Legionella pneumophila in Municipal Shower Systems in Stavanger, Norway; A Longitudinal Surveillance Study Using Whole Genome Sequencing in Risk Management
Abstract
:1. Introduction
2. Materials and Methods
2.1. Surveillance Program
2.2. Sample Collection
2.3. Culture Analysis
2.4. Serotyping
2.5. Sequence-Based Typing
2.6. Whole Genome Sequencing
2.7. Assembly and Quality Control
2.8. Hybrid Assembly and Annotation of Reference Genomes
2.9. Phylogenetic Analysis
2.10. Data Availability
3. Results
3.1. Prevalence of L. pneumophila in Shower Systems in Different Municipal Building Categories
3.2. Culture Status and Characterisation of L. pneumophila Isolates
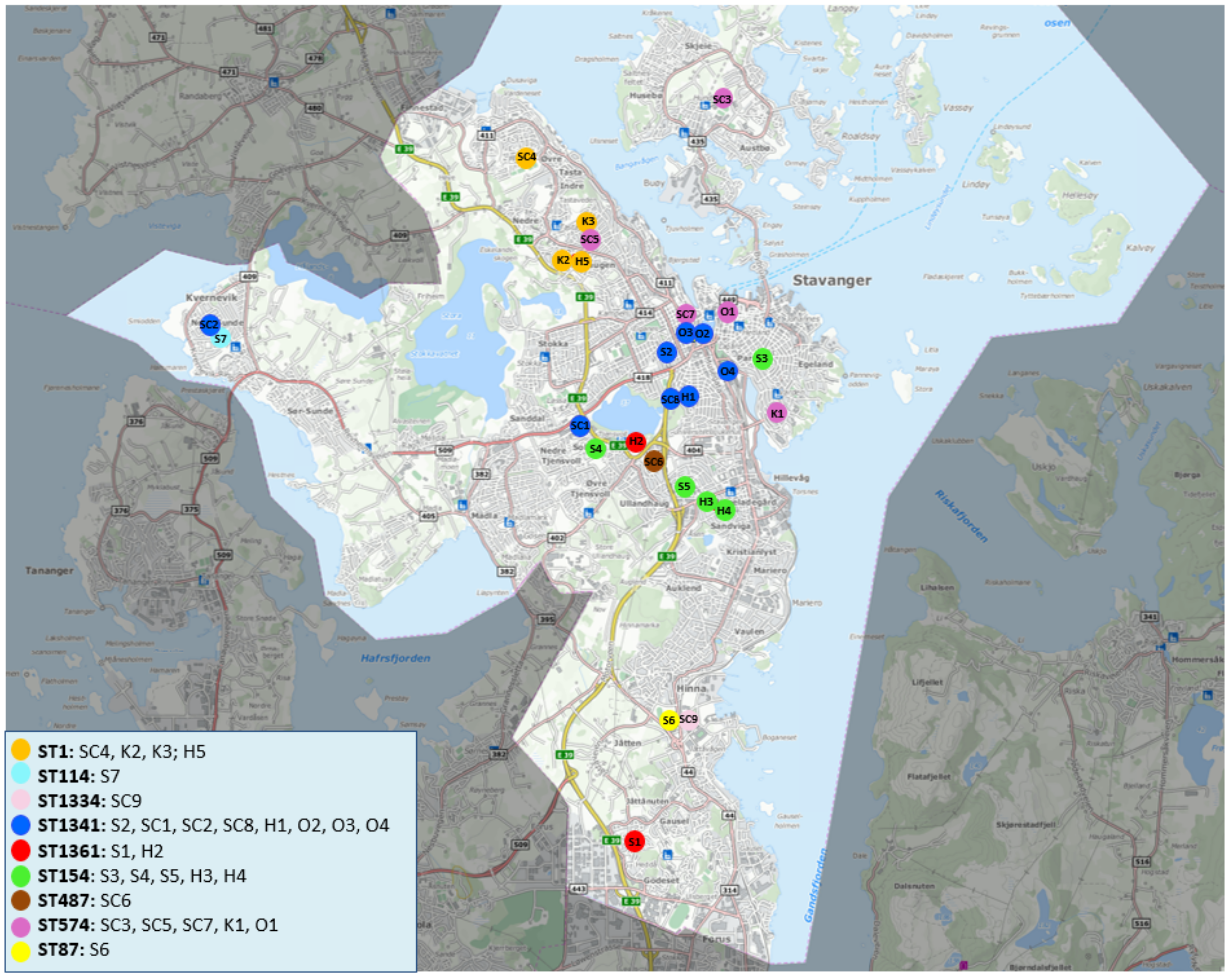
3.3. Geographical Distribution and Phylogenetic Analyses of L. pneumophila Isolated from Municipal Shower Systems
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bartram, J.; Chartier, Y.; Lees, J.V.; Bond, K. Legionella and the Prevention of Legionellosis. Emerg. Infect. Dis. 2008, 14, 1006. [Google Scholar]
- Whiley, H.; Keegan, A.; Fallowfield, H.; Ross, K. Uncertainties associated with assessing the public health risk from Legionella. Front. Microbiol. 2014, 5, 501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, D.; Shames, S.R. Pathogenicity and Virulence of Legionella: Intracellular replication and host response. Virulence 2021, 12, 1122–1144. [Google Scholar] [CrossRef]
- Herwaldt, L.A.; Marra, A.R. Legionella. Curr. Opin. Infect. Dis. 2018, 31, 325–333. [Google Scholar] [CrossRef]
- Gaia, V.; Fry, N.K.; Afshar, B.; Lück, P.C.; Meugnier, H.; Etienne, J.; Peduzzi, R.; Harrison, T.G. Consensus Sequence-Based Scheme for Epidemiological Typing of Clinical and Environmental Isolates of Legionella pneumophila. J. Clin. Microbiol. 2005, 43, 2047–2052. [Google Scholar] [CrossRef] [Green Version]
- Ratzow, S.; Gaia, V.; Helbig, J.H.; Fry, N.K.; Lück, P.C. Addition of neuA, the Gene Encoding N-Acylneuraminate Cytidylyl Transferase, Increases the Discriminatory Ability of the Consensus Sequence-Based Scheme for Typing Legionella pneumophila Serogroup 1 Strains. J. Clin. Microbiol. 2007, 45, 1965–1968. [Google Scholar] [CrossRef] [Green Version]
