Microbial Population Succession and Community Diversity and Its Correlation with Fermentation Quality in Soybean Meal Treated with Enterococcus faecalis during Fermentation and Aerobic Exposure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Microorganisms
2.2. Optimization of Growth Conditions for ZZUPF95
2.3. Optimization of Fermented Soybean Meal (FSBM) Conditions for ZZUPF95
2.4. Preparation of FSBM
2.5. Analysis of FSBM
2.5.1. Evaluation of Sensory
2.5.2. Fermentation Quality
2.5.3. Chemical Analysis
2.5.4. Microbiological Analysis
2.5.5. Scanning Electron Microscopy (SEM) of SBM and FSBM
2.5.6. SDS-PAGE ((Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis) Profile
2.6. Bacterial and Fungi Community Analyses
2.6.1. DNA Extraction
2.6.2. PCR Amplification
2.7. Statistical Analysis
3. Results
3.1. The Optimization of Inorganic Salt, Carbon and Nitrogen Source on Growth Conditions for ZZUPF95
3.2. Determination of Optimal Growth Conditions for ZZUPF95
3.3. Optimization of FSBM Conditions for ZZUPF95
3.4. Fermentation Quality, Chemical Composition and Microbial Population of FSBM
3.4.1. Sensory Evaluation
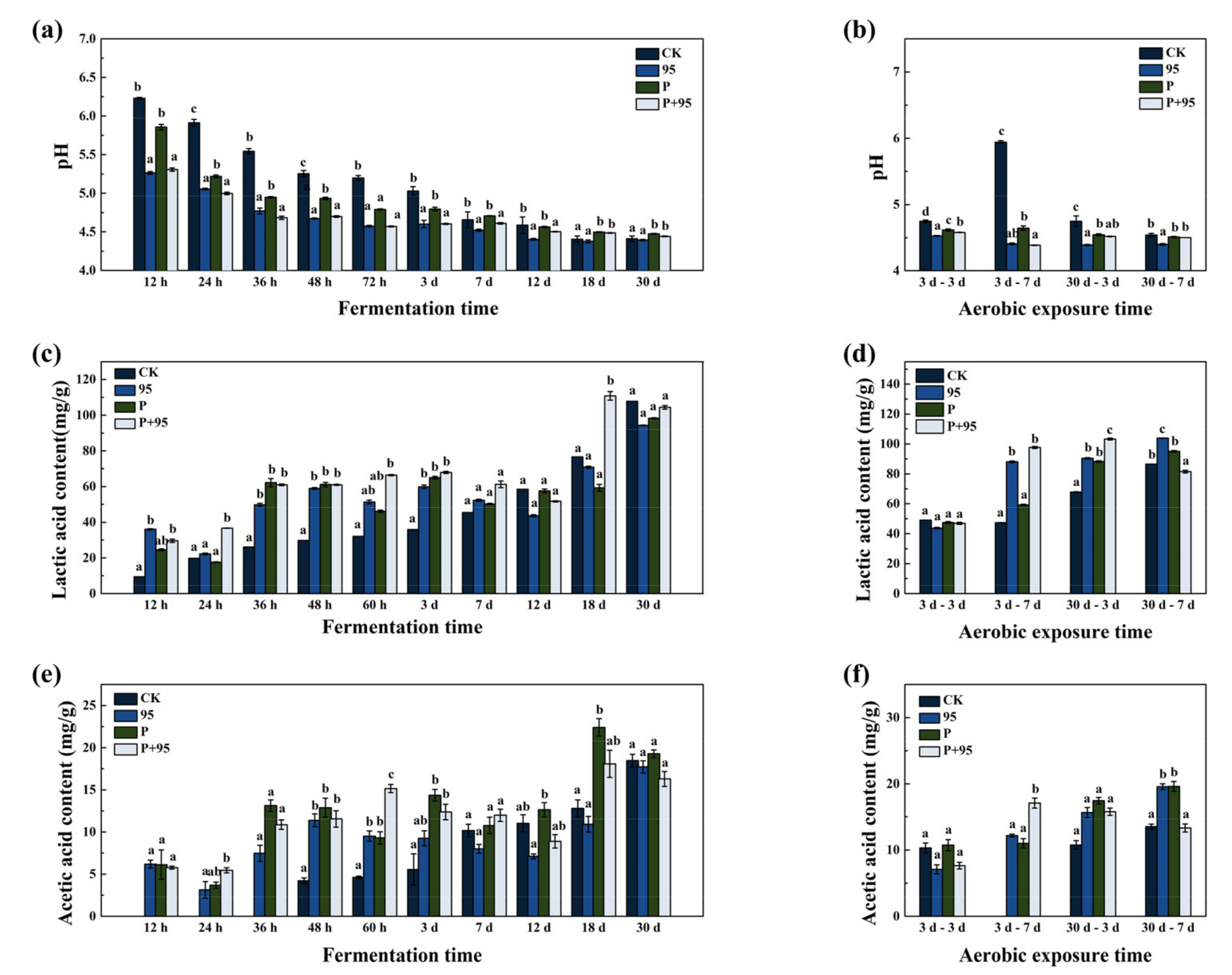
3.4.2. Fermentation Quality of FSBM
3.4.3. Chemical Composition of FSBM
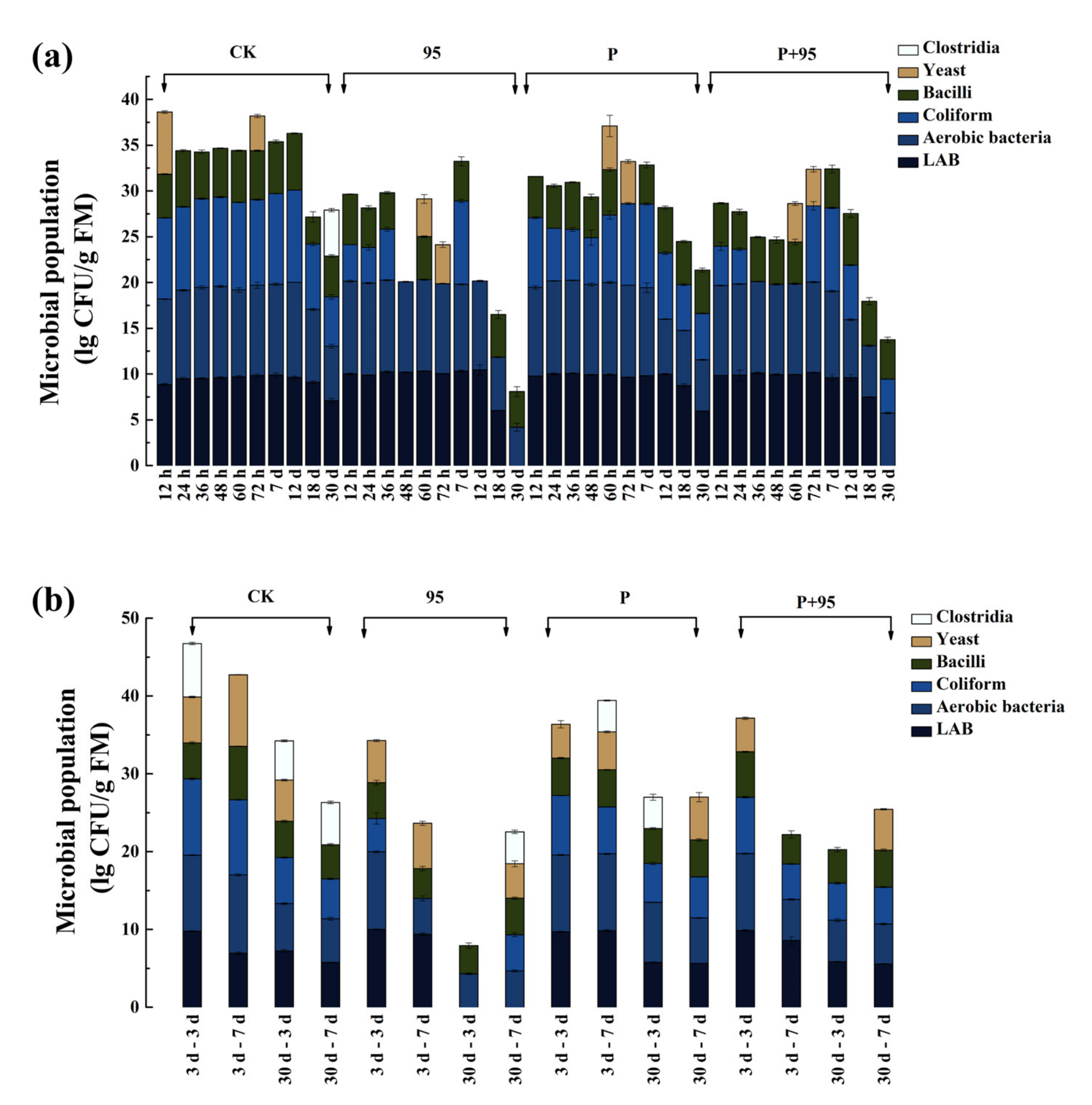
3.4.4. Microbial Population of FSBM
3.4.5. SEM
3.4.6. SDS-PAGE Profile
3.5. Microbial Community Dynamics of FSBM
3.5.1. Alpha Diversity Indices of Microbial Community
3.5.2. Principal Coordinate Analysis
3.5.3. Abundance of Microbial Community
3.6. Correlation Analyses of the Bacterial and Fungi Community with Fermentation Properties
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, Y.; Sun, D.; Zhao, M.; Zhao, Q.; Qiu, C.; Su, G. Effects of solid-state fermentation and proteolytic hydrolysis on defatted soybean meal. LWT-Food Sci. Technol. 2018, 97, 496–502. [Google Scholar] [CrossRef]
- Ruan, S.; Luo, J.; Li, Y.; Wang, Y.; Huang, S.; Lu, F.; Ma, H. Ultrasound-assisted liquid-state fermentation of soybean meal with Bacillus subtilis: Effects on peptides content, ACE inhibitory activity and biomass. Process. Biochem. 2020, 91, 73–82. [Google Scholar] [CrossRef]
