Abstract
We investigated the vaginal microbiota (VMB) composition, prevalence of genital pathogens and their association among pregnant and post-delivery women in Pemba Island, Tanzania. Vaginal swabs were collected from 90 women, at two time points during pregnancy (<20 weeks of gestational age [GA] and ≥20 weeks GA) and once after delivery, when possible. IS-pro assay was used for VMB characterization. Chlamydia trachomatis (CT), Neisseria gonorrhea (NG), Trichomonas vaginalis (TV), Mycoplasma genitalium (MG) and human papillomavirus (HPV) were detected by qPCRs. VMB were mostly Lactobacillus dominant during pregnancy and non-Lactobacillus dominant post-delivery. A significant decrease in VMB richness was observed during pregnancy among paired and unpaired samples. Shannon diversity was significantly lower during pregnancy than post-delivery among unpaired samples. Klebsiella species and Streptococcus anginosus were the most commonly identified pathobionts at all timepoints. A high abundance of pathobionts was mostly seen in women with non-Lactobacillus dominant VMB. At ≥20 weeks GA timepoint during pregnancy, 63.0% of the women carrying one or more genital pathogen (either HPV, CT, TV, or MG) had L. iners dominant VMB. NG was not detected pre-delivery. This study contributes evidence on VMB composition, its changes during pregnancy and post-delivery, and their association with pathobionts and genital pathogens.
1. Introduction
The vaginal microbiota (VMB) consists of commensal microorganisms that exist in a mutually beneficial relationship with the host environment [1,2]. The human VMB are mostly dominated by protective lactic acid-producing Lactobacillus species in the majority of Caucasian women. These bacteria create an acidic environment with anti-microbial properties which hinders the growth and colonization of pathogenic microbial species [3,4,5]. Common Lactobacillus species in the vagina include Lactobacillus (L.) iners, L. crispatus, L. gasseri, or L. jensenii [2,6]. Jesper et al., also showed that next to the common Lactobacillus species, L. vaginalis plays a vital role in the VMB in African women [7]. However, when Lactobacillus species are in low abundance, women can carry higher levels of facultative anaerobic bacteria species, such as Atopobium, Gardnerella, Ureaplasma, Bacteroides, and Prevotella [8]. Furthermore, if the vaginal microbial community is dominated by non-lactic acid-producing species, it is less stable and tends to shift over time [9]. The polymicrobial anaerobic overgrowth disrupts the ecological balance of the VMB and is linked to vaginal anaerobic dysbiotic conditions like bacterial vaginosis (BV) [10,11,12,13]. BV is a common vaginal disorder in African women or women with a sub-Saharan African ethnic background [10,11] and has been associated with adverse outcomes such as miscarriage, premature rupture of membranes, preterm birth, and low birth weight [14,15,16]. The presence of pathobionts in the VMB, defined as potentially pathological organisms that generally live in a non-harming symbiosis, such as Streptococcus agalactiae (Group B streptococcus or GBS), Staphylococcus aureus, and species in the Enterobacteriaceae family, has also been associated with pelvic inflammatory disease or maternal and neonatal infections [17,18,19,20].
In individual women, the VMB composition differs between the non-pregnant and pregnant state. Indeed, the increased oestrogen and progesterone levels during pregnancy lead to physiological changes that also affect VMB composition [21]. Compared to the non-pregnant state, the VMB remain relatively stable during pregnancy, with an overall decrease in richness (number of species), abundance and evenness (relative abundance) of aerobic commensal bacteria and an increase in the abundance of Lactobacillus species from first to third trimester [7,8,22,23,24]. VMB changes in pregnancy mostly occur as transitions between species within the Lactobacillus genus, with rarely a shift to a polymicrobial state [22,25,26,27]. However, towards the end of the pregnancy and, especially, after delivery, following a decrease in oestrogen, a switch to a non-Lactobacillus dominant and more diverse VMB community is common [8,22]. This switch can persist up to one year postpartum [8,22]. Lactobacillus species-poor VMB and an increase in richness and diversity of VMB between 2nd and 3rd trimester have been associated with adverse pregnancy outcomes such as preterm birth, low birth weight, and miscarriage [28,29,30,31,32,33,34]. However, it is important to note that, although low abundances of pathobionts occur more often with lactobacilli than with BV-associated anaerobes, also women with Lactobacillus dominant VMB might have risk of developing adverse outcomes [35,36,37]. Thus, it might be that in certain individuals low abundance microorganisms might influence pregnancy progression more than dominant VMB species. To date, the role of VMB in preterm birth or other adverse pregnancy outcomes is controversial and still under investigation, especially since it seems to vary based on the ethnic background of the population studied. The composition of the VMB of women with African and non-African ancestry differs during pregnancy probably because of host genomics, immunological factors, microbial physiology and environmental influences [26,38,39,40]. Studies conducted on women with African ancestry observed that their VMB is less dominated by Lactobacillus species and more by BV-associated bacteria [6,26,30,33,41,42,43,44]. Several studies have investigated the VMB composition, irrespective of pregnancy status, across African populations, such as in Kenyan, Rwandan, South African, and Tanzanian women [45,46,47]. During pregnancy, it seems that Lactobacillus-dominant VMB (mostly L. iners and L. crispatus) are the most prevalent in women living in sub-Saharan Africa, followed by a more diverse VMB composition [48]. However, in the study conducted in mainland Tanzania, the VMB bacteria were only characterized on genus level and not species level [49].
Due to the possible impact that certain VMB and genital microorganisms have on health (reproductive and pregnancy outcomes) it is important to investigate them also in populations where the burden of disease is highest. In sub-Saharan Africa there is not only a high burden of vaginal dysbiotic conditions (such as BV), but also genital infections (such as sexually transmitted infections) and adverse pregnancy outcomes [50,51,52,53,54]. The interaction between VMB, genital pathogens and pathobionts during pregnancy is complex and remains mostly unclear [20,55]. Moreover, data about the VMB composition and presence of pathobionts in the sub-Saharan African population, especially Tanzania, are still limited. In 2014, a biobanking effort was initiated in Pemba Island, Tanzania with the support of the Bill and Melinda Gates Foundation [56]. Within this previously established (AMANHI) biobanking effort, adverse pregnancy outcomes and vaginal samples were collected during pregnancy and after parturition. The aim of this study was to characterize, in a small sub-set of samples, the VMB composition and its changes, including the presence of pathobionts and genital infections, across two timepoints during pregnancy and once after delivery using the previously collected biobank data and samples. It was hypothesized that the most prevalent VMB composition would be Lactobacillus-dominant VMB, with high frequency of a diverse VMB composition and presence of pathobionts. Insights from this first attempt to longitudinally characterize the VMB composition in Pemban women will contribute evidence on the role of microorganism in maternal and neonatal health among sub-Saharan African women.
2. Materials and Methods
2.1. Samples and Study Design
Vaginal sample collection was performed in the context of a biobanking effort established with the support of the Bill and Melinda Gates Foundation and initiated in 2014 as part of the Alliance for Maternal and Newborn Health Improvement (AMANHI) [56]. All women included in this study gave their prior consent, and data collection was conducted as per protocol [56]. Vaginal swabs collection was performed at 2 timepoints during pregnancy and once after delivery under health care staff supervision in health care facilities in Pemba Island [56,57]. Early pregnancy dating ultrasound determined the gestational age (GA) [52]. The first timepoint of sample collection during pregnancy was between 8–19 weeks and 6 days GA (<20 weeks), the other timepoint during pregnancy was between 20–40 GA weeks (≥20 weeks GA), and samples were collected between 42–60 days post-delivery. The cut-off of 20 weeks GA also refers to a clinically relevant time during pregnancy, after which spontaneous pregnancy loss, referred to as miscarriage, is usually less frequent. The timing of collection was not standardized; women were not sampled at all timepoints—this largely depended on practical limitations in rural clinics. Baseline sociodemographic and previous health care information was collected by health care staff at first antenatal contact. In addition, at each later timepoint further health information was collected by health care staff following the previous published protocol [56]. Swabs were stored in 1 mL eNAT buffer (Copan Italia, Brescia, Italy) at −20 °C at the Public Health Laboratory—Ivo de Carneri in Pemba Island. The collection tubes with eNAT buffer were later transported in dry-ice to Amsterdam Medical Centre in the Netherlands, where they were stored at −20 °C until further processing [58]. In this study, vaginal swabs collected at one or more of the 3 collection timepoints up until January 2019 were analysed. The Zanzibar Medical Research and Ethics Committee (ZAMREC) approved this study (protocol ZAMREC/0002/OCTOBER/013 amended 04/02/18).
2.2. Dna Extraction and Vaginal Microbiota Analysis
DNA from the vaginal swabs was extracted with the Chemagen (Perkin-Elmer, Baesweiler, Germany) automated DNA extraction machine according to the buccal swab extraction kit manufacturer’s instructions, as previously described elsewhere [59]. Elution volume was 200 μL. The IS-pro Microbiota assay (inBiome, Amsterdam, The Netherlands) was used for the VMB analysis [59]. The assay is based on length polymorphisms of the 16S–23S interspace (IS) region combined with sequence polymorphisms of the 16S rDNA. The IS-pro assay consists of two multiplex PCRs and was performed according to the manufacturer’s protocol as previously described [59,60,61]. In short, the first PCR used two different fluorescent-labelled primers, one for the phyla Bacteroidetes, and Fusobacteria, Actinobacteria, Firmicutes, and Verrucomicrobia (FAFV), while the second PCR included primers for the phylum Proteobacteria. An internal amplification control (IAC) was used for quality control of the process and downstream software analyses. After DNA amplification with the GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA, USA), 5 μL of PCR product was mixed with 20 μL formamide and 0.5 μL MapMaker 1500 ROX-labelled size maker (BioVentures, Murfreesboro, TN, USA). The ABI Prism 3500 Genetic Analyzer (Thermo-Fisher, Waltham, MA, USA) was used for the DNA fragment analysis via high-resolution capillary electrophoresis. Species were assigned to resulting amplicon length and colour using a reference database compiled of IS-pro fragments obtained from in silico and in vitro IS-pro PCRs of known vagina associated bacterial species (species calling). The IAC tested positive for all samples. TIBCO Spotfire 7.6 (TIBCO Spotfire Inc., Palo Alto, CA, USA) software was used to visualize the IS-pro colour labelled nucleotide peaks, species and to cluster the sample profiles according to the unweighted pair-group with the arithmetic mean (UPGMA) method. To cluster the microbiome profiles based on similarity column correlation, the process was performed with the UPGMA on a similarity matrix based on cosine similarity of bacterial profiles. Row hierarchical clustering was done by UPGMA on a distance matrix based on Euclidean distance. It identifies and orders the most frequent IS-fragments or bacterial taxa related to the microbiome profiles. For the purpose of this study, six bacterial genera (Streptococcus, Staphylococcus, Enterococcus, Escherichia/Shigella, Haemophilus, and Campylobacter) are considered pathobionts, as suggested by Wijgert et al. [20]. Pathobionts with a relative abundance higher than 20% are considered to be of substantial presence in the VMB profile [20]. After the hierarchical clustering of vaginal microbiome profiles, five microbial communities in line with the previously defined Community State Types (CST) were identified [6]. Alpha diversity indices of the VMB were measured by calculating the richness (number of species) and the Shannon diversity (the richness and relative abundance of bacterial species) index of each sample on the Spotfire software [62].
2.3. Genital Pathogens Analysis
Presence of C. trachomatis, n. gonorrhoeae and T. vaginalis was detected by their respective CE-IVD certified Presto and Real-Time quantitative polymerase chain reaction (qPCR) with ABI Taqman 7500 (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions, and as previously described [63,64]. For M. genitalium detection, a M. genitalium assay targeting the mg219 gene was used on the LightCycler 480 II PCR machine (Roche Diagnostics, Basel, Switzerland) [58,65]. The presence of high-risk human papillomavirus (hrHPV), which might be associated with different degrees of pathophysiology and local inflammation, was detected. To do that, AmpFire® HPV assay was used according to the user manual instruction, to simultaneously identify either HPV 16 genotype, HPV 18 genotype or fifteen other hrHPV genotypes (31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66 and 68, not individually identified and further referred in this manuscript as HPV others) [66].
2.4. Statistical Analysis
Data were analysed using IBM SPSS statistical software version 26 (SPSS Inc., Chicago, IL, USA). Fisher’s exact test was performed to compare dichotomous data in order to test whether the presence of genital infections associated with CST III compared to other CSTs (I, II, IV, V) [67,68]. To test whether alpha diversity indices were significantly different across time points, the Wilcoxon signed rank test (for matched samples) or Mann-Whitney U-test (for unmatched samples) was used. Whitney U-test (for unmatched samples) was used to calculate significant differences in relative abundances for pathobionts across timepoints. A two-tailed p-value of < 0.05 was considered statistically significant.
