Viral Ejection Proteins: Mosaically Conserved, Conformational Gymnasts
Abstract
:1. Introduction
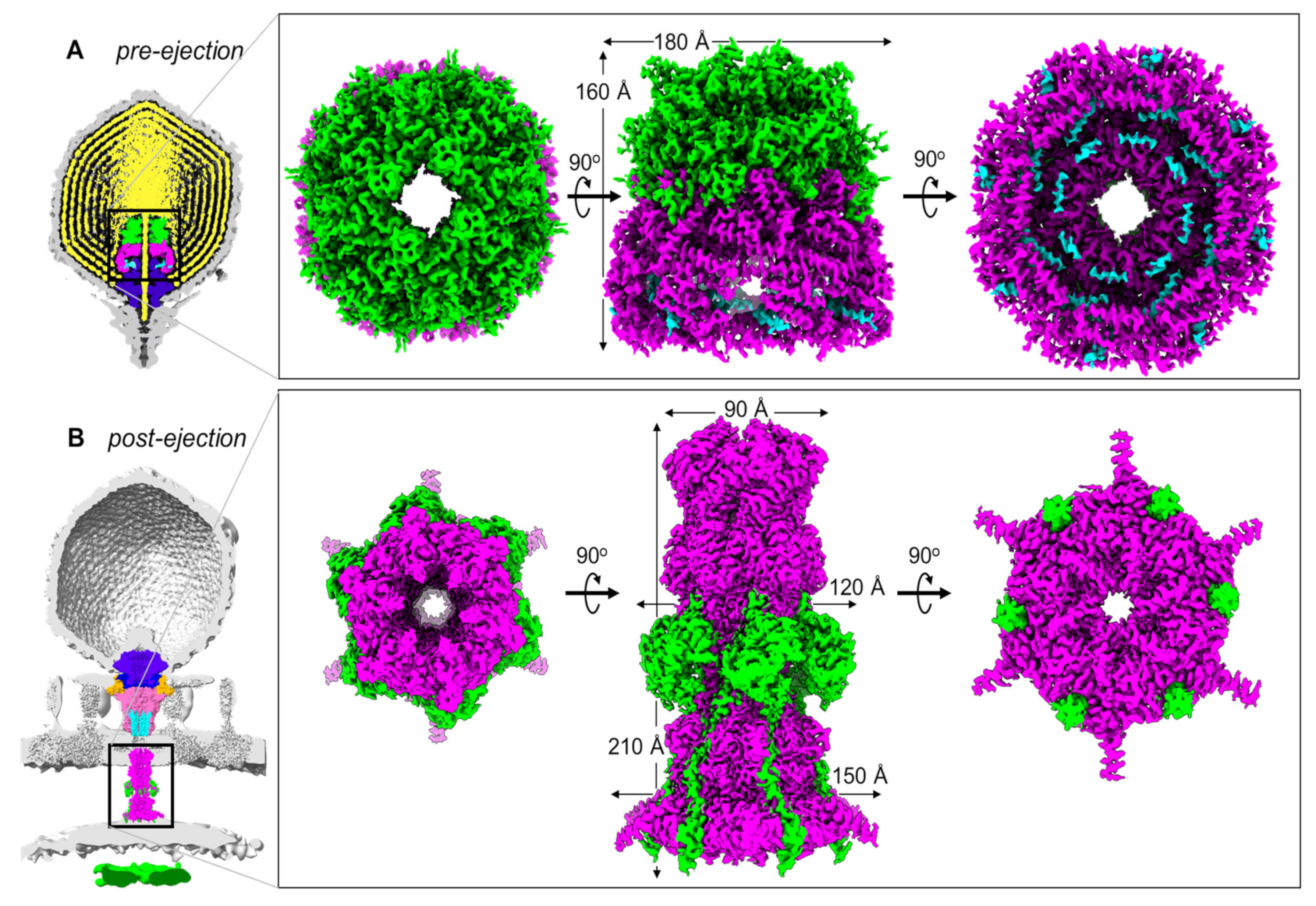
2. The T7 DNA Ejectosome
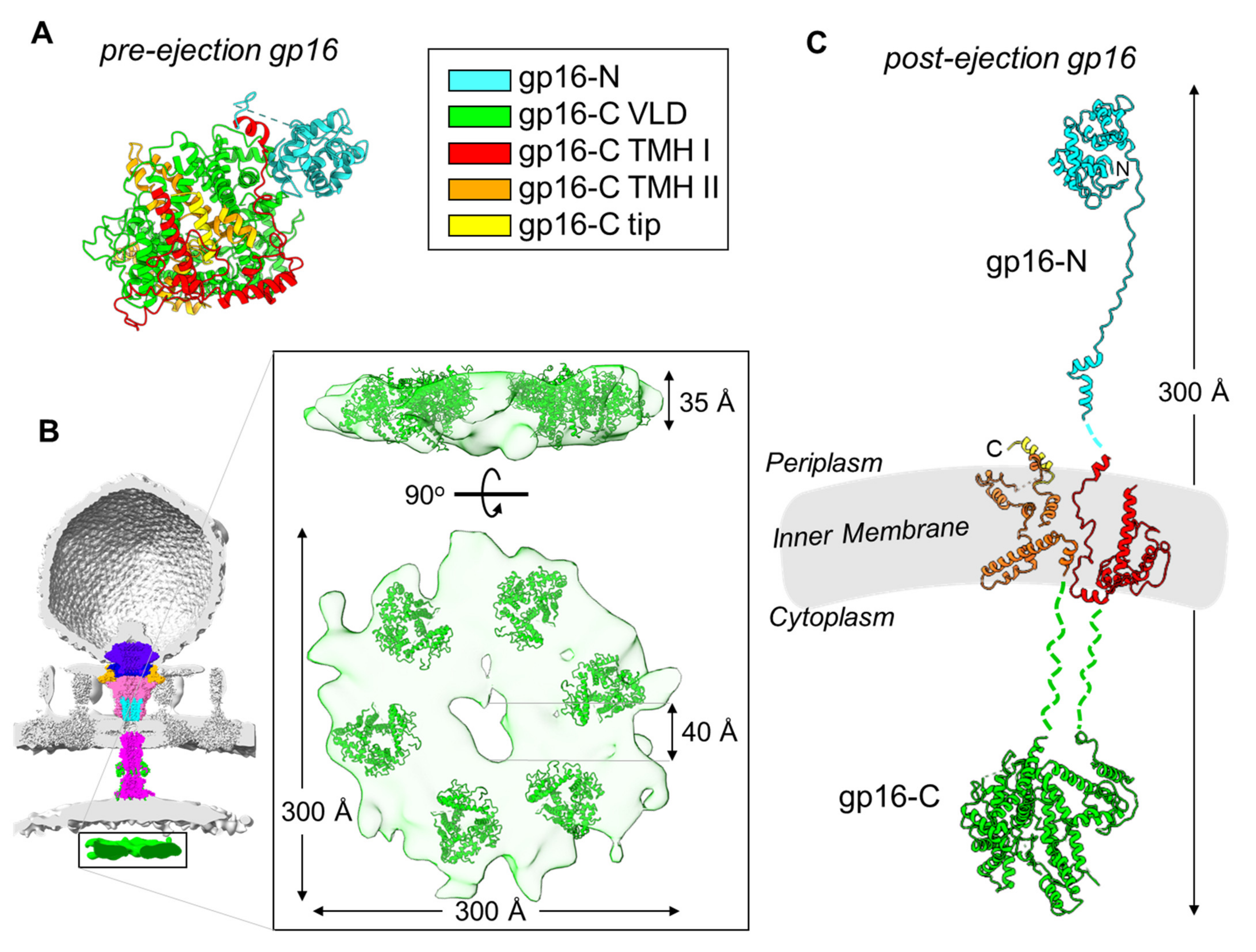
3. Conformational Gymnastics of T7 Ejection Proteins
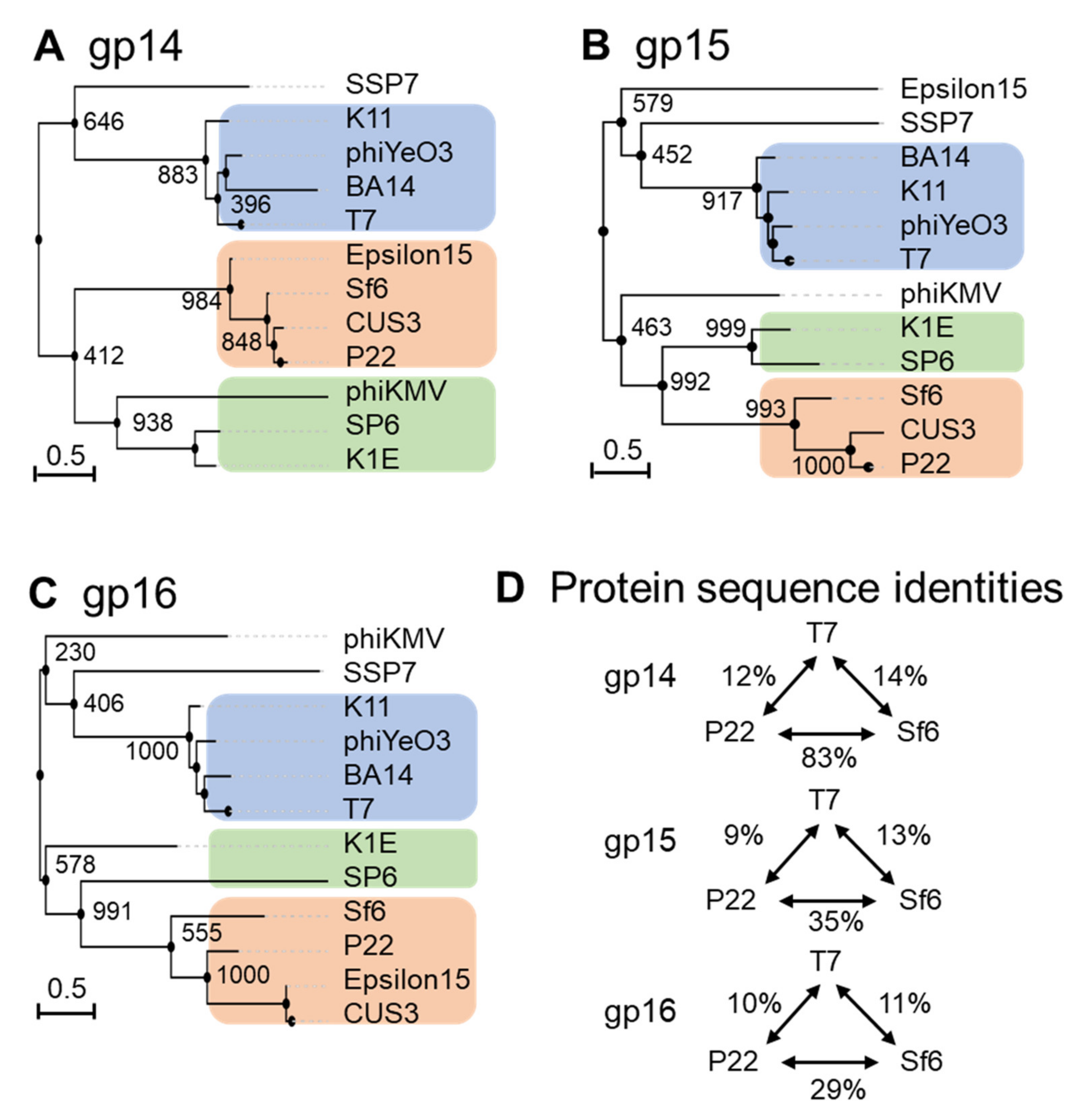
4. Conservation of Ejection Proteins
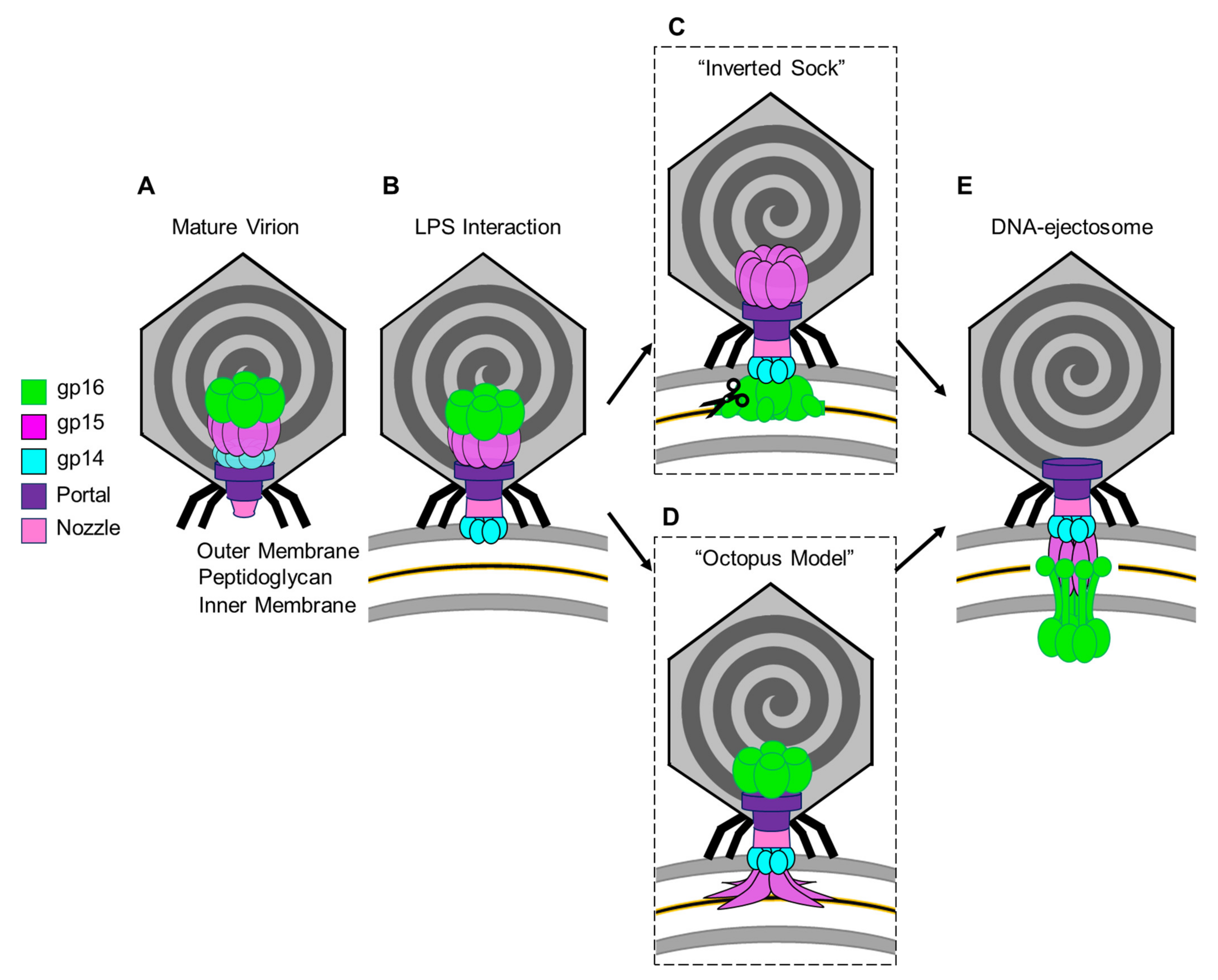
5. Models for Ejection-Protein Assembly into a DNA Ejectosome
6. Conclusive Remarks
- (i)
- Gp14 is the first factor to be ejected into the host, where it folds into a hexameric, constitutively open channel embedded in the host OM.
- (ii)