- Su, L.; Cheng, Y.; Hsiao, F.S.H.; Han, J.; Yu, Y. Optimization of mixed solid-state fermentation of soybean meal by Lactobacillus Species and Clostridium butyricum. Pol. J. Microbiol. 2018, 67, 297–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhou, R.; Ren, Z.; Fan, Y.; Hu, S.; Zhuo, C.; Deng, Z. Improvement of protein quality and degradation of allergen in soybean meal fermented by Neurospora crassa. LWT-Food Sci. Technol. 2019, 101, 220–228. [Google Scholar] [CrossRef]
- Hajimohammadi, A.; Mottaghitalab, M.; Hashemi, M. Effects of microbial fermented sesame meal and enzyme supplementation on the intestinal morphology, microbiota, pH, tibia bone and blood parameters of broiler chicks. Ital. J. Anim. Sci. 2020, 19, 457–467. [Google Scholar] [CrossRef] [Green Version]
- Fisher, H.; Shapiro, R. Counteracting the growth retardation of raw soybean meal with extra protein and calories. J. Nutr. 1963, 80, 425–430. [Google Scholar] [CrossRef]
- Fasina, Y.O.; Classen, H.L.; Garlich, J.D.; Swaisgood, H.E.; Clare, D.A. Investigating the possibility of monitoring lectin levels in commercial soybean meals intended for poultry feeding using steam-heated soybean meal as a model. Poult. Sci. 2003, 82, 648–656. [Google Scholar] [CrossRef]
- Friedman, M.; Gumbmann, M.R. Nutritional improvement of legume proteins through disulfide interchange. Adv. Exp. Med. Biolo. 1986, 199, 357–389. [Google Scholar] [CrossRef]
- Dawood, M.A.O.; Koshio, S. Application of fermentation strategy in aquafeed for sustainable aquaculture. Rev. Aquacult. 2020, 12, 987–1002. [Google Scholar] [CrossRef]
- Cotarlet, M.; Stanciuc, N.; Bahrim, G.E. Yarrowia lipolyticaand Lactobacillus paracasei solid state fermentation as a valuable biotechnological tool for the pork lard and okara's biotransformation. Microorganisms 2020, 8, 1098. [Google Scholar] [CrossRef]
- Li, Y.; Wei, X.; Wang, J.; Ding, W.; Zheng, F.; Hu, X.; Kou, L. Isolation and identification of lactic acid bacteria inhibiting penicillium and analysis of their antimicrobial components. Food Sci. 2015, 36, 1501155. [Google Scholar] [CrossRef]