3. Results
In total, 170 vaginal samples from 90 Afro Shirazi women were included for this analysis. Forty-four samples were collected between 8–20 weeks gestational age (GA), eighty-two samples between 20–40 weeks GA, and forty-four samples between 42–60 days post-delivery (Supplementary Figure S1). Seventy-eight women underwent sample collection at two timepoints (either both during pregnancy or once during pregnancy and the other post-delivery) and two women at three timepoints. Questionnaires have been filled by most participants at baseline and other collection timepoints (Table 1, Table 2 and Table 3).

Table 1.
Description of the sociodemographic, clinical and pregnancy outcomes across cohort.

Table 2.
Mean gestational age at each swab collection.

Table 3.
Delivery information and pregnancy outcomes.
3.1. Sociodemographic Characteristics and Birth Data
The sociodemographic and health-related questions were filled out by more than 85% of the 90 participants (Table 1). The mean maternal age of the participants was 29.9 ± 6.6 years, mean gravidity 5.2 ± 2.6, and mean parity of 4.0 ± 2.4. The majority of the women were multiparous (93.2%). Almost half of the participants had a healthy weight (44.4%). None of the participants smoked at the time of enrolment, and 96.6% were not on any dietary restrictions (n = 88). At enrolment, the most common obstetric history complication reported was miscarriage/abortion (29.3%), followed by stillbirth (12.2%), preterm birth (3.8%) and premature rupture of membranes (PROM) (2.5%). Two of the 88 women (2.3%) reported they had malaria at the time of enrolment. Other medical problems or infections (diabetes, thyroid disease infections with human immunodeficiency virus (HIV), tuberculosis, hepatitis B or C, and urinary tract infection) were not reported.
The information collected might have been different per timepoint, depending on the number of women that responded to the questionnaire item (Table 1, Table 2 and Table 3). At the time of ≥ 20 weeks GA sampling during pregnancy, 34 (43%) women self-reported the use of multivitamins or iron and other minerals, and four (5.1%) women self-reported that they had taken antibiotics during their pregnancy. Some participants reported gums or teeth complaints (2.5% and 6.3%, respectively), but did not report any other health condition (haemorrhage, bleeding, diabetes, thyroid disease, HIV, malaria, tuberculosis, jaundice, hepatitis B or C, and urinary tract symptoms) (n = 79). At the time of sampling post-delivery, 78 women filled in the summary and one (1.3%) woman self-reported she had diabetes during her pregnancy. No other medical conditions or infections that might have occurred during the current pregnancy were reported (thyroid disease, cancer, cardiac problems, epilepsy, mental illness, hypertension HIV, malaria, tuberculosis, jaundice, hepatitis B or C and urinary tract symptoms). Five women (6.4%) reported that they had received antibiotics and 26 (33.3%) women reported they had received other types of medicines (not specified). The gestational time at vaginal swab collection, pregnancy data, pregnancy outcomes and neonatal outcomes are described in Table 2. The majority of the women (89%) received assistance at birth either by a doctor, nurse or midwife (n = 90). One participant experienced a stillbirth, four women a miscarriage, six women had a preterm delivery (<37 GA weeks); the rest had an uncomplicated birth (Table 3). Two women reported that their neonates were ill within the first 48 h of life; however, the type of illness was not further specified. The pregnancy outcomes of the women that had vaginal samples collected post-delivery are described in Table 3.
3.2. Species in the Vaginal Microbiota
The fluorescent colour and the length of the IS region in units of nucleotides per sample were identified in three groups at the phyla level, namely Bacteroidetes, FAFV, and Proteobacteria (Supplementary Figures S2–S4). In the included samples from 90 women, 554 different bacterial species were detected. Across the combined vaginal samples, Lactobacillus species (L. crispatus, L. iners, and L. jensenii) or Klebsiella species were the most identified species of the VMB during pregnancy (Supplementary Figure S4). In contrast, in post-delivery samples, the most identified species were L. iners, Gardnerella vaginalis, L. crispatus, and Klebsiella species (Supplementary Figure S5).
3.3. Shannon Index and Diversity
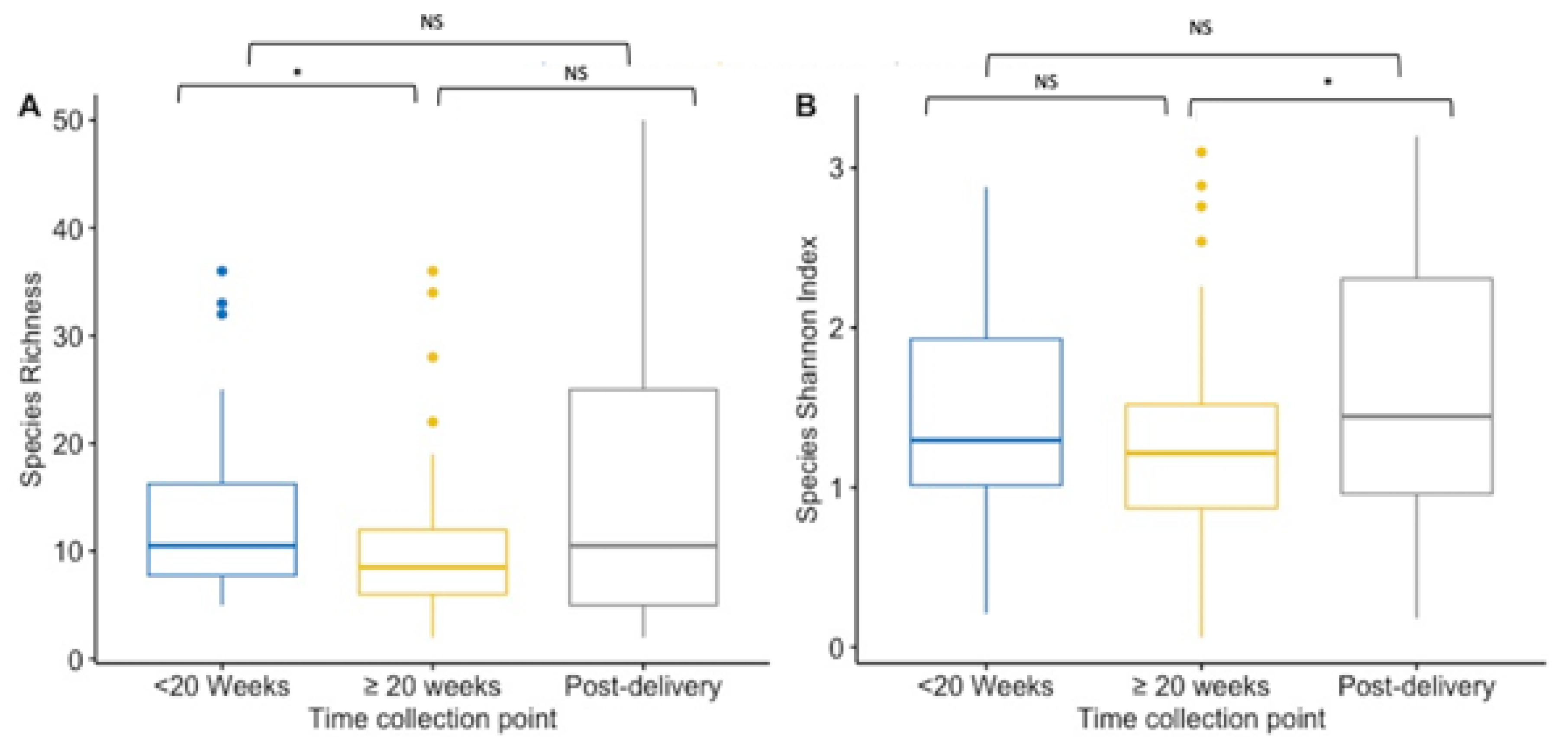
Alpha-diversity indices (richness and Shannon diversity index) of the VMB community were compared between paired and unpaired vaginal samples collected at first and ≥20 weeks GA pregnancy timepoint, and post-delivery for species and IS-fragment count. Data on IS-fragment count are shown in the Supplementary Figures S7 and S9. When performing unpaired analysis, species richness was significantly lower during pregnancy (from mean 10 species to mean 13 species) (p = 0.02) (Figure 1A). While the mean richness (15.39 species) was the highest post-delivery, there were no significant differences in the mean richness in the unpaired vaginal samples collected at enrolment or at the ≥20 weeks GA pregnancy sampling collection (Figure 1A). During pregnancy, the Shannon index did not differ significantly between the two collection points in the unpaired vaginal samples (mean Shannon diversity index was 1.42 at <20 weeks GA timepoint compared to 1.23 at ≥20 weeks GA timepoint) (Figure 1B). However, the mean Shannon diversity index was significantly higher in post-delivery collected unpaired vaginal samples (mean Shannon diversity index = 1.62) compared to the ≥20 weeks GA pregnancy collection point (p = 0.03), but not significantly higher compared to the <20 weeks GA timepoint (Figure 1B).
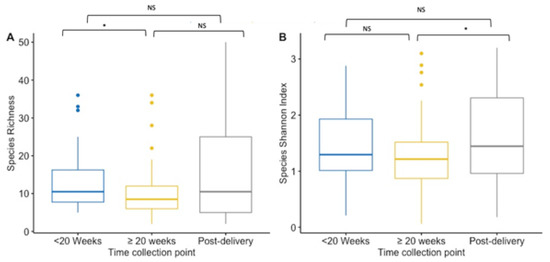
Figure 1.
Boxplot for the richness (A) and Shannon diversity index (B) at the species level for each collection point. Results of the <20 weeks GA pregnancy collection are in blue (n = 44), ≥20 weeks GA pregnancy collection in yellow (n = 82), and post-delivery in grey (n = 44). (A) Species richness is lower at ≥20 weeks GA pregnancy collection compared to the pregnancy collection at <20 weeks GA (p = 0.02). Between the other timepoints there was no significance difference. (B) The Shannon diversity index is higher at post-delivery compared to the index at ≥20 weeks GA pregnancy collection (p = 0.03), but not compared to the diversity index at <20 weeks GA pregnancy collection. During the pregnancy, the index did not differ significantly. NS = non significant. * p-value < 0.05.
