranes. Comp. Immunol. Microbiol. Infect. Dis. 1981, 4, 329–342. [Google Scholar] [CrossRef]
- Schroten, H.; Bosch, M.; Nobis-Bosch, R.; Köhler, H.; Hanisch, F.G.; Plogmann, R. Secretory Immunoglobulin A Is a Component of the Human Milk Fat Globule Membrane. Pediatr. Res. 1999, 45, 82–86. [Google Scholar] [CrossRef] [Green Version]
- Korhonen, H.; Marnila, P.; Gill, H.S. Bovine Milk Antibodies for Health. Br. J. Nutr. 2000, 84, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Mitra, A.K.; Mahalanabis, D.; Ashraf, H.; Unicomb, L.; Eeckels, R.; Tzipori, S. Hyperimmune Cow Colostrum Reduces Diarrhoea Due to Rotavirus: A Double-Blind, Controlled Clinical Trial. Acta Paediatr. 1995, 84, 996–1001. [Google Scholar] [CrossRef]
- Tawfeek, H.I.; Najim, N.H.; Al-Mashikhi, S. Efficacy of an Infant Formula Containing Anti-Escherichia Coli Colostral Antibodies from Hyperimmunized Cows in Preventing Diarrhea in Infants and Children: A Field Trial. Int. J. Infect. Dis. 2003, 7, 120–128. [Google Scholar] [CrossRef] [Green Version]
- Adachi, E.; Tanaka, H.; Toyoda, N.; Takeda, T. Detection of bactericidal antibody in the breast milk of a mother infected with enterohemorrhagic Escherichia Coli O157:H7. Kansenshogaku Zasshi 1999, 73, 451–456. [Google Scholar] [CrossRef] [Green Version]
- Noguera-Obenza, M.; Ochoa, T.J.; Gomez, H.F.; Guerrero, M.L.; Herrera-Insua, I.; Morrow, A.L.; Ruiz-Palacios, G.; Pickering, L.K.; Guzman, C.A.; Cleary, T.G. Human Milk Secretory Antibodies against Attaching and Effacing Escherichia Coli Antigens. Emerg. Infect. Dis. 2003, 9, 545–551. [Google Scholar] [CrossRef]
- Funatogawa, K.; Ide, T.; Kirikae, F.; Saruta, K.; Nakano, M.; Kirikae, T. Use of Immunoglobulin Enriched Bovine Colostrum against Oral Challenge with Enterohaemorrhagic Escherichia Coli O157:H7 in Mice. Microbiol. Immunol. 2002, 46, 761–766. [Google Scholar] [CrossRef] [Green Version]
- Funatogawa, K.; Tada, T.; Kuwahara-Arai, K.; Kirikae, T.; Takahashi, M. Enriched Bovine IgG Fraction Prevents Infections with Enterohaemorrhagic Escherichia Coli O157:H7, Salmonella Enterica Serovar Enteritidis, and Mycobacterium Avium. Food Sci. Nutr. 2019, 7, 2726–2730. [Google Scholar] [CrossRef] [Green Version]
- Rabinovitz, B.C.; Gerhardt, E.; Tironi Farinati, C.; Abdala, A.; Galarza, R.; Vilte, D.A.; Ibarra, C.; Cataldi, A.; Mercado, E.C. Vaccination of Pregnant Cows with EspA, EspB, γ-Intimin, and Shiga Toxin 2 Proteins from Escherichia Coli O157:H7 Induces High Levels of Specific Colostral Antibodies That Are Transferred to Newborn Calves. J. Dairy Sci. 2012, 95, 3318–3326. [Google Scholar] [CrossRef]
- de Oliveira, I.R.; Bessler, H.C.; Bao, S.N.; del Lima, R.; Giugliano, L.G. Inhibition of Enterotoxigenic Escherichia Coli (ETEC) Adhesion to Caco-2 Cells by Human Milk and Its Immunoglobulin and Non-Immunoglobulin Fractions. Braz. J. Microbiol. 2007, 38, 86–92. [Google Scholar] [CrossRef] [Green Version]
- Atroshi, F.; Alaviuhkola, T.; Schildt, R.; Sandholm, M. Fat Globule Membrane of Sow Milk as a Target for Adhesion of K88-Positive Escherichia Coli. Comp. Immunol. Microbiol. Infect. Dis. 1983, 6, 235–245. [Google Scholar] [CrossRef]
- Mulder, H.; Walstra, P. The Milk Fat Globule; Commonwealth Agricultural Bureaux Farnham Royal: Slough, UK, 1974; ISBN 0-85198-289-1. [Google Scholar]