- Shriver, J.A.; Carter, S.D.; Sutton, A.L.; Richert, B.T.; Senne, B.W.; Pettey, L.A. Effects of adding fiber sources to reduced-crude protein, amino acidsupplemented diets on nitrogen excretion, growth performance, and carcass traits of finishing pigs. J. Anim. Sci. 2003, 81, 492–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges, S.; Barbosa, J.; Silva, J.; Gomes, A.M.; Pintado, M.; Silva, C.L.M.; Morais, A.M.M.B.; Teixeira, P. A feasibility study of Lactobacillus plantarum in fruit powders after processing and storage. Int. J. Food Sci. Technol. 2016, 51, 381–388. [Google Scholar] [CrossRef]
- Mukerji, P.; Roper, J.M.; Stahl, B.; Smith, A.B.; Burns, F.; Rae, J.C.; Yeung, N.; Lyra, A.; Svard, L.; Saarinen, M.T. Safety evaluation of AB-LIFE (R) (Lactobacillus plantarum CECT 7527, 7528 and 7529): Antibiotic resistance and 90-day repeated-dose study in rats. Food Chem. Toxicol. 2016, 92, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, N.; Kamra, D.N.; Chaudhary, L.C.; Agarwal, I.; Sahoo, A.; Pathak, N.N. Microbial status and rumen enzyme profile of crossbred calves fed on different microbial feed additives. Lett. Appl. Microbiol. 2002, 34, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Xin, C.; Zhao, Y.; Feng, B.; He, C.; Dong, Y.; Fang, Y.; Wei, S. Study of active ingredients in black soybean sprouts and their safety in cosmetic use. Molecules 2012, 17, 11669–11679. [Google Scholar] [CrossRef] [PubMed]
- Meinlschmidt, P.; Ueberham, E.; Lehmann, J.; Schweiggert-Weisz, U.; Eisner, P. Immunoreactivity, sensory and physico-chemical properties of fermented soy protein isolate. Food Chem. 2016, 205, 229–238. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis of the Association of Official Analytical Chemists, 15th ed.; Association of Official Analytical Chemists: Arlington, VA, USA, 1990. [Google Scholar]
- Pang, H.; Qin, G.; Tan, Z.; Li, Z.; Wang, Y.; Cai, Y. Natural populations of lactic acid bacteria associated with silage fermentation as determined by phenotype, 16S ribosomal RNA and recA gene analysis. Syst. Appl. Microbiol. 2011, 34, 235–241. [Google Scholar] [CrossRef]
- Qiao, W.; Tao, J.; Luo, Y.; Tang, T.; Miao, J.; Yang, Q. Microbial oil production from solid-state fermentation by a newly isolated oleaginous fungus, Mucor circinelloides Q531 from mulberry branches. R. Soc. Open Sci. 2018, 5, 180551. [Google Scholar] [CrossRef] [Green Version]