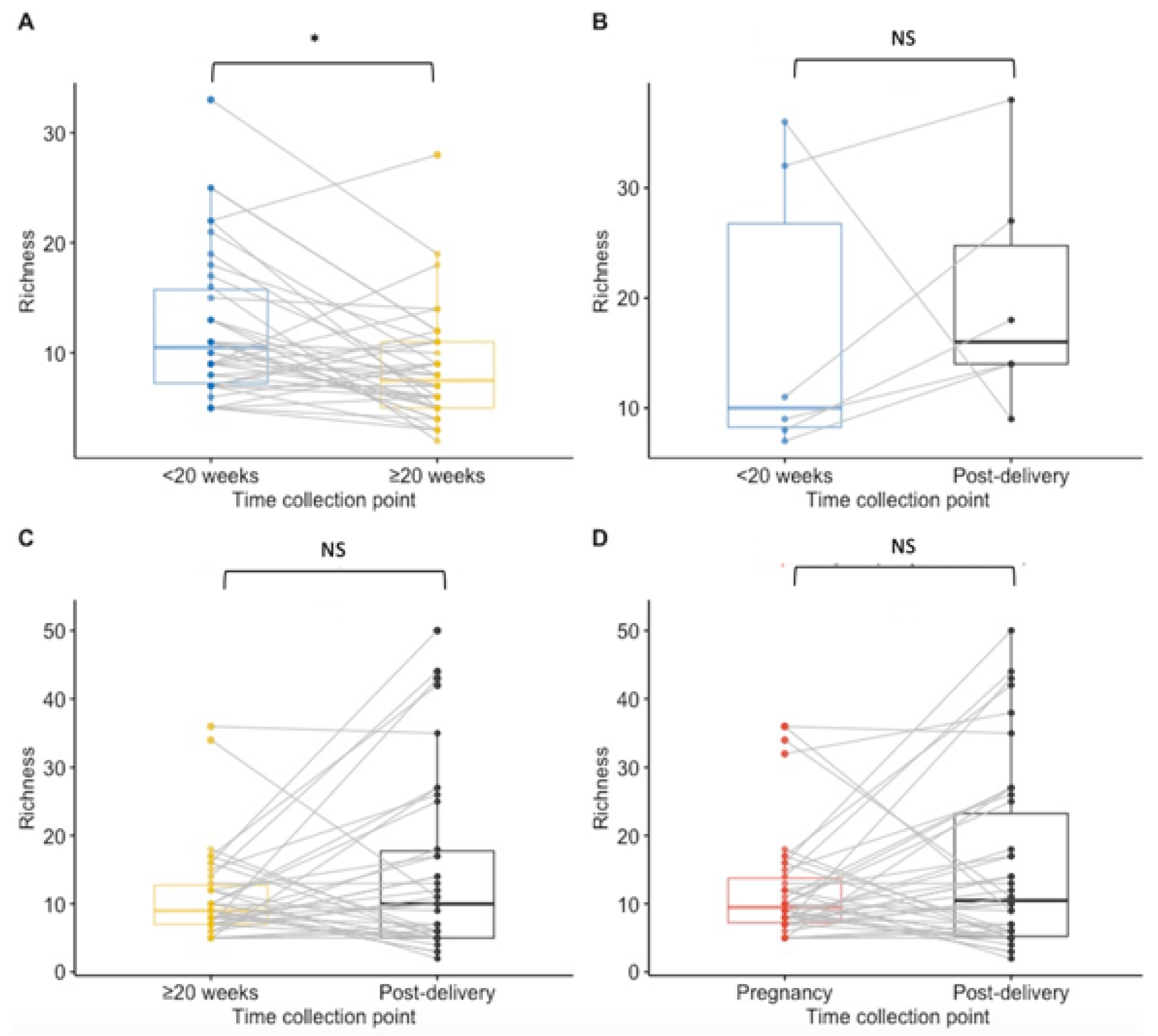
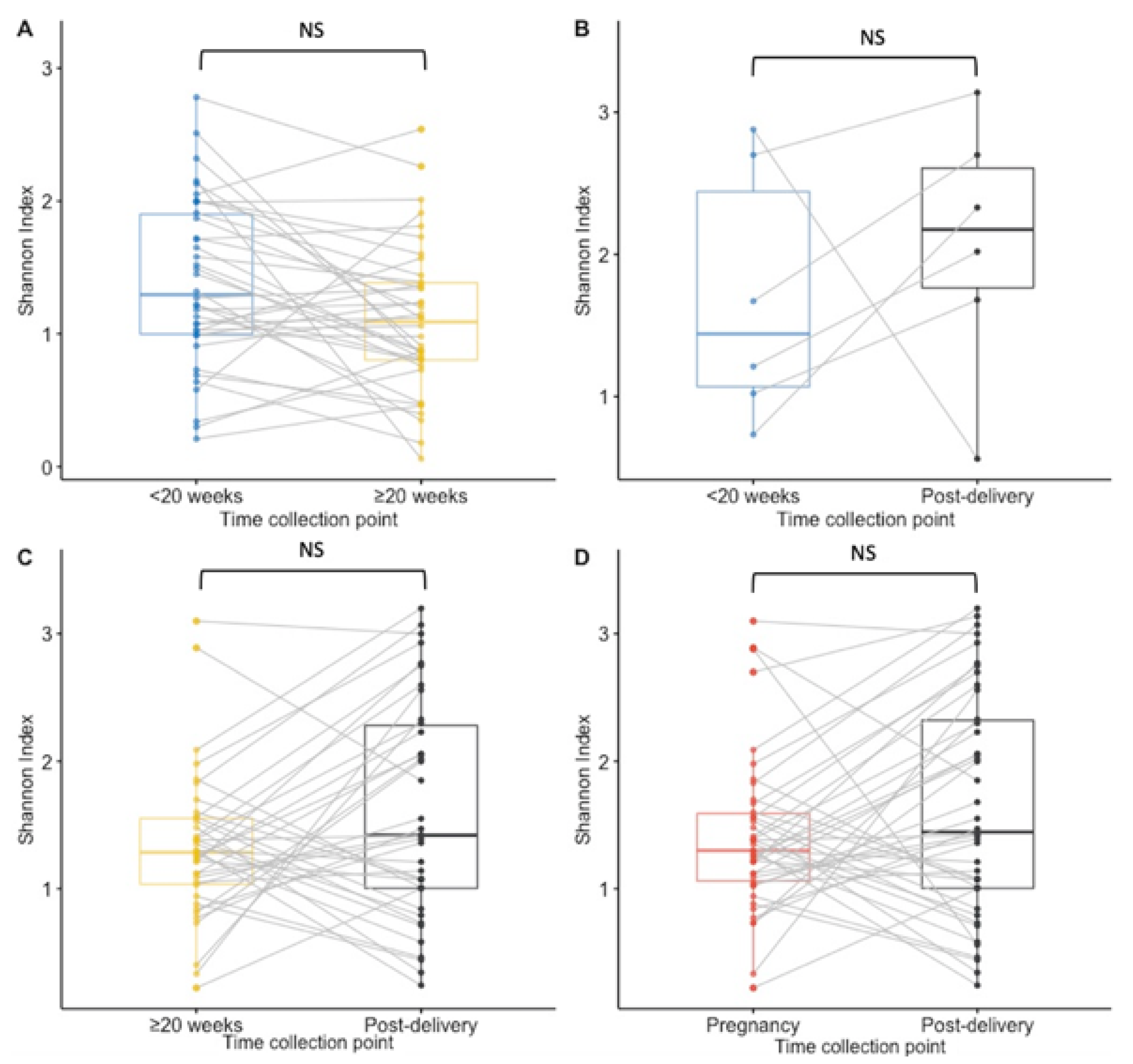
Paired analysis of samples from 38 women tested at both timepoints during pregnancy showed a significant decrease in richness during pregnancy (median < 20 weeks GA timepoint = 10.5 species; median ≥ 20 weeks GA collection pregnancy point = 7.5, p = 0.02) and no statistically significant difference between other time points (Figure 2A–D). The Shannon index findings across timepoints using paired analysis are similar to the Richness findings, with only a significant decrease in diversity observed during pregnancy (Figure 3A,B,D). Unlike in the unpaired analysis, for the paired analysis the Shannon diversity index did not significantly increase when analysing data of a subset of 38 other women that were tested both at the ≥20 weeks GA timepoint during pregnancy and post-delivery (p-value = 0.068) (Figure 3C).
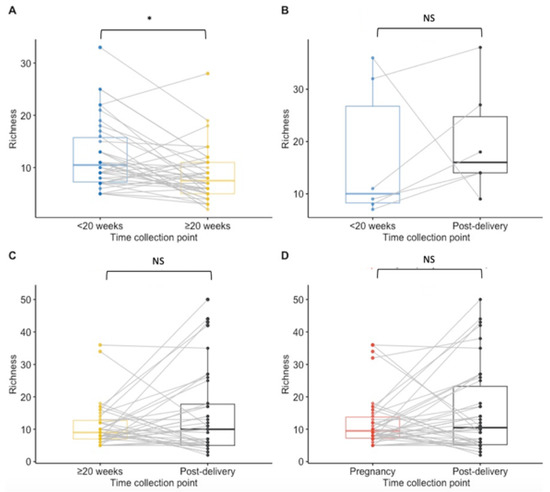
Figure 2.
Boxplots for species richness at each collection point for paired samples. Results of <20 weeks GA pregnancy collection are in blue, ≥20 weeks GA pregnancy collection in yellow, post-delivery in black and overall during pregnancy in red. (A) The richness was significantly lower at the ≥20 weeks GA pregnancy collection point than at <20 weeks GA pregnancy collection (p = 0.02) in matched samples from 38 women. (B) There was no significant difference in the richness between <20 weeks GA pregnancy collection and post-delivery matched samples from 6 women. (C) No significant difference was calculated in the richness between ≥20 weeks GA pregnancy collection and post-delivery matched samples from 38 women. (D) For 42 women that had samples collected at least once during pregnancy and post-delivery, no significant difference in the species richness was calculated. NS = non significant. * p-value < 0.05.
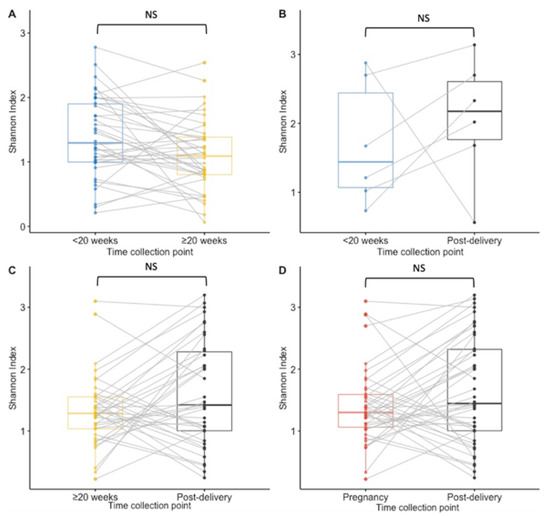
Figure 3.
Boxplots for the Shannon diversity index at species level for paired samples. Results of <20 weeks GA pregnancy collection are in blue, ≥20 weeks GA pregnancy collection in yellow, post-delivery in black and pregnancy in red. (A) The Shannon diversity index was not significantly higher at the ≥20 weeks GA pregnancy collection point than at the <20 weeks GA pregnancy collection in matched samples from 38 women. (B) There was no significant difference in the Shannon diversity index between the <20 weeks GA pregnancy collection and post-delivery matched samples from 6 women. (C) No significant difference was calculated in the Shannon diversity between the ≥20 weeks GA pregnancy collection and post-delivery matched samples from 38 women. (D) For 42 women who had samples collected at least once during pregnancy and post-delivery, no significant difference in the Shannon diversity index was calculated. NS = non significant.
3.4. Community State Types
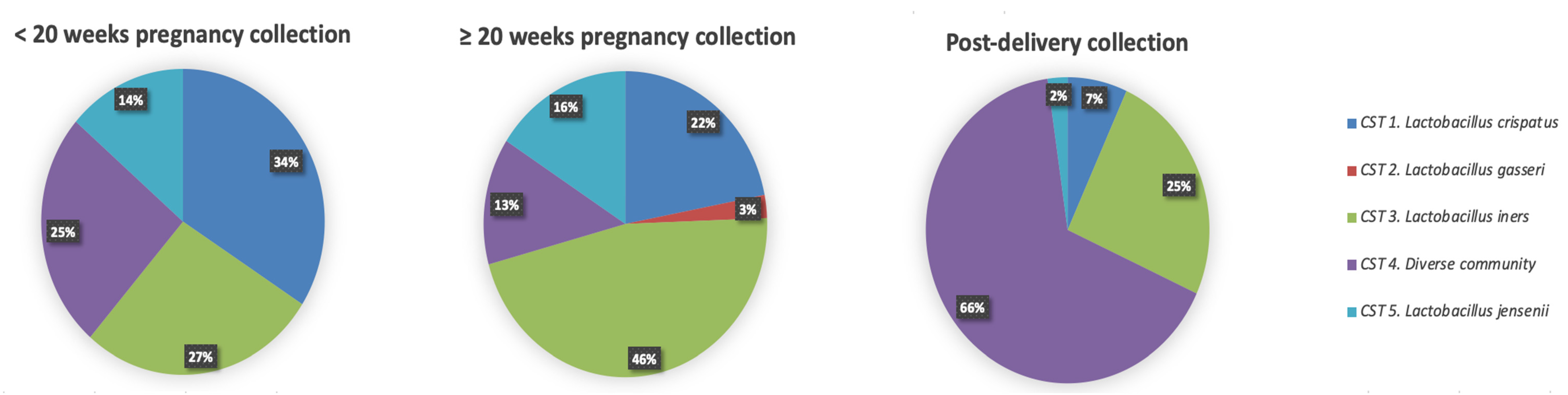
Hierarchical clustering of VMB profiles from samples collected during pregnancy and post-delivery yielded five different CSTs as previously described by Ravel et al. [6] (Figure 4). The VMB profiles of the vaginal samples collected at the <20 weeks GA timepoint during pregnancy (n = 44), were dominated by Lactobacillus species (33/44; 75%) belonging to three different CSTs: CST I (L. crispatus, 15/44; 34%), CST III (L. iners, 12/44; 27%), and CST V (L. jensenii, 6/44; 14%), respectively (Figure 4). The rest of the women (11/44; 25%) had a diverse VMB clustered as CST IV. Compared to samples collected at the < 20 weeks GA timepoint, the VMB profiles of the vaginal samples collected at the ≥ 20 weeks GA pregnancy timepoint (n = 82) were less diverse (CST IV; 11/82; 13% compared to 25%), and more vaginal samples (71/82; 87% compared to 75%) had a Lactobacillus dominant profile belonging to: CST I (18/82;22%), CST II (L. gasseri, 2/82; 2%), CST III (38/82; 46%), and CST V (L. jensenii, 13/82; 16%) (Figure 5), respectively. Due to the limitations of small sample size, the mentioned comparison between the < 20 weeks GA and the ≥ 20 weeks GA timepoint is purely descriptive and has not been statistically tested. Most of the vaginal samples collected post-delivery had a diverse VMB profile (CST IV; 29/44; 66%) followed by a L. iners dominated VMB (CST III; 11/44; 25%) (Figure 4). The remaining vaginal samples were clustered in CST I (3/44; 7%) and CST V (1/44; 2%).