- Geer, S.R.; Barbano, D.M. The Effect of Immunoglobulins and Somatic Cells on the Gravity Separation of Fat, Bacteria, and Spores in Pasteurized Whole Milk1. J. Dairy Sci. 2014, 97, 2027–2038. [Google Scholar] [CrossRef] [Green Version]
- Hansen, S.F.; Larsen, L.B.; Wiking, L. Thermal Effects on IgM-Milk Fat Globule-Mediated Agglutination. J. Dairy Res. 2019, 86, 108–113. [Google Scholar] [CrossRef]
- Turula, H.; Wobus, C.E. The Role of the Polymeric Immunoglobulin Receptor and Secretory Immunoglobulins during Mucosal Infection and Immunity. Viruses 2018, 10, 237. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Xu, L.; Park, H.-B.; Hwang, J.; Kwak, M.; Lee, P.C.W.; Liang, G.; Zhang, X.; Xu, J.; Jin, J.-O. Escherichia Coli Adhesion Portion FimH Functions as an Adjuvant for Cancer Immunotherapy. Nat. Commun. 2020, 11, 1187. [Google Scholar] [CrossRef] [Green Version]
- Rapsinski, G.J.; Newman, T.N.; Oppong, G.O.; van Putten, J.P.M.; Tükel, Ç. CD14 Protein Acts as an Adaptor Molecule for the Immune Recognition of Salmonella Curli Fibers. J. Biol. Chem. 2013, 288, 14178–14188. [Google Scholar] [CrossRef] [Green Version]
- Lenehan, D.; Murray, A.; Smith, S.; Uhlin, B.E.; Mitchell, J. Characterisation of E. Coli Lipopolysaccharide Adherence to Platelet Receptors. Blood 2016, 128, 4906. [Google Scholar] [CrossRef]
- Li, B.; Liu, H.; Wang, W. Multiplex Real-Time PCR Assay for Detection of Escherichia Coli O157:H7 and Screening for Non-O157 Shiga Toxin-Producing E. Coli. BMC Microbiol. 2017, 17, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miszczycha, S.D.; Ganet, S.; Duniere, L.; Rozand, C.; Loukiadis, E.; Thevenot-Sergentet, D. Novel Real-Time PCR Method to Detect Escherichia Coli O157:H7 in Raw Milk Cheese and Raw Ground Meat. J. Food Prot. 2012, 75, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Martens, E.C.; Neumann, M.; Desai, M.S. Interactions of Commensal and Pathogenic Microorganisms with the Intestinal Mucosal Barrier. Nat. Rev. Microbiol. 2018, 16, 457. [Google Scholar] [CrossRef]
- Stinson, L.F.; Ma, J.; Rea, A.; Dymock, M.; Geddes, D.T. Centrifugation Does Not Remove Bacteria from the Fat Fraction of Human Milk. Sci. Rep. 2021, 11, 572. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Lim, M.-C.; Park, K.; Ok, G.; Chang, H.-J.; Lee, N.; Park, T.J.; Choi, S.-W. Detection of E. Coli O157:H7 in Food Using Automated Immunomagnetic Separation Combined with Real-Time PCR. Processes 2020, 8, 908. [Google Scholar] [CrossRef]
- Yang, D.; Wang, Y.; Zhao, L.; Rao, L.; Liao, X. Extracellular PH Decline Introduced by High Pressure Carbon Dioxide Is a Main Factor Inducing Bacteria to Enter Viable but Non-Culturable State. Food Res. Int. 2022, 151, 110895. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. A Comparison of Methods Used to Extract Bacterial DNA from Raw Milk and Raw Milk Cheese. J. Appl. Microbiol. 2012, 113, 96–105. [Google Scholar] [CrossRef]
- Giffel, M.T.; Horst, H.V.D. Comparison between Bactofugation and Microfiltration Regarding Efficiency of Somatic Cell and Bacteria Removal. Bull. Int. Dairy Fed. 2004, 389, 49–53. [Google Scholar]
- Bagel, A.; Université de Lyon, Lyon, France; Douëllou, T.; Université de Lyon, Lyon, France; Sergentet, D.; Université de Lyon, Lyon, France. Distribution of Escherichia coli in bovine raw milk by creaming assay. Unpublished work. 2019. [Google Scholar]