- Niba, A.T.; Beal, J.D.; Kudi, A.C.; Brooks, P.H. Bacterial fermentation in the gastrointestinal tract of non-ruminants: Influence of fermented feeds and fermentable carbohydrates. Trop. Anim. Health Prod. 2009, 41, 1393–1407. [Google Scholar] [CrossRef]
- Dalié, D.K.D.; Deschamps, A.M.; Richard-Forget, F. Lactic acid bacteria—Potential for control of mould growth and mycotoxins: A review. Food Control 2010, 21, 370–380. [Google Scholar] [CrossRef]
- Cai, Y.; Benno, Y.; Ogawa, M.; Kumai, S. Effect of applying lactic acid bacteria isolated from forage crops on fermentation characteristics and aerobic deterioration of silage. J. Dairy Sci. 1999, 82, 520–526. [Google Scholar] [CrossRef]
- Yang, L.; Yuan, X.; Li, J.; Dong, Z.; Shao, T. Dynamics of microbial community and fermentation quality during ensiling of sterile and nonsterile alfalfa with or without Lactobacillus plantarum inoculant. Bioresour. Technol. 2018, 275, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.H.; Lee, N.K.; Paik, H.D. Optimization of medium composition for biomass production of Lactobacillus plantarum 200655 using response surface methodology. J. Microbiol. Biotechnol. 2021, 31, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.; Kim, S.; Ye, S.; Kim, S.R.; Seol, J.; Dooyum, U.D.; Kim, J.; Hong, D.H.; Kim, J.N.; Ha, Y. Effect of temperature on single- and mixed-strain fermentation of ruminant feeds. J. Anim. Sci. Technol. 2020, 62, 227–238. [Google Scholar] [CrossRef] [Green Version]
- Sun, F.; Li, Q.; Liu, H.; Kong, B.; Liu, Q. Purification and biochemical characteristics of the protease from Lactobacillus brevis R4 isolated from Harbin dry sausages. LWT-Food Sci. Technol. 2019, 113, 108287. [Google Scholar] [CrossRef]
- Bujnak, L.; Maskal’ova, I.; Vajda, V. Determination of buffering capacity of selected fermented feedstuffs and the effect of dietary acid-base status on ruminal fluid pH. Acta Vet. Brno 2011, 80, 269–273. [Google Scholar] [CrossRef] [Green Version]
- Varsha, K.K.; Priya, S.; Devendra, L.; Nampoothiri, K.M. Control of spoilage fungi by protective lactic acid bacteria displaying probiotic properties. Appl. Biochem. Biotechnol. 2014, 172, 3402–3413. [Google Scholar] [CrossRef]