- Gp15 forms the periplasmic tunnel that extends the phage tail to cross the periplasm. The protein undergoes dramatic tertiary- and quaternary-structure conformational changes upon ejection, characterized by straightening of the C-terminal domain that swings by ~128° and assembles into a hexameric DNA tunnel wide enough to accommodate hydrated DNA.
- (iii)
- Gp16, the most complex of the three ejection proteins, has two functions: transglycosylase activity (gp16-N) and cytoplasmic DNA-binding activity (gp16-C). The former is phage-specific (not present in P22-like phages), whereas the latter is universally conserved. Gp16 refolds upon ejection, unbundles, and inserts into the IM to form a dual-ring structure. One ring containing gp16-N is part of the periplasmic tunnel with gp15, while the second ring projects into the host cytoplasm, is active in DNA binding, and takes part in DNA ejection.
- (iv)
- The stoichiometry of the assembly changes upon ejection, with the loss of at least two subunits of gp14 and gp15, which are octameric (or larger) during pre-ejection and become hexameric in the post-ejection state. It is unclear if the additional subunits in the pre-ejection conformation are not ejected from the virion or lost in the periplasm. The gp16 post-ejection conformation is also hexameric, implying additional copies of this protein must exist in the virion but are not visible in the core stack due to the limited volume available in the portal, which accommodates only four copies. These additional copies are likely loosely bound to the portal, as in P22-like phages.
- (v)
- Ejection-protein genes tend to be more variable than other virion-assembly proteins with conservations of under 10% in protein sequences, even in phages that infect the same bacterium. There does not appear to be conservation based on hosts, and ejection proteins that cluster into a core stack in the pre-ejection conformation are not necessarily more similar to one another than those diffused inside the capsid, as in P22-like phages.
- (vi)
- Membrane-spanning secondary-structure elements are universally conserved in gp14 and gp16 homologs, suggesting these two ejection proteins provide anchoring and penetrate the host OM and IM, respectively.