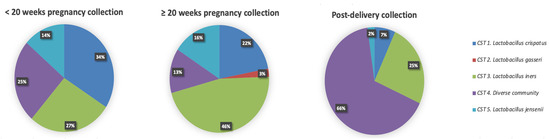
Figure 4.
Frequency of five community state type (CST) identified each collection timepoint (n = 44 at <20 weeks GA and post-delivery timepoints; n = 82 at the ≥20 weeks GA timepoint). In the pie charts the CST cluster is given in colour: CST I, blue; CST II, green; CST III, red; CST IV, yellow; CST V, purple.
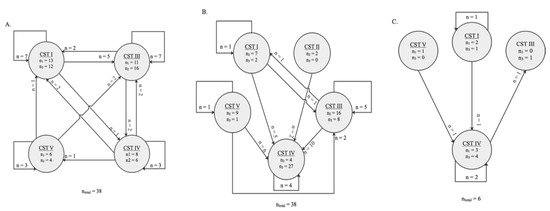
Figure 5.
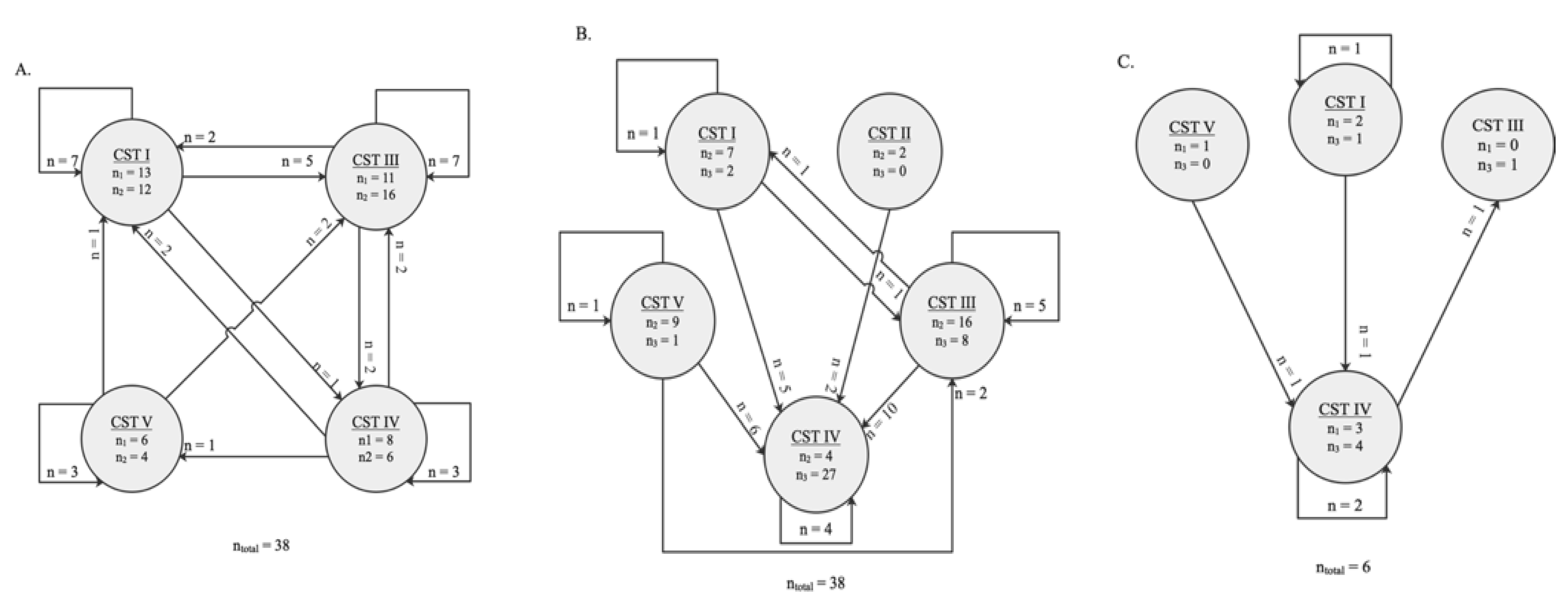
Schematic representation of the switch between community state types (CSTs) between two sampling timepoints. The numbers in the circles indicate how many vaginal samples were clustered in a certain CST by the time collection (during pregnancy). Arrows represent the direction of the switch. The numbers by the arrows represent the number of vaginal microbiota tested belonging to the same women who switched to a given CST at a later timepoint. (A) Thirty-eight women were clustered with a specific CST at timepoint <20 weeks GA and ≥20 weeks GA during pregnancy, of which 18 CST changed type during pregnancy. (B) Among the paired vaginal swabs tested at ≥20 weeks GA and post-delivery, 27 of the 38 CST changed. (C) The CST of three of the vaginal swabs from the same six women tested at <20 weeks GA and post-delivery changed.
Among the six women who had preterm deliveries, during pregnancy two of them had CST I (33%), two CST IV (33%), one CST V (17%) and another had CST III (17%). The two women carrying twins both had vaginal samples belonging to CST I at the <20 weeks GA timepoint during pregnancy. One of the women with multiple gestation also had CST I at the ≥20 weeks GA timepoint; after parturition the VMB profile switched to CST IV. The other participant switched from CST I to CST III at the ≥20 weeks GA timepoint; unfortunately there was no vaginal sample available post-delivery. The only woman who reported a stillbirth delivery had a vaginal sample belonging to CST III at the second timepoint. The vaginal profile at the first timepoint and post-delivery could not have been analysed due to the unavailability of these vaginal swabs.
For the four women who had a miscarriage, two vaginal samples at the <20 weeks GA pregnancy collection belonged to CST IV, one to CST III and another to CST I. Vaginal profiles from three women were also analysed at the post-delivery timepoint; one woman had CST I, another women CST IV, and another women CST III.
Furthermore, four women reported antibiotics usage at the ≥20 weeks GA pregnancy collection timepoint. At that timepoint, their vaginal samples belonged to CST III (n = 2), CST I (n = 1), CST IV (n = 1).
Overall, in the tested samples, changes of vaginal profiles occurred during pregnancy and after delivery (Figure 5). During pregnancy, the VMB communities shifted from one CST to another in 18 of the 38 (47%) vaginal samples that were analysed between the <20 weeks GA and the ≥20 weeks GA timepoint during pregnancy (Figure 5A). Between the two timepoints during pregnancy, a switch to CST III accounted for 9 (50%) of these changes. The CST shifted in 27 of the 38 women (71%) whose vaginal profiles were analysed at the ≥20 weeks GA timepoint during pregnancy and post-delivery (Figure 5B). Moreover, the CST shifted in 3 out of 6 (50%) vaginal profiles that were analysed at the <20 weeks GA timepoint in pregnancy and post-delivery (Figure 5C). From pregnancy to post-pregnancy timepoints, a switch to CST IV accounted for 23 (85%) of these changes. While CST I and CST III were the most interconnected states across longitudinal timepoints, with bidirectional transitions with each other and other CSTs (mostly CST IV) (Figure 5A–C).
3.5. Vaginal Pathobionts and Genital Pathogens
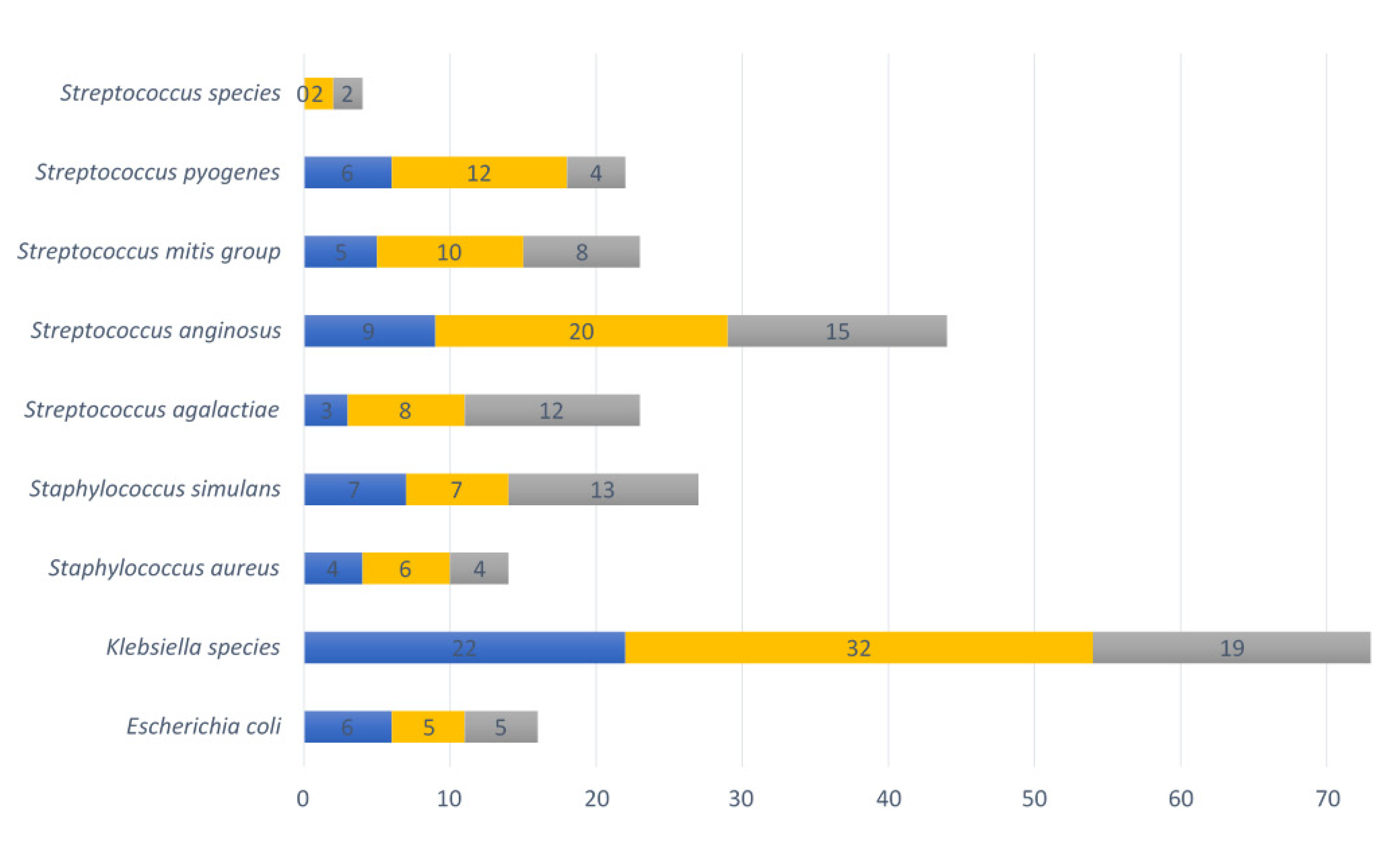
The following bacterial species of the VMB with pathogenic potential were identified: Escherichia coli, Klebsiella species, Staphylococcus aureus, Staphylococcus simulans, Streptococcus agalactiae, Streptococcus anginosus, Streptococcus mitis group, Streptococcus pyogenes, and Streptococcus species (other than already mentioned). In total, 73% (32/44) of vaginal swabs contained at least one such pathobiont at the <20 weeks GA timepoint during pregnancy, 68% (58/82) at ≥20 weeks GA, and 86% (38/44) post-delivery. The most common pathobiont at all three timepoints was the Klebsiella species, followed by Streptococcus anginosus, Streptococcus pyogenes, the Streptococcus mitis group, Staphylococcus simulans and Streptococcus agalactiae (Figure 6).