- Refstie, S.; Sahlstrom, S.; Brathen, E.; Baeverfjord, G.; Krogedal, P. Lactic acid fermentation eliminates indigestible carbohydrates and antinutritional factors in soybean meal for Atlantic salmon (Salmo salar). Aquaculture 2005, 246, 331–345. [Google Scholar] [CrossRef]
- Mohd-Setapar, S.H.; Abd-Talib, N.; Aziz, R. Review on crucial parameters of silage quality. Apcbee Procedia 2012, 3, 99–103. [Google Scholar] [CrossRef] [Green Version]
- Tabacco, E.; Piano, S.; Revello-Chion, A.; Borreani, G. Effect of Lactobacillus buchneri LN4637 and Lactobacillus buchneri LN40177 on the aerobic stability, fermentation products, and microbial populations of corn silage under farm conditions. J. Dairy Sci. 2011, 94, 5589–5598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautam, N.; Sharma, N. Evaluation of probiotic potential of new bacterial strain, Lactobacillus spicheri G2 isolated from gundruk. Proc. Natl. Acad. Sci. India Sect. B—Biol. Sci. 2015, 85, 979–986. [Google Scholar] [CrossRef]
- Saelim, K.; Jampaphaeng, K.; Maneerat, S. Functional properties of Lactobacillus plantarum S0/7 isolated fermented stinky bean (Sa Taw Dong) and its use as a starter culture. J. Funct. Foods 2017, 38, 370–377. [Google Scholar] [CrossRef]
- Suiryanrayna, M.V.A.N.; Ramana, J.V. A review of the effects of dietary organic acids fed to swine. J. Anim. Sci. Biotechnol. 2015, 6, 45. [Google Scholar] [CrossRef] [Green Version]
- Santos, V.A.Q.; Nascimento, C.G.; Schimidt, C.A.P.; Mantovani, D.; Dekker, R.F.H.; da Cunha, M.A.A. Solid-state fermentation of soybean okara: Isoflavones biotransformation, antioxidant activity and enhancement of nutritional quality. LWT-Food Sci. Technol. 2018, 92, 509–515. [Google Scholar] [CrossRef]
- Olude, O.; George, F.; Alegbeleye, W. Utilization of autoclaved and fermented sesame (Sesamum indicum L.) seed meal in diets for Til-aqua natural male tilapia. Anim. Nutr. 2016, 2, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.; Kim, E.B.; Han, G.G.; Kim, B.G. The Influence of dietary supplementation of bacteriophages on energy and nutrient digestibility and intestinal microbiota of pigs. FASEB J. 2017, 31, lb401. [Google Scholar] [CrossRef]