- (vii)
- The N-terminal peptidoglycan-hydrolase domain of T7 gp16 can swap to the gp15 homolog, suggesting a mosaically modular organization and an evolution of ejection proteins whereby the individual components may diverge as long as all parts are present in the final molecular machine.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suttle, C.A. Marine viruses — Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, T.; Kallmeyer, J.; Cypionka, H.; Engelen, B. High virus-to-cell ratios indicate ongoing production of viruses in deep subsurface sediments. ISME J. 2014, 8, 1503–1509. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.; São-José, C. Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef] [Green Version]
- Veesler, D.; Cambillau, C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev. 2011, 75, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, H.W. Tailed bacteriophages: The order caudovirales. Adv. Virus Res. 1998, 51, 135–201. [Google Scholar]
- Rixon, F.J.; Schmid, M.F. Structural similarities in DNA packaging and delivery apparatuses in Herpesvirus and dsDNA bacteriophages. Curr. Opin. Virol. 2014, 5, 105–110. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef] [Green Version]
- Veesler, D.; Spinelli, S.; Mahony, J.; Lichière, J.; Blangy, S.; Bricogne, G.; Legrand, P.; Ortiz-Lombardia, M.; Campanacci, V.; van Sinderen, D.; et al. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 8954–8958. [Google Scholar] [CrossRef] [Green Version]
- Zinke, M.; Schroder, G.F.; Lange, A. Major tail proteins of bacteriophages of the order Caudovirales. J. Biol. Chem. 2021, 298, 101472. [Google Scholar] [CrossRef]
- Casjens, S.R.; Molineux, I.J. Short noncontractile tail machines: Adsorption and DNA delivery by podoviruses. Adv. Exp. Med. Biol. 2012, 726, 143–179. [Google Scholar] [CrossRef] [PubMed]
- Fokine, A.; Rossmann, M.G. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage 2014, 4, e28281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostyuchenko, V.A.; Chipman, P.R.; Leiman, P.G.; Arisaka, F.; Mesyanzhinov, V.V.; Rossmann, M.G. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat. Struct. Mol. Biol. 2005, 12, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Leiman, P.G.; Shneider, M.M. Contractile tail machines of bacteriophages. Adv. Exp. Med. Biol. 2012, 726, 93–114. [Google Scholar] [CrossRef] [PubMed]
- Cumby, N.; Reimer, K.; Mengin-Lecreulx, D.; Davidson, A.R.; Maxwell, K.L. The phage tail tape measure protein, an inner membrane protein and a periplasmic chaperone play connected roles in the genome injection process of E. coli phage HK97. Mol. Microbiol. 2015, 96, 437–447. [Google Scholar] [CrossRef]
- Vinga, I.; Baptista, C.; Auzat, I.; Petipas, I.; Lurz, R.; Tavares, P.; Santos, M.A.; São-José, C. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol. Microbiol. 2012, 83, 289–303. [Google Scholar] [CrossRef]
- Plisson, C.; White, H.E.; Auzat, I.; Zafarani, A.; Sao-Jose, C.; Lhuillier, S.; Tavares, P.; Orlova, E.V. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 2007, 26, 3720–3728. [Google Scholar] [CrossRef]
- Molineux, I.J.; Panja, D. Popping the cork: Mechanisms of phage genome ejection. Nat. Rev. Microbiol. 2013, 11, 194–204. [Google Scholar] [CrossRef]
- Molineux, I.J. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 2001, 40, 1–8. [Google Scholar] [CrossRef]
- Molineux, I.J.; Lopez, L.L.; Roznowski, A.P. Phage Genome and Protein Ejection In Vivo. In Encyclopedia of Virology, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2021; Volume 4, pp. 206–218. [Google Scholar] [CrossRef]
- Parent, K.N.; Schrad, J.R.; Cingolani, G. Breaking Symmetry in Viral Icosahedral Capsids as Seen through the Lenses of X-ray Crystallography and Cryo-Electron Microscopy. Viruses 2018, 10, 67. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, A.; Olia, A.S.; Cingolani, G. Architecture of viral genome-delivery molecular machines. Curr. Opin. Struct. Biol. 2014, 25, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Wilkowska, K.; Targonska, M.; Smarz, A.; Sęktas, M. Periplasmic expression of a restriction endonuclease in Escherichia coli and its effect on the antiviral activity of the host. Acta Biochim. Pol. 2019, 66, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.I.; Salama, N.R. The gram-negative bacterial periplasm: Size matters. PLOS Biol. 2018, 16, e2004935. [Google Scholar] [CrossRef] [PubMed]