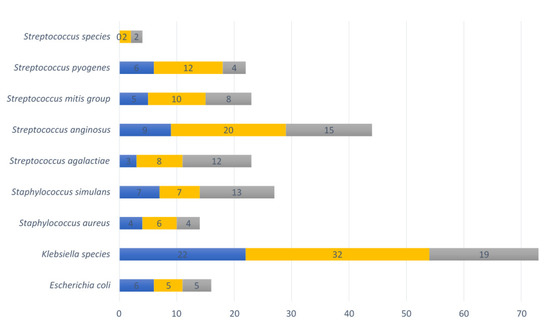
Figure 6.
Nine pathobionts observed in vaginal samples per collection point. Numbers of vaginal samples detected with pathobionts at <20 weeks GA pregnancy collection (n = 44), ≥20 weeks GA pregnancy collection (n = 82), and post-delivery (n = 44). Blue refers to the first timepoint during pregnancy, yellow to the second timepoint during pregnancy and grey to the post-delivery timepoint.
Three samples containing pathobionts at the <20 weeks GA timepoint (3/58) also had a substantial presence (>20% relative abundance, range between 22 and 35%) of pathobionts (two samples with Klebsiella species, and one sample with Streptococcus mitis group and Streptococcus anginosus). Among these samples, the sample with a relative abundance above 30% with the pathobiont Klebsiella species belonged to CST III, while the other two samples belonged to CST IV. At the ≥20 weeks GA time point, seven vaginal samples (7/38) had a substantial (relative abundance range: 20–44%) presence of pathobionts at the ≥20 weeks GA timepoint (Klebsiella species [n = 2], Streptococcus agalactiae [n = 1], Streptococcus anginosus [n = 3] and Staphylococcus simulans [n = 1]). Among those samples with high pathobionts load, two had a VMB belonging to CST III, while the others belonged to CST IV (n = 4) or CST V (n = 1).
Nine vaginal samples (9/38) had a substantial presence of pathobionts (relative abundance 24–89%) at the post-delivery collection point. Two different vaginal samples had a combination of a high relative abundance of Staphylococcus simulans (>30%) and Streptococcus anginosus (>44%). The rest of the samples had a high relative abundance of Klebsiella species (42%), Streptococcus anginosus (26% and 32%) and Streptococcus agalactiae (24%, 69%, 76%, and 89%). All nine post-delivery samples with a substantial presence of pathobionts belonged to CST IV. The overall relative abundance of pathobionts was significantly higher in the samples collected post-delivery (mean 10.6%) than in samples collected during pregnancy (<20 weeks GA pregnancy collection mean 6.0%; p-value = 0.04, ≥20 weeks GA pregnancy collection mean 4.9%; p-value = 0.01).
The known genital pathogens C. trachomatis, T. vaginalis, M. genitalium, N. gonorrhoeae and hrHPV other than genotypes 16 or 18 were also detected in the samples (Table 4). Nine samples (20.5%) were positive for at least one genital pathogen at the <20 weeks GA timepoint (Table 4). C. trachomatis was detected in four vaginal samples, each belonging to CST I, CST III, CST IV, and CST V, respectively. The two vaginal samples that were positive for T. vaginalis at the <20 weeks GA timepoint belonged to CST I and CST III, while three others were positive for hrHPV genotypes and belonged to CST I, CST III or CST V, respectively (Table 4). The presence of urogenital pathogens at the <20 weeks GA timepoint during pregnancy did not cluster significantly more within CST III (Table 4).

Table 4.
Number of vaginal samples positive for urogenital pathogen within a community state type.
At the ≥20 weeks GA timepoint during pregnancy, 18 (22.8%) vaginal samples tested positive for one or more genital pathogens (Table 4), most of them belonged to CST III (n = 12), followed by CST I (n = 2), CST V (n = 2), CST II (n = 1), and CST IV (n = 1). Most of the samples with a genital pathogen at the ≥ 20 weeks GA timepoint belonged to CST III (12/38); this difference was statistically significant (p = 0.01) when comparing all combined pathogens across CSTs (6/38).
Of the 44 post-delivery samples, five (11.4%) were positive for C. trachomatis (n = 1), three (6.8%) for T. vaginalis, one (2.3%) for N. gonorrhoeae, or four (9.1%) for hrHPV genotypes. The sample positives for a genital infection were clustered either in CST III along or CST IV (Table 4). The results of the persistence of genital infection during and after pregnancy in a larger cohort of this population is described elsewhere [57].
4. Discussion
Using vaginal samples and questionnaire data previously collected from a biobank in Pemba Island, this study analysed VMB and genital infections among 90 local Afro-Shirazi women during pregnancy and post-delivery. This study strived to comprehensively characterize the vaginal microbial environment, by identifying the bacterial species present (including pathobionts), by using this information to cluster the VMB into CSTs, and by identifying the presence of selected genital pathogens. During pregnancy, the VMB communities were mostly Lactobacillus-dominated with most VMB profiles belonging to CST I, II, III, and V (65% of the vaginal samples at the <20 weeks GA timepoint and 81% of the vaginal samples at ≥20 weeks GA [8]). These results are in concordance with previous (longitudinal) observations where a high relative abundance of Lactobacillus species was observed during pregnancy, and L. crispatus (CST I) and L. iners (CST III) were the most commonly identified species of the VMB community during pregnancy [23,25,69,70]. In particular, alike to previous studies among African and African-American pregnant women, this study also showed the presence of L. jensenii (CST V) or L. gasseri (CST II), though most often not as dominant Lactobacillus species [7,23,45]. These observations during pregnancy in this and other studies are in discordance with the studies of non-pregnant Tanzanian women and another cohort of sub-Saharan African women [7,23,45,47].
Across the two timepoints in pregnancy, Shannon diversity index, a measure of alpha diversity, of VMB was stable, and the richness was significantly lower (p = 0.02) than after pregnancy, as previously observed by other longitudinal studies [25]. Similar to a study in North American women from African ancestry, the prevalence of Lactobacillus-dominated VMB in this current study was lower at the <20 weeks GA timepoint than in later trimesters of pregnancy, and the switch in VMB in pregnancy was most commonly (50% [9/18] of cases) towards L. iners dominant VMB [26]. The high frequency of a Lactobacillus dominant VMB and the stability of the alpha diversity in pregnancy has been attributed to the high levels of oestrogens that indirectly promote glycogen production and support Lactobacillus species colonization, which in turn metabolize lactic acid and promote a healthy low vaginal pH [23]. This hypothesis might explain the predominant role of high oestrogens levels during pregnancy, independent of ethnicity, in promoting Lactobacillus abundance. In the non-pregnant state, on the other hand, when oestrogens’ role is not as important, the frequency of the non-Lactobacillus dominated CST IV has been reported to be higher in women from sub-Saharan Africa or with African ancestry [26,45,71,72].
After delivery, a 100- to 1000-fold drop of oestrogens and resulting disruption of the vaginal microbial environment is expected [8,43,73,74], including the oestrogen-driven Lactobacillus species dominance [43]. In agreement with these data, the study results showed that, in paired samples, most vaginal profiles shifted to CST IV post-delivery. In 74% of the vaginal samples collected post-delivery the VMB profile were more diverse, belonging to CST IV, and were less Lactobacillus dominant than VMB during pregnancy. This observation agrees with previous ones among British women of different ethnic backgrounds and among American women. Such changes have been reported to persist up to one year, independent of ethnicity [8,43]. Mean and median richness and Shannon diversity index were higher in the post-delivery samples compared to samples collected during pregnancy, though this difference was only statistically significant for the mean Shannon diversity index calculated for unpaired samples (p = 0.03). The median Shannon diversity index calculated for the paired samples collected at the ≥20 weeks GA pregnancy timepoint and post-delivery was close to statistical significance (p = 0.07). Nevertheless, these findings combined suggest that the VMB diversity and richness during pregnancy is lower than post-delivery in women from Pemba Island, Tanzania. These findings are also in concordance with previous observations in post-delivery VMB analysis from mainland Tanzanian, British and American women, in whom the authors also described a more diverse and richer VMB throughout pregnancy [8,43,49]. Also similar to other studies, an increase in BV-associated bacteria such as Gardnerella vaginalis, Prevotella species and Anaerococcus species, was also observed post-delivery [8,43,49]. The presence of Prevotella species and Anaerococcus species have been associated with a higher vaginal pH and BV [6,75]. In this present study, the presence of clinical BV could not be assessed, as this was not addressed in the questionnaire. However, in the questionnaires, none of the women reported any urinary tract infections symptoms, which often present as symptoms of the urogenital tract [58]. Thus, it is possible that most women in this cohort with a dysbiotic (CST IV) VMB were asymptomatic [11]. However further studies should investigate the exact prevalence of BV and BV-symptomatology in this Tanzanian population. Nonetheless, even though mostly asymptomatic, the role of certain BV-associated species during pregnancy is gaining increasing interest as BV-associated species have been associated with preterm birth in Caucasian cohorts [8,30,76]. However, a more extensive African-American study found no specific taxon, including Gardnerella, as a significant marker for preterm birth [23,30]. Further research is needed to investigate the role of BV-associated bacteria in sub-Saharan African cohorts, especially since the burden of preterm birth is high in this world region [77].
The presence of certain vaginal commensal microorganisms (pathobionts), such as S. agalactiae, Staphylococcus aureus and species in the Enterobacteriaceae family under certain circumstances can be also of clinical relevance in various maternal, pregnancy-related and neonatal health conditions [17,18,78,79]. In the samples tested in this study, pathobionts were more present post-delivery (86%) than during pregnancy (68% at the <20 weeks GA timepoint and 73% at the ≥20 weeks GA timepoint in pregnancy). Moreover, post-delivery, more women (20%) had a VMB containing a substantial presence of pathobionts (relative abundance more than 20%) compared to VMB from samples collected during pregnancy (6.8% at the <20 weeks GA and 8.5% at the ≥20 weeks GA timepoint during pregnancy). However, it is for debate whether the risk of pathogenic potential increases only when the VMB contains a substantial presence of a pathobiont (higher abundance) [20]. As previously observed in the non-pregnant cohort studies by Wijgert et al., in this study, pathobionts co-occur with both lactobacilli and BV-associated bacterial species during pregnancy [20]. However, post-delivery most pathobionts were detected in women with a dysbiotic vaginal profile (CST IV). The causal association between CST IV and pathobionts’ abundance, along with their role in pregnancy complications, such as preterm birth, should be further investigated.
Klebsiella species and Streptococcus anginosus were the most prevalent pathobionts during pregnancy and post-delivery, and in most cases, they were also the most prevalent pathobionts with a substantial presence in the VMB. The prevalence of Klebsiella species during pregnancy (2.3% at <20 weeks GA and 4.5% at the ≥20 weeks GA timepoint) were similar in this study compared to previous findings in pregnant Nigerian or Nepalese women where the point-prevalence was 3% or 5.6%, respectively [80,81]. Streptococcus anginosus is one of most identified pathobionts in patients with aerobic vaginitis, together with other pathobionts (including Escherichia coli, Streptococcus spp., Staphylococcus aureus, Staphylococcus epidermidis) [82]. The exact role of Streptococcus anginosus in the female genital tract and the influence of aerobic vaginitis in pregnancy is still unknown; however, there have been cases where fatal neonatal sepsis was reported to occur due to a specific Streptococcus anginosus biotype [79,83].
There are also indications that K. pneumoniae in particular is associated with neonatal deaths and premature pregnancy loss [84]. Thus, identifications of these pathogens (also in substantial presence) in this cohort indicate the need to further investigate in larger studies the clinical association between Klebsiella species and Streptococcus anginosus and adverse pregnancy outcomes.
Currently, in certain parts of the world, especially high-income countries, Streptococcus agalactiae, also known as GBS, is tested in pregnancy as part of antenatal care to minimize the risk of vertical infection to the foetus and new-born [85]. GBS infection can lead to preterm birth and can be fatal for neonates, causing severe cases of pneumonia, meningitis or sepsis [17,18,86]. The prevalence of GBS observed in this study (6.8% at <20 weeks GA, 10% at the ≥20 weeks GA timepoint in pregnancy, and 27% post-delivery) are in line with the pooled prevalence of a recent meta-analysis report on the prevalence of GBS in pregnant and post-delivery women in Tanzania (16.14%; 95% CI 2.9, 29.4) [87]. It should be noted that about 1 in 100 infants born from mothers carrying GBS develop invasive GBS infection disease in studies performed on other populations [88,89]. Thus, as evidence on the involvement of pathobionts in maternal and new-born health, and their possible vertical transmission, it is essential to further investigate the association between abundance of GBS, and other pathobionts, and perinatal health. It is even more so, now that some of these pathobionts strains, like Klebsiella and E. coli, are becoming increasingly resistance to a broad range of antibiotics [81,89].