- Goodarzi Boroojeni, F.; Senz, M.; Kozłowski, K.; Boros, D.; Wisniewska, M.; Rose, D. The effects of fermentation and enzymatic treatment of pea on nutrient digestibility and growth performance of broilers. Animal 2017, 11, 1698–1707. [Google Scholar] [CrossRef] [Green Version]
- Ni, K.; Wang, F.; Zhu, B.; Yang, J.; Zhou, G.; Pan, Y.; Tao, Y.; Zhong, J. Effects of lactic acid bacteria and molasses additives on the microbial community and fermentation quality of soybean silage. Bioresour. Technol. 2017, 238, 706–715. [Google Scholar] [CrossRef]
- Wang, T.; Teng, K.; Cao, Y.; Shi, W.; Xuan, Z.; Zhou, J.; Zhang, J.; Zhong, J. Effects of Lactobacillus hilgardii 60TS-2, with or without homofermentative Lactobacillus plantarum B90, on the aerobic stability, fermentation quality and microbial community dynamics in sugarcane top silage. Bioresour. Technol. 2020, 312, 123600. [Google Scholar] [CrossRef]
- Emerstorfer, F.; Hein, W.; Resch, R.; Poetsch, E.; Zitz, U.; Kneifel, W. Application of plant-based antimicrobials for the growth inhibition of clostridia in pressed beet pulp silage. J. Sci. Food Agric. 2011, 91, 2038–2044. [Google Scholar] [CrossRef] [PubMed]
- Keshri, J.; Chen, Y.R.; Pinto, R.; Kroupitski, Y.; Weinberg, Z.G.; Sela, S. Microbiome dynamics during ensiling of corn with and without Lactobacillus plantarum inoculant. Appl. Microbiol. Biotechnol. 2018, 102, 4025–4037. [Google Scholar] [CrossRef] [PubMed]
- Wijayawardene, N.N.; Hyde, K.D.; Lumbsch, H.T.; Jian, K.L. Outline of Ascomycota: 2017. Fungal Divers. 2017, 88, 167–263. [Google Scholar] [CrossRef]
- Roncero, I.G.M.; Concepción, H.; Manuel, R.R.; Maceira, G.; Marta, M. Fusarium as a model for studying virulence in soilborne plant pathogens. Physiol. Mol. Plant Pathol. 2003, 62, 87–98. [Google Scholar] [CrossRef]
- Carbonetto, B.; Nidelet, T.; Guezenec, S.; Perez, M.; Segond, D.; Sicard, D. Interactions between Kazachstania humilis yeast species and lactic acid bacteria in sourdough. Microorganisms 2020, 8, 240. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Long, L.; An, Y.; Jing, Y. The thioredoxin reductase-encoding gene Actrxr1 is involved in the cephalosporin C production of Acremonium chrysogenum in methionine-supplemented medium. Appl. Microbiol. Biotechnol. 2013, 97, 2551–2562. [Google Scholar] [CrossRef]