- Anderson, J.F. The Relative Proportion Of Bacteria In Top Milk (Cream Layer) And Bottom Milk (Skim Milk), and Its Bearing On Infant Feeding. J. Infect. Dis. 1909, 6, 392–400. [Google Scholar] [CrossRef]
- Bird, J. Cream Separation. Int. J. Dairy Technol. 1991, 44, 61–63. [Google Scholar] [CrossRef]
- Kahane, I.; Ofek, I. Toward Anti-Adhesion Therapy for Microbial Diseases; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; ISBN 978-1-4613-0415-9. [Google Scholar]
- Orth, K.; Krachler, A.-M. Made to Stick: Anti-Adhesion Therapy for Bacterial Infections: A Major Advantage in Targeting Adhesion Is That the Body Clears Invading Pathogens Instead of Killing Them. Microbe Mag. 2013, 8, 286–290. [Google Scholar] [CrossRef] [Green Version]
- Sharon, N. Carbohydrates as Future Anti-Adhesion Drugs for Infectious Diseases. Biochim. Biophys. Acta 2006, 1760, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Horemans, T.; Kerstens, M.; Clais, S.; Struijs, K.; van den Abbeele, P.; Assche, T.V.; Maes, L.; Cos, P. Evaluation of the Anti-Adhesive Effect of Milk Fat Globule Membrane Glycoproteins on Helicobacter Pylori in the Human NCI-N87 Cell Line and C57BL/6 Mouse Model. Helicobacter 2012, 17, 312–318. [Google Scholar] [CrossRef]
- Kvistgaard, A.S.; Pallesen, L.T.; Arias, C.F.; López, S.; Petersen, T.E.; Heegaard, C.W.; Rasmussen, J.T. Inhibitory Effects of Human and Bovine Milk Constituents on Rotavirus Infections. J. Dairy Sci. 2004, 87, 4088–4096. [Google Scholar] [CrossRef] [Green Version]
- Martín-Sosa, S.; Martín, M.-J.; Hueso, P. The Sialylated Fraction of Milk Oligosaccharides Is Partially Responsible for Binding to Enterotoxigenic and Uropathogenic Escherichia Coli Human Strains. J. Nutr. 2002, 132, 3067–3072. [Google Scholar] [CrossRef] [PubMed]
- Schroten, H.; Hanisch, F.G.; Plogmann, R.; Hacker, J.; Uhlenbruck, G.; Nobis-Bosch, R.; Wahn, V. Inhibition of Adhesion of S-Fimbriated Escherichia Coli to Buccal Epithelial Cells by Human Milk Fat Globule Membrane Components: A Novel Aspect of the Protective Function of Mucins in the Nonimmunoglobulin Fraction. Infect. Immun. 1992, 60, 2893–2899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, P.M.; Goode, P.L.; Mobasseri, A.; Zopf, D. Inhibition of Helicobacter Pylori Binding to Gastrointestinal Epithelial Cells by Sialic Acid-Containing Oligosaccharides. Infect. Immun. 1997, 65, 750–757. [Google Scholar] [CrossRef] [Green Version]
- Sprong, R.C.; Hulstein, M.F.E.; Lambers, T.T.; van der Meer, R. Sweet Buttermilk Intake Reduces Colonisation and Translocation of Listeria Monocytogenes in Rats by Inhibiting Mucosal Pathogen Adherence. Br. J. Nutr. 2012, 108, 2026–2033. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Hirmo, S.; Wadström, T.; Willén, R. Inhibition of Helicobacter Pylori Infection by Bovine Milk Glycoconjugates in a BALB/cA Mouse Model. J. Med. Microbiol. 2001, 50, 430–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manoni, M.; Di Lorenzo, C.; Ottoboni, M.; Tretola, M.; Pinotti, L. Comparative Proteomics of Milk Fat Globule Membrane (MFGM) Proteome across Species and Lactation Stages and the Potentials of MFGM Fractions in Infant Formula Preparation. Foods 2020, 9, 1251. [Google Scholar] [CrossRef]