In addition to pathobionts, this study also investigated the presence of the pathogens C. trachomatis, hrHPV, M. genitalium, and T. vaginalis in this cohort. In total, 21%, 23%, and 11% of vaginal samples tested positive for a genital pathogen at the <20 weeks GA and ≥20 weeks GA timepoints during pregnancy and post-delivery, respectively. It has been proposed that a VMB characterized by low diversity of species has a protective role against ascending infections of the genital tract and adverse pregnancy outcomes, and that infections in early pregnancy may hinder the transition of VMB to a beneficial state associated to a lower risk of pregnancy complication, i.e., CST I [23,67,90]. T. vaginalis has been associated with reduced Lactobacillus species in the VMB, but not BV-related species [87,88]. In contrast, L. iners has been associated with susceptibility to genital infections (HIV and other sexually transmitted infections) [67,68,91,92]. In this study, The VMB samples of most women carrying a genital pathogen belonged to CST III (L. iners dominant) at the ≥ 20 weeks GA pregnancy timepoint. However, at the other timepoints, the presence of none of the genital pathogens clustered with VMB belonging this specific CST. To this date, the role of L. iners in the VMB equilibrium is not fully understood, as it has been associated with both eubiotic and dysbiotic VMB states [67,92,93]. Thus, the role of L. iners in VMB’s health and the association between L. iners dominant VMB (CST III) and genital pathogens during pregnancy should be further evaluated.
5. Limitations
The main limitation of this study was the limited availability of longitudinal paired samples. This was mainly due to practical challenges in the sample collection process. Currently, only vaginal samples from two-woman sampled at all three-collection timepoints were available for testing. Nevertheless, the findings of this first analysis in pregnant Pemban women raise many questions (regarding the role of VMB with pathobionts, genital infections and pregnancy outcomes) that could be further investigated in a bigger cohort. In this study, the number of participants was limited to 90 and, as such, the prevalence of adverse pregnancy outcomes was low. However, pregnancy complications are prevalent in Pemba, and the role of VMB in pregnancy outcomes should be further investigated in this population in a bigger cohort [94]. This study does nonetheless provide baseline characteristics, including clinically relevant ones, and insights on pregnancy outcomes among women in Pemba Island. Unfortunately, due to the use of pre-existing biobank data, some information, such as BV-related clinical data, that would have been relevant to our aim was not among the information available from the questionnaire.
With regards to the methodological approaches used in this study, the IS-pro assay gives comparable results to next-generation sequencing (NGS) and has been shown to even outperform NGS when using samples with lower bacterial loads [61,95]. 16S RNA gene sequencing is currently the most used method to the determine the microbiota of vaginal samples. Singer et al., compared vaginal samples of IS-pro and 16S RNA gene sequencing and showed that the vast majority of samples had a similar profile in terms of alpha diversity [96]. Not all bacterial taxa to species level have been identified due to restrictions of the available database. However, this mainly related to low-abundant organisms. For instance, L. vaginalis could not be identified yet in this analysis.
The use of antibiotics and twin pregnancies, both factors of which can interfere with VMB composition, were only reported by a small number of women and therefore were not excluded from this analysis.
Finally, the use of the term “pathobiont” in this study and other studies should be taken carefully, as it is not a well-defined concept [97]. It has been shown in some studies that the conditions under which pathobionts exhibit virulence relate to impaired host immune defences or altered microbiota compositions [97]. This in turn suggests an agreement on what a balanced VMB state or an alter VMB state are [55]. On the other hand, the clinical significance of VMB compositions remains debated, especially among women from sub-Saharan Africa or African ancestry [48,97,98].
6. Conclusions
The VMB was generally Lactobacillus dominant (65% at <20 weeks GA and 81% at the ≥20 weeks GA timepoint) during pregnancy and non-Lactobacillus dominant (73.9%) post-delivery in women from Pemba Island. The VMB richness significantly decreased during pregnancy in paired and unpaired samples, whereas the Shannon diversity significantly increased post-delivery solely in unpaired samples. A substantial presence of pathobionts was observed in a diverse (or L. iners-dominant) VMB at all three timepoints. Solely at the ≥20 weeks GA pregnancy collection, genital infection positively associated with L. iners dominant VMB. Future studies should further investigate the temporal and directional microbial changes during pregnancy, the role of CSTs with adverse pregnancy outcomes and how pathobionts and genital pathogens interfere with the VMB composition, especially with L. iners dominant VMB.
Investigating the causality of particular VMB characteristics in reproductive health might be essential to improve maternal and child health, particularly in populations where the burden of VMB dysbiosis, genital infections and adverse pregnancy outcomes are high, such as in certain parts of sub-Saharan Africa. Once clarity is provided on beneficial and non-beneficial VMB in pregnant women of different ethnicities, better and cheaper diagnostics tools and treatment options (probiotics or other biotherapeutics products) can be investigated for more personalized clinical solutions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10030509/s1. Figure S1: Number of vaginal samples at each collection point; Figure S2: Cluster analysis 44 vaginal samples collected during pregnancy at fist pregnancy collection; Figure S3: Cluster analysis 82 vaginal samples collected during pregnancy at ≥20 weeks GA pregnancy collection; Figure S4: Cluster analysis 44 vaginal samples collected post-delivery; Figure S5: The frequency of bacterial species or unknown genus/family bacteria observed per collection time point; Figure S6: Heatmap of all samples of the cohort in Pemba Island showing the 29 species with the highest relative abundance; Figure S7: Boxplot for the richness (A) and Shannon diversity index (B) at nucleotide level for each collection point; Figure S8: Boxplots for nucleotide richness at each collection point for paired samples; Figure S9: Boxplots for Shannon diversity index at nucleotide level for paired samples.
Author Contributions
Conceptualization, S.D., S.A.M. and E.A.; Formal analysis, N.C.A.J., A.E.B. and S.O.; Funding acquisition, S.A.M., S.S. and E.A.; Methodology, N.C.A.J., M.H.J., L.P., A.E.B., A.M. and S.S.; Project administration, S.D. and S.S.; Resources, S.D. and S.M.A.; Supervision, S.D., S.A.M. and E.A.; Writing—original draft, N.C.A.J.; Writing—review & editing, N.C.A.J. and E.A. All authors have read and agreed to the published version of the manuscript.
Funding
This study was in part subsidized by the Otto Kranendonk Fund from the Netherlands Society for Tropical Medicine and International Health (NVTG).
Institutional Review Board Statement
The Zanzibar Medical Research and Ethics Committee (ZAMREC) approved this study (protocol ZAMREC/0002/OCTOBER/013 amended 04/02/18).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data are available from the authors with the permission of the Biobank governing body/local institution and the Principal Investigator of the site.
Acknowledgments
All the women who participated in the biobank study in Pemba Island Tanzania and all staff of the biobank. Sonja N.H. Puljhun, Monique M. Verveer, Roel Heijmans, Jolein Pleijster, Maarten Boon, and Nina B. Uiljdert for their direct technical assistance in the laboratory or with Spotify. H. Wang for his assistance with generating some of the figures.
Conflicts of Interest
Andries E. Budding is a paid advisor to ArtPred BV. Other Authors declare no competing interests.
References
- Marchesi, J.R.; Ravel, J. The vocabulary of microbiome research: A proposal. Microbiome 2015, 3, 31. [Google Scholar] [CrossRef]
- Verstraelen, H.; Vieira-Baptista, P.; De Seta, F.; Ventolini, G.; Lonnee-Hoffmann, R.; Lev-Sagie, A. The Vaginal Microbiome: I. Research Development, Lexicon, Defining “Normal” and the Dynamics Throughout Women’s Lives. J. Low. Genit. Tract Dis. 2022, 26, 73. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.M.; Seferovic, M.; Pace, R.M.; Aagaard, K.M. The microbiome in preterm birth. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 52, 103–113. [Google Scholar] [CrossRef]
- O’Hanlon, D.E.; Moench, T.R.; Cone, R.A. Vaginal pH and microbicidal lactic acid when lactobacilli dominate the microbiota. PLoS ONE 2013, 8, e80074. [Google Scholar] [CrossRef]
- O’Hanlon, D.E.; Moench, T.R.; Cone, R.A. In vaginal fluid, bacteria associated with bacterial vaginosis can be suppressed with lactic acid but not hydrogen peroxide. BMC Infect. Dis. 2011, 11, 200. [Google Scholar] [CrossRef] [PubMed]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.K.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108, 4680–4687. [Google Scholar] [CrossRef]
- Jespers, V.; van de Wijgert, J.; Cools, P.; Verhelst, R.; Verstraelen, H.; Delany-Moretlwe, S.; Mwaura, M.; Ndayisaba, G.F.; Mandaliya, K.; Menten, J.; et al. The significance of Lactobacillus crispatus and L. vaginalis for vaginal health and the negative effect of recent sex: A cross-sectional descriptive study across groups of African women. BMC Infect. Dis. 2015, 15, 115. [Google Scholar] [CrossRef] [PubMed]
- DiGiulio, D.B.; Callahan, B.J.; McMurdie, P.J.; Costello, E.K.; Lyell, D.J.; Robaczewska, A.; Sun, C.L.; Goltsman, D.S.A.; Wong, R.J.; Shawa, G.; et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. USA 2015, 112, 11060–11065. [Google Scholar] [CrossRef] [PubMed]
- Bayigga, L.; Kateete, D.P.; Anderson, D.J.; Sekikubo, M.; Nakanjako, D. Diversity of vaginal microbiota in sub-Saharan Africa and its effects on HIV transmission and prevention. Am. J. Obstet. Gynecol. 2019, 220, 155–166. [Google Scholar] [CrossRef]
- Brotman, R.M. Vaginal microbiome and sexually transmitted infections: An epidemiologic perspective. J. Clin. Investig. 2011, 121, 4610–4617. [Google Scholar] [CrossRef] [PubMed]
- Woodman, Z. Can one size fit all? Approach to bacterial vaginosis in sub-Saharan Africa. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 16. [Google Scholar] [CrossRef] [PubMed][Green Version]
- González-Beiras, C.; Marks, M.; Chen, C.Y.; Roberts, S.; Mitjà, O. Epidemiology of Haemophilus ducreyi infections. Emerg. Infect. Dis. 2016, 22, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lev-Sagie, A.; De Seta, F.; Verstraelen, H.; Ventolini, G.; Lonnee-Hoffmann, R.; Vieira-Baptista, P. The Vaginal Microbiome: II. Vaginal Dysbiotic Conditions. J. Low. Genit. Tract Dis. 2022, 26, 79. [Google Scholar] [CrossRef] [PubMed]