- Su, W.; Jiang, Z.; Hao, L.; Li, W.; Gong, T.; Zhang, Y.; Du, S.; Wang, C.; Lu, Z.; Jin, M. Variations of soybean meal and corn mixed substrates in physicochemical characteristics and microbiota during two-stage solid-state fermentation. Front. Microbiol. 2021, 12, 688839. [Google Scholar] [CrossRef]

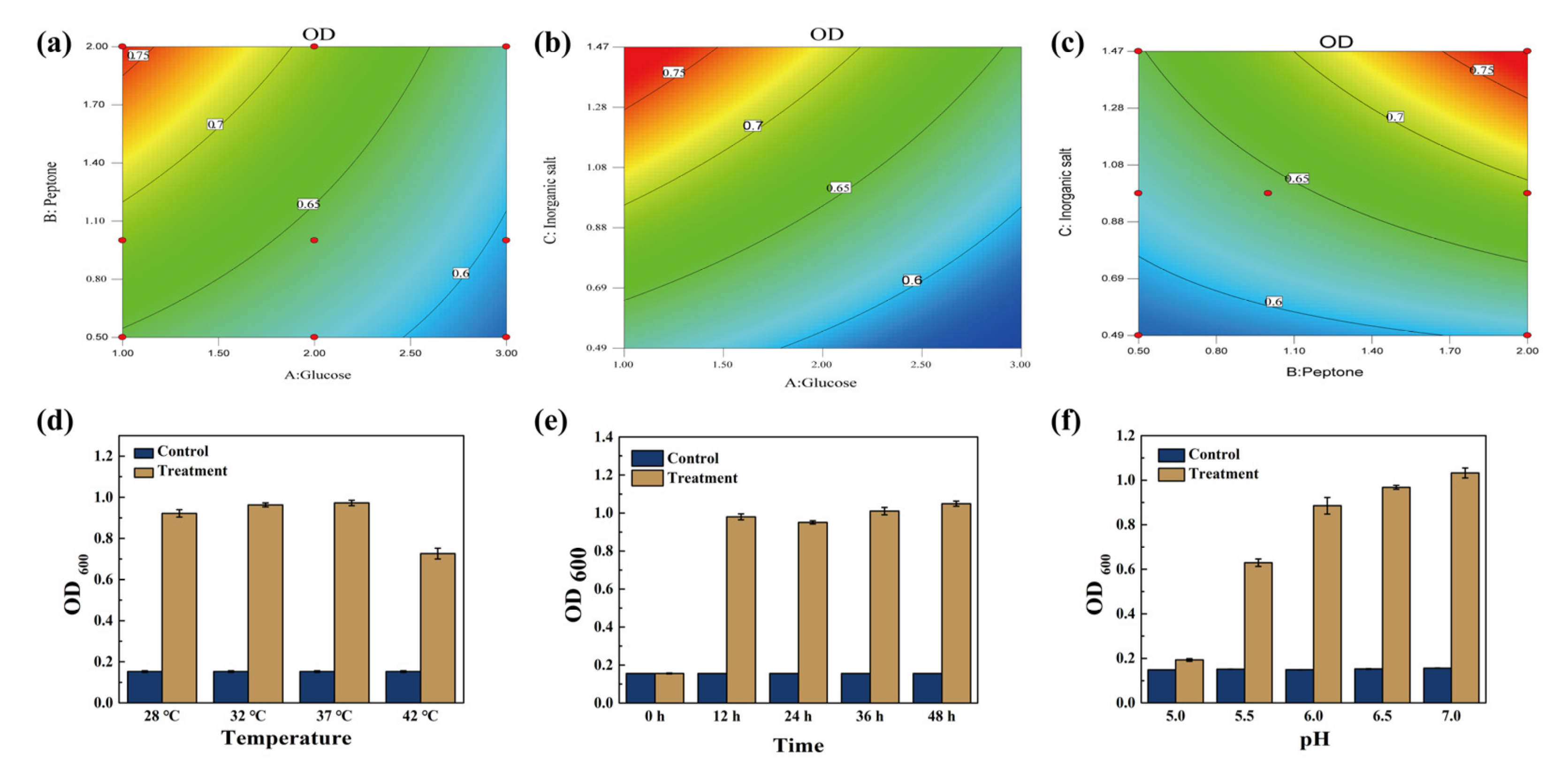
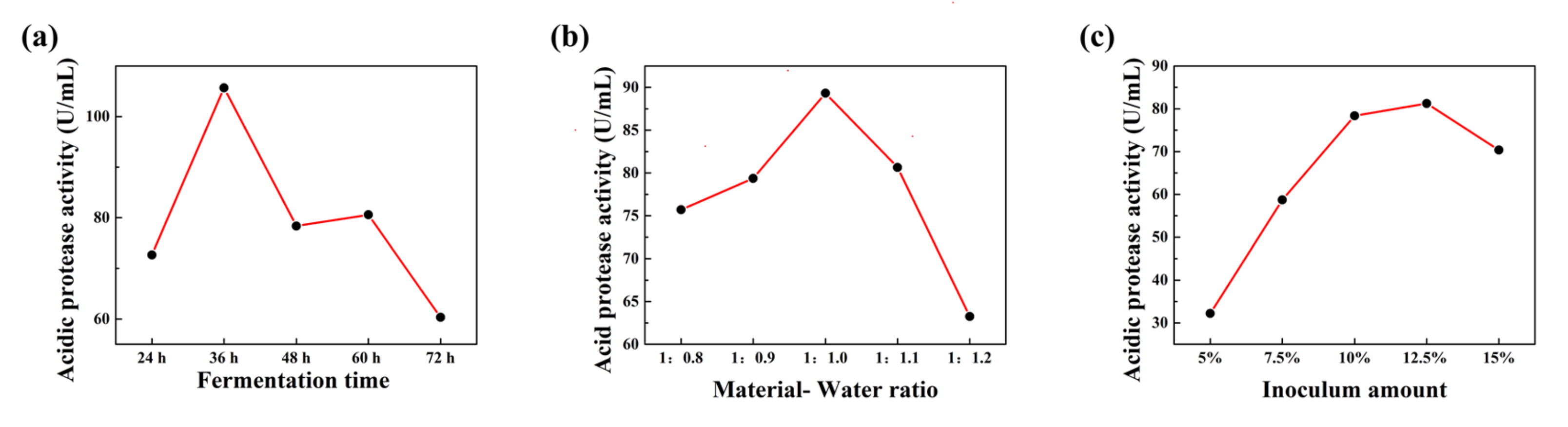

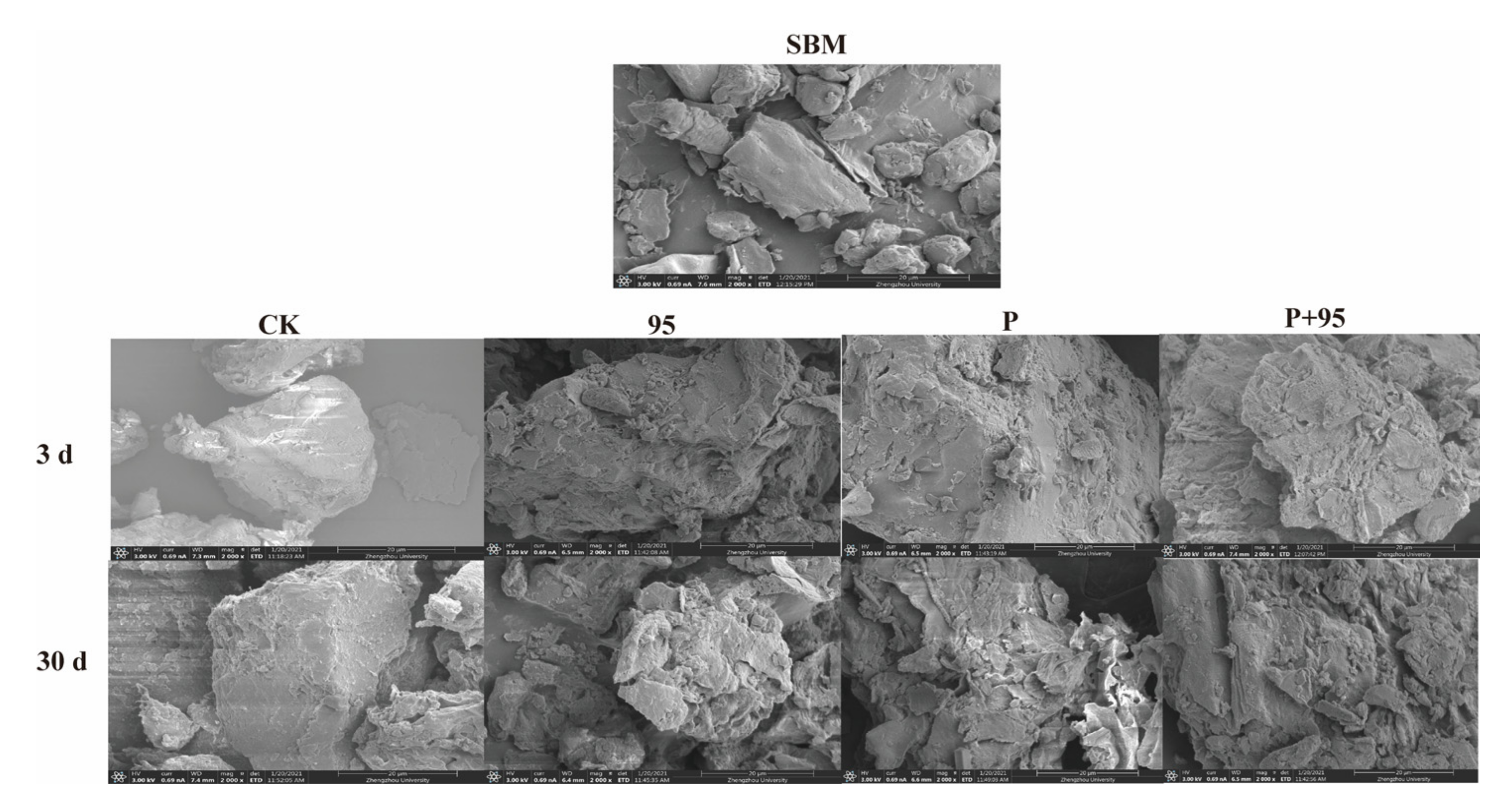


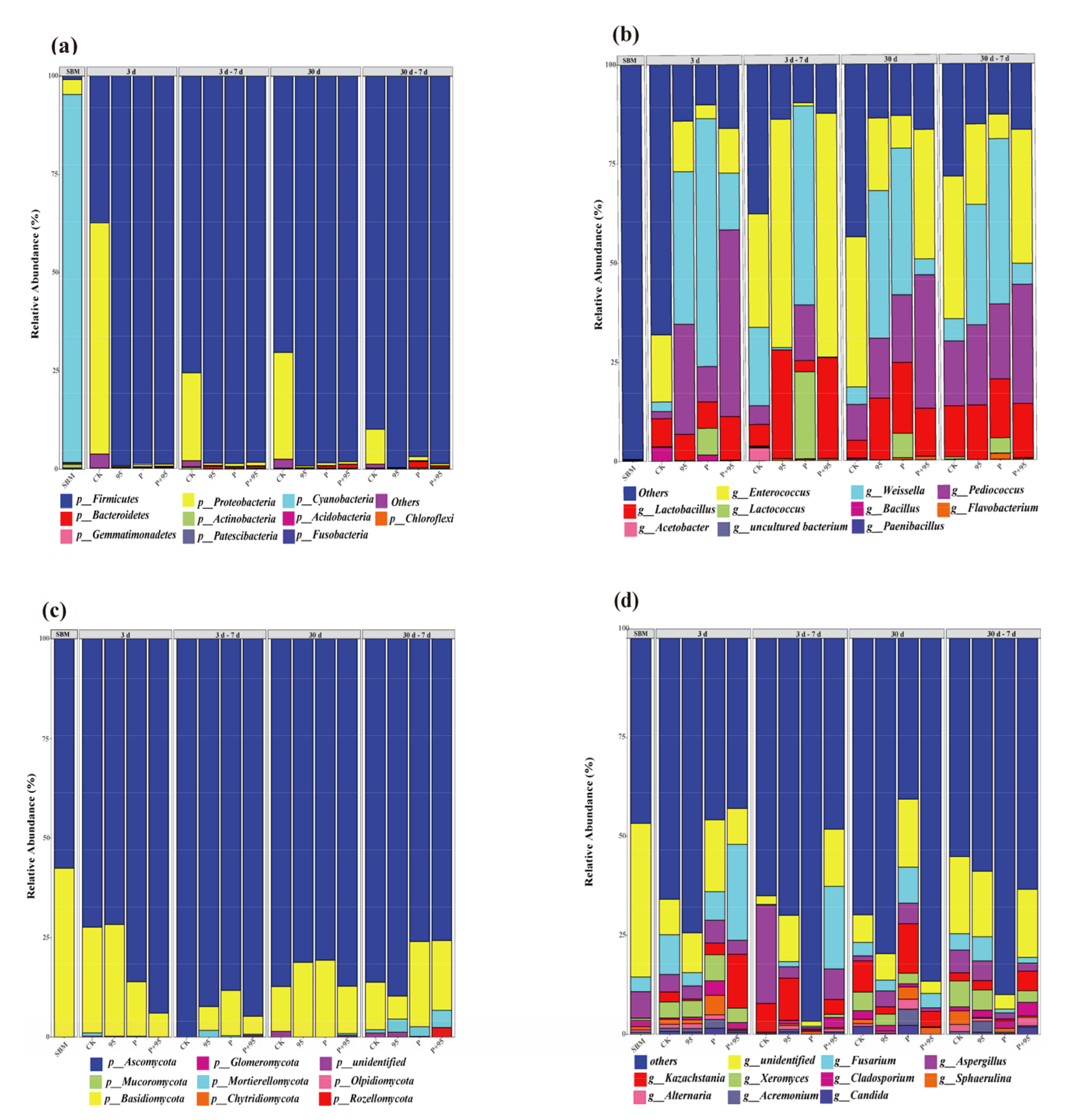
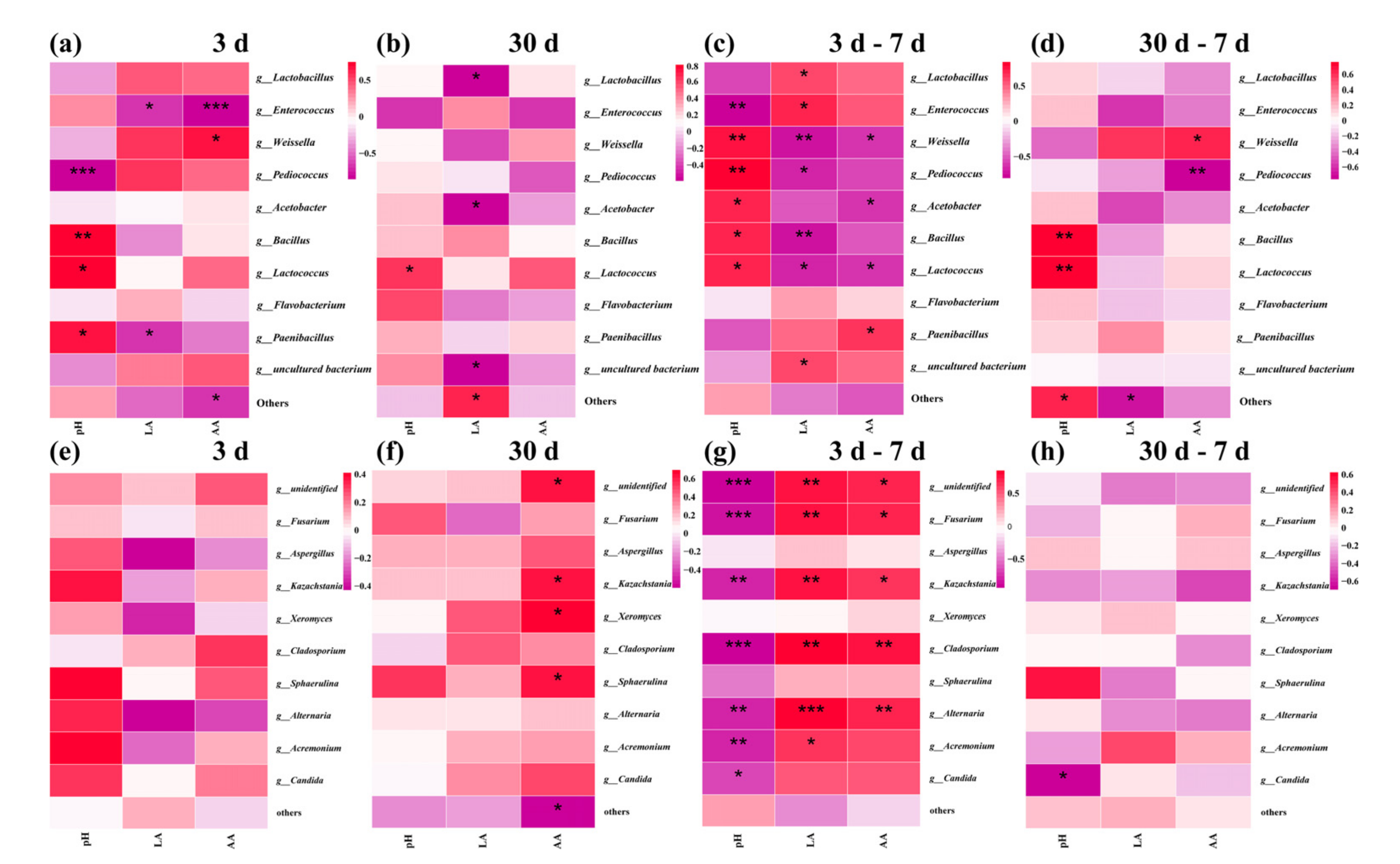
| Source | Sum of Squares | DF | Mean Square | F Value | p-Value |
|---|---|---|---|---|---|
| Model | 0.068 | 6 | 0.011 | 13.74 | 0.0003 |
| A | 0.023 | 1 | 0.023 | 28.39 | 0.0003 |
| B | 0.016 | 1 | 0.016 | 18.82 | 0.0015 |
| C | 0.027 | 1 | 0.027 | 33.04 | 0.0002 |
| AB | 1.53 × 10−3 | 1 | 1.53 × 10−3 | 1.85 | 0.2037 |
| AC | 1.44 × 10−3 | 1 | 1.44 × 10−3 | 1.75 | 0.2159 |
| BC | 3.04 × 10−3 | 1 | 3.04 × 10−3 | 3.68 | 0.0841 |
| Residual | 8.27 × 10−3 | 10 | 8.27 × 10−4 | ||
| Cor Total | 0.076 | 16 | |||
| Std. Dev. | 0.029 | R-Squared | 0.8918 | ||
| Mean | 0.65 | Adj R-Squared | 0.827 | ||
| C.V. % | 4.44 | Pred R-Squared | 0.6249 | ||
| PRESS | 0.029 | Adeq Precision | 11.631 |
| Treatment | Fermentation Time | Inoculum Mount | Material-Water Ratio | Acid Protease Activity |
|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 136.84 |
| 2 | 2 | 2 | 1 | 171.05 |
| 3 | 3 | 3 | 1 | 110.67 |
| 4 | 3 | 2 | 2 | 92.71 |
| 5 | 2 | 1 | 2 | 127.77 |
| 6 | 1 | 3 | 2 | 148.30 |
| 7 | 1 | 2 | 3 | 147.27 |
| 8 | 2 | 3 | 3 | 129.14 |
| 9 | 3 | 1 | 3 | 168.82 |
| K1 | 144.13 | 144.50 | 139.52 | |
| K2 | 142.65 | 137.01 | 122.93 | |
| K3 | 124.07 | 129.37 | 148.41 | |
| R | 20.07 | 15.11 | 25.49 |
| Days | Treatments | Bacteria | Fungi | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Shannon | Chao 1 | OTUS | Good’s Coverage | Shannon | Chao 1 | OTUs | Good’s Coverage | ||
| 3 d | CK | 5.83 | 3521.64 | 1785 | 0.99 | 4.03 | 99.67 | 115 | 0.98 |
| 95 | 4.76 | 3009.98 | 1301 | 0.99 | 4.01 | 81.72 | 83 | 0.99 | |
| P | 4.78 | 3278.14 | 1545 | 0.99 | 5.01 | 225.69 | 321 | 0.97 | |
| P + 95 | 4.32 | 2577.36 | 1244 | 0.99 | 3.45 | 91.07 | 103 | 0.99 | |
| 3–7 d | CK | 5.8 | 3516.52 | 1753 | 0.99 | 1.69 | 11.98 | 32 | 0.99 |