- Letellier, L.; Plançon, L.; Bonhivers, M.; Boulanger, P. Phage DNA transport across membranes. Res. Microbiol. 1999, 150, 499–505. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, J.C.; Gilcrease, E.B.; Woodbury, B.M.; Teschke, C.M.; Casjens, S.R. Intravirion DNA Can Access the Space Occupied by the Bacteriophage P22 Ejection Proteins. Viruses 2021, 13, 1504. [Google Scholar] [CrossRef]
- Agirrezabala, X.; Martin-Benito, J.; Caston, J.R.; Miranda, R.; Valpuesta, J.M.; Carrascosa, J.L. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 2005, 24, 3820–3829. [Google Scholar] [CrossRef] [Green Version]
- Guo, F.; Liu, Z.; Vago, F.; Ren, Y.; Wu, W.; Wright, E.T.; Serwer, P.; Jiang, W. Visualization of uncorrelated, tandem symmetry mismatches in the internal genome packaging apparatus of bacteriophage T7. Proc. Natl. Acad. Sci. USA 2013, 110, 6811–6816. [Google Scholar] [CrossRef] [Green Version]
- Cerritelli, M.E.; Trus, B.L.; Smith, C.S.; Cheng, N.; Conway, J.F.; Steven, A.C. A second symmetry mismatch at the portal vertex of bacteriophage T7: 8-fold symmetry in the procapsid core. J. Mol. Biol. 2003, 327, 1–6. [Google Scholar] [CrossRef]
- Chen, W.; Xiao, H.; Wang, L.; Wang, X.; Tan, Z.; Han, Z.; Li, X.; Yang, F.; Liu, Z.; Song, J.; et al. Structural changes in bacteriophage T7 upon receptor-induced genome ejection. Proc. Natl. Acad. Sci. USA 2021, 118, e2102003118. [Google Scholar] [CrossRef]
- Cheng, N.; Wu, W.; Watts, N.R.; Steven, A.C. Exploiting radiation damage to map proteins in nucleoprotein complexes: The internal structure of bacteriophage T7. J. Struct. Biol. 2014, 185, 250–256. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Leavitt, J.C.; Cheng, N.; Gilcrease, E.B.; Motwani, T.; Teschke, C.M.; Casjens, S.R.; Steven, A.C. Localization of the Houdinisome (Ejection Proteins) inside the Bacteriophage P22 Virion by Bubblegram Imaging. MBio 2016, 7, e01152-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Tu, J.; Liu, J.; Molineux, I.J. Structural dynamics of bacteriophage P22 infection initiation revealed by cryo-electron tomography. Nat. Microbiol. 2019, 4, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Sdao, S.M.; Dover, J.A.; Porcek, N.B.; Knobler, C.M.; Gelbart, W.M.; Parent, K.N. Bacteriophage P22 ejects all of its internal proteins before its genome. Virology 2015, 485, 128–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Margolin, W.; Molineux, I.J.; Liu, J. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science 2013, 339, 576–579. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Young, L.N.; Zhang, X.; Boudko, S.P.; Fokine, A.; Zbornik, E.; Roznowski, A.P.; Molineux, I.J.; Rossmann, M.G.; Fane, B.A. Icosahedral bacteriophage PhiX174 forms a tail for DNA transport during infection. Nature 2014, 505, 432–435. [Google Scholar] [CrossRef]
- Dreiseikelmann, B. Translocation of DNA across bacterial membranes. Microbiol. Rev. 1994, 58, 293–316. [Google Scholar] [CrossRef]
- Chang, J.T.; Schmid, M.F.; Haase-Pettingell, C.; Weigele, P.R.; King, J.A.; Chiu, W. Visualizing the structural changes of bacteriophage Epsilon15 and its Salmonella host during infection. J. Mol. Biol. 2010, 402, 731–740. [Google Scholar] [CrossRef] [Green Version]
- Stewart, M.P.; Langer, R.; Jensen, K.F. Intracellular Delivery by Membrane Disruption: Mechanisms, Strategies, and Concepts. Chem. Rev. 2018, 118, 7409–7531. [Google Scholar] [CrossRef]
- Dimitrov, D.S. Virus entry: Molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2004, 2, 109–122. [Google Scholar] [CrossRef]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef]
- Molineux, I.J. Fifty-three years since Hershey and Chase; Much ado about pressure but which pressure is it? Virology 2006, 344, 221–229. [Google Scholar] [CrossRef] [Green Version]