- Hernell, O.; Timby, N.; Domellöf, M.; Lönnerdal, B. Clinical Benefits of Milk Fat Globule Membranes for Infants and Children. J. Pediatr. 2016, 173, S60–S65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Padhi, E.; Hasegawa, Y.; Larke, J.; Parenti, M.; Wang, A.; Hernell, O.; Lönnerdal, B.; Slupsky, C. Compositional Dynamics of the Milk Fat Globule and Its Role in Infant Development. Front. Pediatr. 2018, 6, 313. [Google Scholar] [CrossRef] [Green Version]
- da Silva, R.C.; Colleran, H.L.; Ibrahim, S.A. Milk Fat Globule Membrane in Infant Nutrition: A Dairy Industry Perspective. J. Dairy Res. 2021, 88, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Timby, N.; Domellöf, M.; Lönnerdal, B.; Hernell, O. Supplementation of Infant Formula with Bovine Milk Fat Globule Membranes12. Adv. Nutr. 2017, 8, 351–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Bovine MFGM Components | Bacterial Components | References |
|---|---|---|
| Adipophilin * (ADPH) | F4ac (E. coli) fimbria | [142] |
| Alpha 1-antichymotrypsin (serpin) | - | [149] |
| Annexins A1, A2, A5 | LPS (lipid A), OmpB, YadC (tip adhesin of Yad fimbriae) | [150,151,152] |
| Apolipoprotein serum amyloid A protein | OmpA | [153] |
| Apolipoproteins | LPS | [154,155] |
| Butyrophilin * | F4ac (E. coli) fimbria | [29,142] |
| Calnexin | LPS, peptidoglycan | [156] |
| Cathelicidin 1 | LPS, LTA | [157] |
| CD36 * | LPS, LTA | [29,158] |
| CD5L protein | - | [159] |
| Elongation factor thermal unstable Tu (EF-Tu) | - | [148] |
| Fatty acid-binding protein * | F4ac (E. coli) fimbria | [142] |
| Fibrinogen | Fibrinogen-binding protein (MSCRAMMs), curli | [160,161,162,163,164,165] |
| Galectin 7 | LPS | [166] |
| Gelsolin | LPS, LTA | [167] |
| Immunoglobulins | Many bacterial proteins | - |
| Integrin | Many bacterial proteins | [52,165,168,169,170,171] |
| Lactadherin * | F4ac (E. coli) fimbria | [142,172] |
| Lactoferrin | OMPs | [173] |
| Macrophage scavenger receptor | LPS, LTA | [174] |
| MUC1 *, MUC15 * | Many bacterial proteins | [175] |
| Polymeric immunoglobulin receptor (PIgR) | Ig-mediated adhesion, direction interaction via adhesin | [176,177] |
| Prolactin-inducible protein (mPIP) | - | [178,179] |
| Peptidoglycan recognition protein 1 | - | [180] |
| Protein disulfide-isomerase (PDI) | - | [181] |
| Toll-like receptor 4, 2 | Many bacterial proteins | [182,183,184,185] |
| Uromodulin | Surface layer protein A, FimH | [186,187] |
| Vimentin | Many bacterial proteins | [188] |
| Vitronectin | Many bacterial proteins | [189] |
| Zymogen granule protein 16 homolog B | LTA, peptidoglycan | [190] |
| β-lactoglobulin | Spa pili | [191,192] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bagel, A.; Sergentet, D. Shiga Toxin-Producing Escherichia coli and Milk Fat Globules. Microorganisms 2022, 10, 496. https://doi.org/10.3390/microorganisms10030496
Bagel A, Sergentet D. Shiga Toxin-Producing Escherichia coli and Milk Fat Globules. Microorganisms. 2022; 10(3):496. https://doi.org/10.3390/microorganisms10030496
Chicago/Turabian StyleBagel, Arthur, and Delphine Sergentet. 2022. "Shiga Toxin-Producing Escherichia coli and Milk Fat Globules" Microorganisms 10, no. 3: 496. https://doi.org/10.3390/microorganisms10030496
APA StyleBagel, A., & Sergentet, D. (2022). Shiga Toxin-Producing Escherichia coli and Milk Fat Globules. Microorganisms, 10(3), 496. https://doi.org/10.3390/microorganisms10030496