| 95 | 2.6 | 1923.12 | 741 | 0.99 | 3.45 | 198.05 | 316 | 0.98 | |
| P | 4.11 | 3038.66 | 1461 | 0.99 | 1.01 | 37.93 | 58 | 0.99 | |
| P + 95 | 2.37 | 1761.73 | 708 | 0.99 | 4.04 | 218.7 | 385 | 0.96 | |
| 30 d | CK | 5.41 | 3213.72 | 1456 | 0.99 | 3.97 | 63.65 | 83 | 0.99 |
| 95 | 4.74 | 2062.2 | 1017 | 0.99 | 3.23 | 56.41 | 71 | 0.99 | |
| P | 5.03 | 2610.79 | 1265 | 0.99 | 5.87 | 355.33 | 356 | 0.98 | |
| P + 95 | 3.9 | 1807.25 | 841 | 0.99 | 2.95 | 23.17 | 28 | 0.99 | |
| 30–7 d | CK | 5.2 | 2824.77 | 1332 | 0.99 | 4.82 | 293.78 | 361 | 0.98 |
| 95 | 4.75 | 2340.07 | 993 | 0.99 | 5.34 | 290.42 | 399 | 0.97 | |
| P | 4.89 | 3252.91 | 1571 | 0.99 | 4.26 | 79.27 | 97 | 0.99 | |
| P + 95 | 3.99 | 2469.96 | 1064 | 0.99 | 4.2 | 346.15 | 369 | 0.98 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, H.; Wang, W.; Wang, Z.; Tan, Z.; Qin, G.; Wang, Y.; Pang, H. Microbial Population Succession and Community Diversity and Its Correlation with Fermentation Quality in Soybean Meal Treated with Enterococcus faecalis during Fermentation and Aerobic Exposure. Microorganisms 2022, 10, 530. https://doi.org/10.3390/microorganisms10030530
Ma H, Wang W, Wang Z, Tan Z, Qin G, Wang Y, Pang H. Microbial Population Succession and Community Diversity and Its Correlation with Fermentation Quality in Soybean Meal Treated with Enterococcus faecalis during Fermentation and Aerobic Exposure. Microorganisms. 2022; 10(3):530. https://doi.org/10.3390/microorganisms10030530
Chicago/Turabian StyleMa, Hao, Weiwei Wang, Zhenyu Wang, Zhongfang Tan, Guangyong Qin, Yanping Wang, and Huili Pang. 2022. "Microbial Population Succession and Community Diversity and Its Correlation with Fermentation Quality in Soybean Meal Treated with Enterococcus faecalis during Fermentation and Aerobic Exposure" Microorganisms 10, no. 3: 530. https://doi.org/10.3390/microorganisms10030530
APA StyleMa, H., Wang, W., Wang, Z., Tan, Z., Qin, G., Wang, Y., & Pang, H. (2022). Microbial Population Succession and Community Diversity and Its Correlation with Fermentation Quality in Soybean Meal Treated with Enterococcus faecalis during Fermentation and Aerobic Exposure. Microorganisms, 10(3), 530. https://doi.org/10.3390/microorganisms10030530