- Garcia, L.R.; Molineux, I.J. Incomplete entry of bacteriophage T7 DNA into F plasmid-containing Escherichia coli. J. Bacteriol. 1995, 177, 4077–4083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, L.R.; Molineux, I.J. Transcription-independent DNA translocation of bacteriophage T7 DNA into Escherichia coli. J. Bacteriol. 1996, 178, 6921–6929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kemp, P.; Garcia, L.R.; Molineux, I.J. Changes in bacteriophage T7 virion structure at the initiation of infection. Virology 2005, 340, 307–317. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.Y.; Kemp, P.; Molineux, I.J. Gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology 2010, 398, 176–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuervo, A.; Fabrega-Ferrer, M.; Machon, C.; Conesa, J.J.; Fernandez, F.J.; Perez-Luque, R.; Perez-Ruiz, M.; Pous, J.; Vega, M.C.; Carrascosa, J.L.; et al. Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism. Nat. Commun. 2019, 10, 3746. [Google Scholar] [CrossRef] [Green Version]
- Leptihn, S.; Gottschalk, J.; Kuhn, A. T7 ejectosome assembly: A story unfolds. Bacteriophage 2016, 6, e1128513. [Google Scholar] [CrossRef] [Green Version]
- Lupo, D.; Leptihn, S.; Nagler, G.; Haase, M.; I, J.M.; Kuhn, A. The T7 ejection nanomachine components gp15-gp16 form a spiral ring complex that binds DNA and a lipid membrane. Virology 2016, 486, 263–271. [Google Scholar] [CrossRef]
- González-García, V.A.; Bocanegra, R.; Pulido-Cid, M.; Martín-Benito, J.; Cuervo, A.; Carrascosa, J.L. Characterization of the initial steps in the T7 DNA ejection process. Bacteriophage 2015, 5, e1056904. [Google Scholar] [CrossRef]
- Gonzalez-Garcia, V.A.; Pulido-Cid, M.; Garcia-Doval, C.; Bocanegra, R.; van Raaij, M.J.; Martin-Benito, J.; Cuervo, A.; Carrascosa, J.L. Conformational changes leading to T7 DNA delivery upon interaction with the bacterial receptor. J. Biol. Chem. 2015, 290, 10038–10044. [Google Scholar] [CrossRef] [Green Version]
- Dai, W.; Fu, C.; Raytcheva, D.; Flanagan, J.; Khant, H.A.; Liu, X.; Rochat, R.H.; Haase-Pettingell, C.; Piret, J.; Ludtke, S.J.; et al. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature 2013, 502, 707–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murata, K.; Zhang, Q.; Gerardo Galaz-Montoya, J.; Fu, C.; Coleman, M.L.; Osburne, M.S.; Schmid, M.F.; Sullivan, M.B.; Chisholm, S.W.; Chiu, W. Visualizing Adsorption of Cyanophage P-SSP7 onto Marine Prochlorococcus. Sci. Rep. 2017, 7, 44176. [Google Scholar] [CrossRef] [PubMed]
- Swanson, N.A.; Lokareddy, R.K.; Li, F.; Hou, C.-F.D.; Leptihn, S.; Pavlenok, M.; Niederweis, M.; Pumroy, R.A.; Moiseenkova-Bell, V.Y.; Cingolani, G. Cryo-EM structure of the periplasmic tunnel of T7 DNA-ejectosome at 2.7 A resolution. Mol. Cell 2021, 81, 3145–3159.e3147. [Google Scholar] [CrossRef]
- Perez-Ruiz, M.; Pulido-Cid, M.; Luque-Ortega, J.R.; Valpuesta, J.M.; Cuervo, A.; Carrascosa, J.L. Assisted assembly of bacteriophage T7 core components for genome translocation across the bacterial envelope. Proc. Natl. Acad. Sci. USA 2021, 118, e2026719118. [Google Scholar] [CrossRef] [PubMed]
- Serwer, P.; Wright, E.T.; Hakala, K.W.; Weintraub, S.T. Evidence for bacteriophage T7 tail extension during DNA injection. BMC Res. Notes 2008, 1, 36. [Google Scholar] [CrossRef] [Green Version]
- Swanson, N.A.; Lokareddy, R.K.; Li, F.; Hou, C.F.; Pavlenok, M.; Niederweis, M.; Cingolani, G. Expression and purification of phage T7 ejection proteins for cryo-EM analysis. STAR Protoc. 2021, 2, 100960. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, A.; Haase, M.; Leptihn, S. Assisted and Unassisted Protein Insertion into Liposomes. Biophys. J. 2017, 113, 1187–1193. [Google Scholar] [CrossRef]
- Veevers, R.; Hayward, S. Methodological improvements for the analysis of domain movements in large biomolecular complexes. Biophys. Physicobiol. 2019, 16, 328–336. [Google Scholar] [CrossRef] [Green Version]
- Moak, M.; Molineux, I.J. Role of the Gp16 lytic transglycosylase motif in bacteriophage T7 virions at the initiation of infection. Mol. Microbiol. 2000, 37, 345–355. [Google Scholar] [CrossRef] [Green Version]
- Moak, M.; Molineux, I.J. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 2004, 51, 1169–1183. [Google Scholar] [CrossRef]
- Struthers-Schlinke, J.S.; Robins, W.P.; Kemp, P.; Molineux, I.J. The internal head protein Gp16 controls DNA ejection from the bacteriophage T7 virion. J. Mol. Biol. 2000, 301, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Kemp, P.; Gupta, M.; Molineux, I.J. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol. Microbiol. 2004, 53, 1251–1265. [Google Scholar] [CrossRef] [PubMed]