- Arif, F. Bacterial Vaginosis: Risk of Adverse Pregnancy Outcome. J. Gynecol. Res. Obstet. 2018, 15–17. [Google Scholar] [CrossRef]
- Kervinen, K.; Kalliala, I.; Glazer-Livson, S.; Virtanen, S.; Nieminen, P.; Salonen, A. Vaginal microbiota in pregnancy: Role in induction of labor and seeding the neonate’s microbiota? J. Biosci. 2019, 44, 1–6. [Google Scholar] [CrossRef]
- Dunlop, A.L.; Satten, G.A.; Hu, Y.J.; Knight, A.K.; Hill, C.C.; Wright, M.L.; Smith, A.K.; Read, T.D.; Pearce, B.D.; Corwin, E.J. Vaginal Microbiome Composition in Early Pregnancy and Risk of Spontaneous Preterm and Early Term Birth Among African American Women. Front. Cell. Infect. Microbiol. 2021, 11. [Google Scholar] [CrossRef]
- Cools, P.; Jespers, V.; Hardy, L.; Crucitti, T.; Delany-Moretlwe, S.; Mwaura, M.; Ndayisaba, G.F.; Van De Wijgert, J.H.H.M.; Vaneechoutte, M. A multi-country cross-sectional study of vaginal carriage of group B streptococci (GBS) and Escherichia coli in resource-poor settings: Prevalences and risk factors. PLoS ONE 2016, 11, e0148052. [Google Scholar] [CrossRef]
- Black, C.G.; Tavares, L.; Stachel, A.; Ratner, A.J.; Randis, T.M. Distribution of Late-Onset Neonatal Sepsis Pathogens Differs in Inpatient and Outpatient Settings. Am. J. Perinatol. 2019, 36, 1136–1141. [Google Scholar] [CrossRef]
- Brunham, R.C.; Gottlieb, S.L.; Paavonen, J. Pelvic inflammatory disease. N. Engl. J. Med. 2015, 372, 2039–2048. [Google Scholar] [CrossRef]
- van de Wijgert, J.H.H.M.; Verwijs, M.C.; Gill, A.C.; Borgdorff, H.; van der Veer, C.; Mayaud, P. Pathobionts in the Vaginal Microbiota: Individual Participant Data Meta-Analysis of Three Sequencing Studies. Front. Cell. Infect. Microbiol. 2020, 10, 129. [Google Scholar] [CrossRef]
- Wira, C.R.; Fahey, J.V.; Rodriguez-Garcia, M.; Shen, Z.; Patel, M.V. Regulation of mucosal immunity in the female reproductive tract: The role of sex hormones in immune protection against sexually transmitted pathogens. Am. J. Reprod. Immunol. 2014, 72, 236–258. [Google Scholar] [CrossRef] [PubMed]
- Doyle, R.; Gondwe, A.; Fan, Y.M.; Maleta, K.; Ashorn, P.; Klein, N.; Harris, K. A Lactobacillus-deficient vaginal microbiota dominates postpartum women in rural Malawi. Appl. Environ. Microbiol. 2018, 84, 6. [Google Scholar] [CrossRef] [PubMed]
- Romero, R.; Hassan, S.S.; Gajer, P.; Tarca, A.L.; Fadrosh, D.W.; Nikita, L.; Galuppi, M.; Lamont, R.F.; Chaemsaithong, P.; Miranda, J.; et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2014, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Stout, M.J.; Conlon, B.; Landeau, M.; Lee, I.; Bower, C.; Zhao, Q.; Roehl, K.A.; Nelson, D.M.; MacOnes, G.A.; Mysorekar, I.U. Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am. J. Obstet. Gynecol. 2013, 208, e1–e226. [Google Scholar] [CrossRef] [PubMed]
- Avershina, E.; Slangsvold, S.; Simpson, M.R.; Storrø, O.; Johnsen, R.; Øien, T.; Rudi, K. Diversity of vaginal microbiota increases by the time of labor onset. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Serrano, M.G.; Parikh, H.I.; Brooks, J.P.; Edwards, D.J.; Arodz, T.J.; Edupuganti, L.; Huang, B.; Girerd, P.H.; Bokhari, Y.A.; Bradley, S.P.; et al. Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat. Med. 2019, 25, 1001–1011. [Google Scholar] [CrossRef]
- Aagaard, K.; Riehle, K.; Ma, J.; Segata, N.; Mistretta, T.A.; Coarfa, C.; Raza, S.; Rosenbaum, S.; van den Veyver, I.; Milosavljevic, A.; et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS ONE 2012, 7, e36466. [Google Scholar] [CrossRef]
- Callahan, B.J.; DiGiulio, D.B.; Aliaga Goltsman, D.S.; Sun, C.L.; Costello, E.K.; Jeganathan, P.; Biggio, J.R.; Wong, R.J.; Druzin, M.L.; Shaw, G.M.; et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc. Natl. Acad. Sci. USA 2017, 114, 9966–9971. [Google Scholar] [CrossRef]
- Kroon, S.J.; Ravel, J.; Huston, W.M. Cervicovaginal microbiota, women’s health, and reproductive outcomes. Fertil. Steril. 2018, 110, 327–336. [Google Scholar] [CrossRef]
- Stout, M.J.; Zhou, Y.; Wylie, K.M.; Tarr, P.I.; Macones, G.A.; Tuuli, M.G. Early pregnancy vaginal microbiome trends and preterm birth. Am. J. Obstet. Gynecol. 2017, 217, 356.e1–356.e18. [Google Scholar] [CrossRef]
- Brown, R.G.; Marchesi, J.R.; Lee, Y.S.; Smith, A.; Lehne, B.; Kindinger, L.M.; Terzidou, V.; Holmes, E.; Nicholson, J.K.; Bennett, P.R.; et al. Vaginal dysbiosis increases risk of preterm fetal membrane rupture, neonatal sepsis and is exacerbated by erythromycin. BMC Med. 2018, 16, 9. [Google Scholar] [CrossRef]
- Romero, R.; Hassan, S.S.; Gajer, P.; Tarca, A.L.; Fadrosh, D.W.; Bieda, J.; Chaemsaithong, P.; Miranda, J.; Chaiworapongsa, T.; Ravel, J. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome 2014, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Hyman, R.W.; Fukushima, M.; Jiang, H.; Fung, E.; Rand, L.; Johnson, B.; Vo, K.C.; Caughey, A.B.; Hilton, J.F.; Davis, R.W.; et al. Diversity of the vaginal microbiome correlates with preterm birth. Reprod. Sci. 2014, 21, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Jayaprakash, T.P.; Wagner, E.C.; Van Schalkwyk, J.; Albert, A.Y.K.; Hill, J.E.; Money, D.M.; Hemmingsen, S.M.; Castillo, E.; Janssen, P.A. High diversity and variability in the vaginal microbiome in women following Preterm Premature Rupture of Membranes (PPROM): A prospective cohort study. PLoS ONE 2016, 11, e0166794. [Google Scholar] [CrossRef] [PubMed]
- van de Wijgert, J.H.H.M.; Jespers, V. The global health impact of vaginal dysbiosis. Res. Microbiol. 2017, 168, 859–864. [Google Scholar] [CrossRef]
- Van De Wijgert, J.H.H.M.; Borgdorff, H.; Verhelst, R.; Crucitti, T.; Francis, S.; Verstraelen, H.; Jespers, V. The vaginal microbiota: What have we learned after a decade of molecular characterization? PLoS ONE 2014, 9, e105998. [Google Scholar] [CrossRef]
- Van De Wijgert, J.H.H.M.; Morrison, C.S.; Cornelisse, P.G.A.; Munjoma, M.; Moncada, J.; Awio, P.; Wang, J.; Van Der Pol, B.; Chipato, T.; Salata, R.A.; et al. Bacterial vaginosis and vaginal yeast, but not vaginal cleansing, increase HIV-1 acquisition in African women. J. Acquir. Immune Defic. Syndr. 2008, 48, 203–210. [Google Scholar] [CrossRef]
- Modi, B.P.; Teves, M.E.; Pearson, L.N.; Parikh, H.I.; Haymond-Thornburg, H.; Tucker, J.L.; Chaemsaithong, P.; Gomez-Lopez, N.; York, T.P.; Romero, R.; et al. Mutations in fetal genes involved in innate immunity and host defense against microbes increase risk of preterm premature rupture of membranes (PPROM). Mol. Genet. Genom. Med. 2017, 5, 720–729. [Google Scholar] [CrossRef]
- York, T.P.; Eaves, L.J.; Neale, M.C.; Strauss, J.F. The contribution of genetic and environmental factors to the duration of pregnancy. Am. J. Obstet. Gynecol. 2014, 210, 398–405. [Google Scholar] [CrossRef]
- Barcelona de Mendoza, V.; Wright, M.L.; Agaba, C.; Prescott, L.; Desir, A.; Crusto, C.A.; Sun, Y.V.; Taylor, J.Y. A Systematic Review of DNA Methylation and Preterm Birth in African American Women. Biol. Res. Nurs. 2017, 19, 308–317. [Google Scholar] [CrossRef]
- Ravel, J.; Brotman, R.M.; Gajer, P.; Ma, B.; Nandy, M.; Fadrosh, D.W.; Sakamoto, J.; Koenig, S.S.K.; Fu, L.; Zhou, X.; et al. Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome 2013, 1, 29. [Google Scholar] [CrossRef] [PubMed]
- Fettweis, J.M.; Paul Brooks, J.; Serrano, M.G.; Sheth, N.U.; Girerd, P.H.; Edwards, D.J.; Strauss, J.F.; Jefferson, K.K.; Buck, G.A. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology 2014, 160, 2272–2282. [Google Scholar] [CrossRef] [PubMed]
- MacIntyre, D.A.; Chandiramani, M.; Lee, Y.S.; Kindinger, L.; Smith, A.; Angelopoulos, N.; Lehne, B.; Arulkumaran, S.; Brown, R.; Teoh, T.G.; et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Brown, C.J.; Abdo, Z.; Davis, C.C.; Hansmann, M.A.; Joyce, P.; Foster, J.A.; Forney, L.J. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 2007, 1, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Gautam, R.; Borgdorff, H.; Jespers, V.; Francis, S.C.; Verhelst, R.; Mwaura, M.; Delany-Moretlwe, S.; Ndayisaba, G.; Kyongo, J.K.; Hardy, L.; et al. Correlates of the molecular vaginal microbiota composition of African women. BMC Infect. Dis. 2015, 15, 86. [Google Scholar] [CrossRef]
- McMillan, A.; Rulisa, S.; Gloor, G.B.; Macklaim, J.M.; Sumarah, M.; Reid, G. Pilot assessment of probiotics for pregnant women in Rwanda. PLoS ONE 2018, 13, e0195081. [Google Scholar] [CrossRef]
- Hummelen, R.; Fernandes, A.D.; Macklaim, J.M.; Dickson, R.J.; Changalucha, J.; Gloor, G.B.; Reid, G. Deep Sequencing of the Vaginal Microbiota of Women with HIV. PLoS ONE 2010, 5, e12078. [Google Scholar] [CrossRef]
- Juliana, N.C.A.; Suiters, M.J.M.; Al-Nasiry, S.; Morré, S.A.; Remco, P.H.R.; Ambrosino, E. The association between vaginal microbiota dysbiosis, bacterial vaginosis and aerobic vaginitis, and adverse pregnancy outcomes of women living in sub-Saharan Africa: A systematic review. Front. Public Health 2020, 8. [Google Scholar] [CrossRef]
- Bisanz, J.E.; Enos, M.K.; PrayGod, G.; Seney, S.; Macklaim, J.M.; Chilton, S.; Willner, D.; Knight, R.; Fusch, C.; Fusch, G.; et al. Microbiota at multiple body sites during pregnancy in a rural tanzanian population and effects of Moringa-supplemented probiotic yogurt. Appl. Environ. Microbiol. 2015, 81, 4965–4975. [Google Scholar] [CrossRef]
- Kyei-Nimakoh, M.; Carolan-Olah, M.; McCann, T.V. Access barriers to obstetric care at health facilities in sub-Saharan Africa-a systematic review. Syst. Rev. 2017, 6, 1–16. [Google Scholar] [CrossRef]
- World Health Organization. World Health Statistics 2019: Monitoring Health for the SDGs, Sustainable Development Goals; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Mullick, S.; Watson-Jones, D.; Beksinska, M.; Mabey, D. Sexually transmitted infections in pregnancy: Prevalence, impact on pregnancy outcomes, and approach to treatment in developing countries. Sex. Transm. Infect. 2005, 81, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Adachi, K.; Nielsen-Saines, K.; Klausner, J.D. Chlamydia trachomatis Infection in Pregnancy: The Global Challenge of Preventing Adverse Pregnancy and Infant Outcomes in Sub-Saharan Africa and Asia. Biomed. Res. Int. 2016, 2016, 9315757. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Incidence and Prevalence of Selected Curable Sexually Transmitted Infections-2008; WHO: Geneva, Switzerland, 2012. [Google Scholar]
- Al-Nasiry, S.; Ambrosino, E.; Schlaepfer, M.; Morré, S.A.; Wieten, L.; Voncken, J.W.; Spinelli, M.; Mueller, M.; Kramer, B.W. The Interplay Between Reproductive Tract Microbiota and Immunological System in Human Reproduction. Front. Immunol. 2020, 11, 378. [Google Scholar] [CrossRef]
- Alliance for Maternal and Newborn Health Improvement; Baqui, A.H.; Khanam, R.; Rahman, M.S.; Ahmed, A.; Rahman, H.H.; Moin, M.I.; Ahmed, S.; Jehan, F.; Nisar, I.; et al. Understanding biological mechanisms underlying adverse birth outcomes in developing countries: Protocol for a prospective cohort (AMANHI bio-banking) study. J. Glob. Health 2017, 7, 021201. [Google Scholar]