- Perez, G.L.; Huynh, B.; Slater, M.; Maloy, S. Transport of phage P22 DNA across the cytoplasmic membrane. J. Bacteriol. 2009, 191, 135–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenstein, J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996, 266, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evolut. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [Green Version]
- Hardies, S.C.; Thomas, J.A.; Black, L.; Weintraub, S.T.; Hwang, C.Y.; Cho, B.C. Identification of structural and morphogenesis genes of Pseudoalteromonas phage phiRIO-1 and placement within the evolutionary history of Podoviridae. Virology 2016, 489, 116–127. [Google Scholar] [CrossRef] [Green Version]
- Dedeo, C.L.; Cingolani, G.; Teschke, C.M. Portal Protein: The Orchestrator of Capsid Assembly for the dsDNA Tailed Bacteriophages and Herpesviruses. Annu. Rev. Virol. 2019, 6, 141–160. [Google Scholar] [CrossRef]
- Casjens, S.R.; Thuman-Commike, P.A. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology 2011, 411, 393–415. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Speir, J.A.; Matsui, T.; Lin, Z.; Liang, L.; Lynn, A.Y.; Varnado, B.; Weiss, T.M.; Tang, L. Structure of a Bacterial Virus DNA-Injection Protein Complex Reveals a Decameric Assembly with a Constricted Molecular Channel. PLoS ONE 2016, 11, e0149337. [Google Scholar] [CrossRef]
- Leiman, P.G.; Battisti, A.J.; Bowman, V.D.; Stummeyer, K.; Muhlenhoff, M.; Gerardy-Schahn, R.; Scholl, D.; Molineux, I.J. The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J. Mol. Biol. 2007, 371, 836–849. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Chang, J.; Jakana, J.; Weigele, P.; King, J.; Chiu, W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 2006, 439, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Lander, G.C.; Olia, A.; Li, R.; Casjens, S.; Prevelige, P., Jr.; Cingolani, G.; Baker, T.S.; Johnson, J.E. Peering down the barrel of a bacteriophage portal: The genome packaging and release valve in p22. Structure 2011, 19, 496–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lokareddy, R.K.; Sankhala, R.S.; Roy, A.; Afonine, P.V.; Motwani, T.; Teschke, C.M.; Parent, K.N.; Cingolani, G. Portal protein functions akin to a DNA-sensor that couples genome-packaging to icosahedral capsid maturation. Nat. Commun. 2017, 8, 14310. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Phage (Host) | gp14-Like (OM) | gp15-Like (Tunnel) | gp16-Like (IM) |
|---|---|---|---|
| T7 (Escherichia coli) | gp14 (196 aa) | gp15 (747 aa) M | gp16 (1318 aa) XL |
| CUS-3 (Escherichia coli) | gp7 (230 aa) | gp20 (449 aa) S | gp16 (719 aa) S |
| 13a (Escherichia coli) | gp14 (196 aa) | gp15 (747 aa) M | gp16 (1318 aa) XL |
| BA14 (Escherichia coli) | gp14 (201 aa) | gp15 (759 aa) M | gp16 (1315 aa) XL |
| K1E (Escherichia coli) | gp34 (240 aa) | gp35 (982 aa) XL | gp36 (1102 aa) M |
| HK620 (Escherichia coli) | gp7 (230 aa) | gp20 (449 aa) S | gp16 (722 aa) S |
| P22 (Salmonella enterica) | gp7 (229 aa) | gp20 (471 aa) S | gp16 (609 aa) XS |
| Epsilon 15 (Salmonella enterica) | gp11 (229 aa) | gp12 (499 aa) S | gp13 (708 aa) S |
| SP6 (Salmonella enterica) | gp35 (239 aa) | gp36 (978 aa) XL | gp37 (1270 aa) L |
| Sf6 (Shigella flexneri) | gp11 (230 aa) | gp12 (431 aa) S | gp13 (665 aa) XS |
| P-SSP7 (Prochlorococcus marinus) | gp14 (200 aa) | gp15 (837 aa) L | gp16 (1245 aa) L |
| K11 (Klebsiella pneumoniae) | gp14 (196 aa) | gp15 (751 aa) M | gp16 (1321 aa) XL |
| phiYeO3-12 (Yersinia enterocolitica) | gp14 (197 aa) | gp15 (747 aa) M | gp16 (1320 aa) XL |
| phiKMV (Psudomonas aeruginosa) | gp35 (181 aa) | gp36 (898 aa) L | gp37 (1337 aa) XL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swanson, N.A.; Hou, C.-F.D.; Cingolani, G. Viral Ejection Proteins: Mosaically Conserved, Conformational Gymnasts. Microorganisms 2022, 10, 504. https://doi.org/10.3390/microorganisms10030504
Swanson NA, Hou C-FD, Cingolani G. Viral Ejection Proteins: Mosaically Conserved, Conformational Gymnasts. Microorganisms. 2022; 10(3):504. https://doi.org/10.3390/microorganisms10030504
Chicago/Turabian StyleSwanson, Nicholas A., Chun-Feng D. Hou, and Gino Cingolani. 2022. "Viral Ejection Proteins: Mosaically Conserved, Conformational Gymnasts" Microorganisms 10, no. 3: 504. https://doi.org/10.3390/microorganisms10030504
APA StyleSwanson, N. A., Hou, C.-F. D., & Cingolani, G. (2022). Viral Ejection Proteins: Mosaically Conserved, Conformational Gymnasts. Microorganisms, 10(3), 504. https://doi.org/10.3390/microorganisms10030504