- Juliana, N.C.A.; Omar, A.M.; Pleijster, J.; Aftab, F.; Uijldert, N.B.; Ali, S.M.; Ouburg, S.; Sazawal, S.; Morré, S.A.; Deb, S.; et al. The Natural Course of Chlamydia trachomatis, Neisseria gonorrhoeae, Trichomonas vaginalis, and Mycoplasma genitalium in Pregnant and Post-Delivery Women in Pemba Island, Tanzania. Microorganisms 2021, 9, 1180. [Google Scholar] [CrossRef] [PubMed]
- Juliana, N.C.A.; Deb, S.; Ouburg, S.; Chauhan, A.; Pleijster, J.; Ali, S.M.; Morré, S.A.; Sazawal, S.; Ambrosino, E. The Prevalence of Chlamydia trachomatis and Three Other Non-Viral Sexually Transmitted Infections among Pregnant Women in Pemba Island Tanzania. Pathogens 2020, 9, 625. [Google Scholar] [CrossRef] [PubMed]
- Dols, J.A.M.; Molenaar, D.; van der Helm, J.J.; Caspers, M.P.M.; de Kat Angelino-Bart, A.; Schuren, F.H.J.; Speksnijder, A.G.C.L.; Westerhoff, H.V.; Richardus, J.H.; Boon, M.E.; et al. Molecular assessment of bacterial vaginosis by Lactobacillus abundance and species diversity. BMC Infect. Dis. 2016, 16, 180. [Google Scholar] [CrossRef]
- Budding, A.E.; Hoogewerf, M.; Vandenbroucke-Grauls, C.M.J.E.; Savelkoul, P.H.M. Automated Broad-Range Molecular Detection of Bacteria in Clinical Samples. Am. Soc. Microbiol. 2016, 54, 934–943. [Google Scholar] [CrossRef]
- Budding, A.E.; Grasman, M.E.; Lin, F.; Bogaards, J.A.; Soeltan-Kaersenhout, D.J.; Vandenbroucke-Grauls, C.M.J.E.; Van Bodegraven, A.A.; Savelkoul, P.H.M. IS-pro: High-throughput molecular fingerprinting of the intestinal microbiota. FASEB J. 2010, 24, 4556–4564. [Google Scholar] [CrossRef]
- Jost, L. The Relation between Evenness and Diversity. Diversity 2010, 2, 207–232. [Google Scholar] [CrossRef]
- de Waaij, D.J.; Ouburg, S.; Dubbink, J.H.; Peters, R.P.H.; Morré, S.A. Evaluation of Prestoplus assay and LightMix kit Trichomonas vaginalis assay for detection of Trichomonas vaginalis in dry vaginal swabs. J. Microbiol. Methods 2016, 127, 102–104. [Google Scholar] [CrossRef][Green Version]
- de Waaij, D.J.; Dubbink, J.H.; Peters, R.P.H.; Ouburg, S.; Morré, S.A. Comparison of GMT presto assay and Roche cobas® 4800 CT/NG assay for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in dry swabs. J. Microbiol. Methods 2015, 118, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Müller, E.E.; Venter, J.M.E.; Magooa, M.P.; Morrison, C.; Lewis, D.A.; Mavedzenge, S.N. Development of a rotor-gene real-time PCR assay for the detection and quantification of Mycoplasma genitalium. J. Microbiol. Methods 2012, 88, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Juliana, N.C.A.; Juma, M.H.; Heijmans, R.; Ouburg, S.; Ali, S.M.; Chauhan, A.S.; Pemba, A.B.; Sazawal, S.; Morré, S.A.; Deb, S.; et al. Detection of high-risk human papillomavirus (HPV) by the novel AmpFire isothermal HPV assay among pregnant women in Pemba Island, Tanzania. Pan Afr. Med. J. 2020, 37, 183. [Google Scholar] [CrossRef] [PubMed]
- Masha, S.C.; Cools, P.; Descheemaeker, P.; Reynders, M.; Sanders, E.J.; Vaneechoutte, M. Urogenital pathogens, associated with Trichomonas vaginalis, among pregnant women in Kilifi, Kenya: A nested case-control study. BMC Infect. Dis. 2018, 18, 549. [Google Scholar] [CrossRef]
- Vaneechoutte, M. Lactobacillus iners, the unusual suspect. Res. Microbiol. 2017, 168, 826–836. [Google Scholar] [CrossRef]
- Freitas, A.C.; Chaban, B.; Bocking, A.; Rocco, M.; Yang, S.; Hill, J.E.; Money, D.M.; Hemmingsen, S.; Reid, G.; Dumonceaux, T.; et al. The vaginal microbiome of pregnant women is less rich and diverse, with lower prevalence of Mollicutes, compared to non-pregnant women. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Nunn, K.L.; Witkin, S.S.; Schneider, G.M.; Boester, A.; Nasioudis, D.; Minis, E.; Gliniewicz, K.; Forney, L.J. Changes in the Vaginal Microbiome during the Pregnancy to Postpartum Transition. Reprod. Sci. 2021, 28, 1996–2005. [Google Scholar] [CrossRef]
- Gudza-Mugabe, M.; Havyarimana, E.; Jaumdally, S.; Garson, K.L.; Lennard, K.; Tarupiwa, A.; Mugabe, F.; Marere, T.; Mavenyengwa, R.T.; Masson, L.; et al. HIV infection is associated with preterm delivery independent of vaginal microbiota in pregnant African women. J. Infect. Dis. 2019, 221, 1194–1203. [Google Scholar] [CrossRef]
- Borgdorff, H.; Verwijs, M.C.; Wit, F.W.N.M.; Tsivtsivadze, E.; Ndayisaba, G.F.; Verhelst, R.; Schuren, F.H.; Van De Wijgert, J.H.H.M. The impact of hormonal contraception and pregnancy on sexually transmitted infections and on cervicovaginal microbiota in african sex workers. Sex. Transm. Dis. 2015, 42, 143–152. [Google Scholar] [CrossRef]
- O’Hara, M.W.; Schlechte, J.A.; Lewis, D.A.; Wright, E.J. Prospective Study of Postpartum Blues: Biologic and Psychosocial Factors. Arch. Gen. Psychiatry 1991, 48, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Nott, P.N.; Franklin, M.; Armitage, C.; Gelder, M.G. Hormonal changes and mood in the puerperium. Br. J. Psychiatry 1976, 128, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Onderdonk, A.B.; Delaney, M.L.; Fichorova, R.N. The human microbiome during bacterial vaginosis. Clin. Microbiol. Rev. 2016, 29, 223–238. [Google Scholar] [CrossRef]
- Parnell, L.A.; Briggs, C.M.; Mysorekar, I.U. Maternal microbiomes in preterm birth: Recent progress and analytical pipelines. Semin. Perinatol. 2017, 41, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Kinney, M.V.; Lawn, J.E.; Howson, C.P.; Belizan, J. 15 million preterm births annually: What has changed this year? Reprod. Health 2012, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Donders, G.G.G.; Bellen, G.; Grinceviciene, S.; Ruban, K.; Vieira-Baptista, P. Aerobic vaginitis: No longer a stranger. Res. Microbiol. 2017, 168, 845–858. [Google Scholar] [CrossRef]
- Kaambo, E.; Africa, C.W.J. The threat of aerobic vaginitis to pregnancy and neonatal morbidity. Afr. J. Reprod. Health 2017, 21, 108–118. [Google Scholar] [CrossRef]
- Akerele, J.; Abhulimen, P.; Okonofua, F. Prevalence of asymptomatic genital infection among pregnant women in Benin City, Nigeria. Afr. J. Reprod. Health 2002, 6, 93–97. [Google Scholar] [CrossRef]
- Shrestha, L.B.; Baral, R.; Poudel, P.; Khanal, B. Clinical, etiological and antimicrobial susceptibility profile of pediatric urinary tract infections in a tertiary care hospital of Nepal. BMC Pediatr. 2019, 19, 36. [Google Scholar] [CrossRef]
- Tao, Z.; Zhang, L.; Zhang, Q.; Lv, T.; Chen, R.; Wang, L.; Huang, Z.; Hu, L.; Liao, Q. The pathogenesis of streptococcus anginosus in aerobic vaginitis. Infect. Drug Resist. 2019, 12, 3745–3754. [Google Scholar] [CrossRef]
- Cox, R.A.; Chen, K.; Coykendall, A.L.; Wesbecher, P.; Hersont, V.C. Fatal infection in neonates of 26 weeks’ gestation due to Streptococcus milleri: Report of two cases. J. Clin. Pathol. 1987, 40, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Omwandho, C.O.A.; Gruessner, S.E.M.; Tinneberg, H.R. Early pregnancy loss and neonatal deaths associated with Klebsiella pneumonia infection: A mini review of possible occupational health risk. Arch. Gynecol. Obstet. 2006, 273, 258–260. [Google Scholar] [CrossRef] [PubMed]
- Le Doare, K.; Heath, P.T.; Plumb, J.; Owen, N.A.; Brocklehurst, P.; Chappell, L.C. Uncertainties in Screening and Prevention of Group B Streptococcus Disease. Clin. Infect. Dis. 2019, 69, 720–725. [Google Scholar] [CrossRef]
- Stevens, D.; Kaplan, E. Streptococcal Infections: Clinical Aspects, Microbiology, and Molecular Pathogenesis; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Gizachew, M.; Tiruneh, M.; Moges, F.; Tessema, B. Streptococcus agalactiae maternal colonization, antibiotic resistance and serotype profiles in Africa: A meta-analysis. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 14. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, C.P.; Lamagni, T.; Patel, D.; Efstratiou, A.; Cunney, R.; Meehan, M.; Ladhani, S.; Reynolds, A.J.; Campbell, R.; Doherty, L.; et al. Group B streptococcal disease in UK and Irish infants younger than 90 days, 2014–2015: A prospective surveillance study. Lancet Infect. Dis. 2019, 19, 83–90. [Google Scholar] [CrossRef]
- Nanayakkara, D.; Liyanapathirana, V.; Kandauda, C.; Gihan, C.; Ekanayake, A.; Adasooriya, D. Maternal vaginal colonization with selected potential pathogens of neonatal sepsis in the era of antimicrobial resistance, a single center experience from Sri Lanka. BMC Infect. Dis. 2018, 18, 351. [Google Scholar] [CrossRef] [PubMed]
- Brabin, L.; Roberts, S.A.; Gies, S.; Nelson, A.; Diallo, S.; Stewart, C.J.; Kazienga, A.; Birtles, J.; Ouedraogo, S.; Claeys, Y.; et al. Effects of long-term weekly iron and folic acid supplementation on lower genital tract infection—A double blind, randomised controlled trial in Burkina Faso. BMC Med. 2017, 15, 206. [Google Scholar] [CrossRef]
- Borgdorff, H.; Armstrong, S.D.; Tytgat, H.L.P.; Xia, D.; Ndayisaba, G.F.; Wastling, J.M.; Van De Wijgert, J.H.H.M. Unique insights in the cervicovaginal Lactobacillus iners and L. crispatus proteomes and their associations with microbiota dysbiosis. PLoS ONE 2016, 11, e0150767. [Google Scholar] [CrossRef]
- Zheng, N.; Guo, R.; Wang, J.; Zhou, W.; Ling, Z. Contribution of Lactobacillus iners to Vaginal Health and Diseases: A Systematic Review. Front. Cell. Infect. Microbiol. 2021, 11. [Google Scholar] [CrossRef]
- Petrova, M.I.; Reid, G.; Vaneechoutte, M.; Lebeer, S. Lactobacillus iners: Friend or Foe? Trends Microbiol. 2017, 25, 182–191. [Google Scholar] [CrossRef]
- Ahmed, I.; Ali, S.M.; Amenga-Etego, S.; Ariff, S.; Bahl, R.; Baqui, A.H.; Begum, N.; Bhandari, N.; Bhatia, K.; Bhutta, Z.A.; et al. Population-based rates, timing, and causes of maternal deaths, stillbirths, and neonatal deaths in south Asia and sub-Saharan Africa: A multi-country prospective cohort study. Lancet Glob. Health 2018, 6, e1297–e1308. [Google Scholar] [CrossRef]
- Singer, M.; Borg, M.; Ouburg, S.; Morré, S.A. The relation of the vaginal microbiota to early pregnancy development during in vitro fertilization treatment—A meta-analysis. J. Gynecol. Obstet. Hum. Reprod. 2019, 48, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.; Koedooder, R.; Bos, M.P.; Poort, L.; Schoenmakers, S.; Savelkoul, P.H.M.; Laven, J.S.E.; de Jonge, J.D.; Morré, S.A.; Budding, A.E. The profiling of microbiota in vaginal swab samples using 16S rRNA gene sequencing and IS-pro analysis. BMC Microbiol. 2021, 21, 100. [Google Scholar] [CrossRef] [PubMed]
- Jochum, L.; Stecher, B. Label or Concept—What Is a Pathobiont? Trends Microbiol. 2020, 28, 789–792. [Google Scholar] [CrossRef] [PubMed]
- McBurney, M.I.; Davis, C.; Fraser, C.M.; Schneeman, B.O.; Huttenhower, C.; Verbeke, K.; Walter, J.; Latulippe, M.E. Establishing What Constitutes a Healthy Human Gut Microbiome: State of the Science, Regulatory Considerations, and Future Directions. J. Nutr. 2019, 149, 1882–1895. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).