- Department of Health. Health Protection Agency 1988–2005. Sequence-Based Typing (SBT) Database for Legionella pneumophila. Available online: https://discovery.nationalarchives.gov.uk/details/r/C14933394 (accessed on 30 January 2022).
- Khodr, A.; Kay, E.; Gomez-Valero, L.; Ginevra, C.; Doublet, P.; Buchrieser, C.; Jarraud, S. Molecular epidemiology, phylogeny and evolution of Legionella. Infect. Genet. Evol. 2016, 43, 108–122. [Google Scholar] [CrossRef]
- Raphael, B.H.; Baker, D.J.; Nazarian, E.; Lapierre, P.; Bopp, D.; Kozak-Muiznieks, N.A.; Morrison, S.S.; Lucas, C.E.; Mercante, J.W.; Musser, K.A.; et al. Genomic Resolution of Outbreak-Associated Legionella pneumophila Serogroup 1 Isolates from New York State. Appl. Environ. Microbiol. 2016, 82, 3582–3590. [Google Scholar] [CrossRef] [Green Version]
- Blystad, H.; Bjorlow, E.; Aavitsland, P.; Holm, J. Outbreak of legionellosis in Stavanger, Norway—Final report. Wkly. Releases 1997–2007 2001, 5, 2059. [Google Scholar] [CrossRef]
- Blystad, H.; Brantsaeter, A.B.; Løvoll, Ø. Outbreak of community-acquired legionnaires’ disease in southeast Norway, May 2005. Wkly. Releases 1997–2007 2005, 10, 2709. [Google Scholar] [CrossRef]
- Folkehelseinstituttet. Vannrapport 123-Forebygging av Legionellasmitte–en Veiledning (4. Utgave 2015)-FHI. Available online: https://www.fhi.no/publ/2015/forebygging-av-legionellasmitte/ (accessed on 22 January 2022). (In Norwegian)
- Lovdata. Forskrift for Miljørettet Helsevern. Kap 3a. Krav om å Hindre Spredning av Legionella via Aerosol. Forskrift om Miljørettet Helsevern-Lovdata. 2008. Available online: https://lovdata.no/dokument/SF/forskrift/2003-04-25-486/kap3a#kap3a (accessed on 22 January 2022). (In Norwegian).
- Wiik, R.; Krøvel, A.V. Necessity and Effect of Combating Legionella pneumophila in Municipal Shower Systems. PLoS ONE 2014, 9, e114331. [Google Scholar] [CrossRef] [Green Version]
- Helbig, J.; Bernander, S.; Pastoris, M.C.; Etienne, J.; Gaia, V.; Lauwers, S.; Lindsay, D.; Lück, P.C.; Marques, T.; Mentula, S.; et al. Pan-European study on culture-proven Legionnaires’ disease: Distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. Infect. Dis. 2002, 21, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, K.A.; Hamilton, M.T.; Johnson, W.; Jjemba, P.; Bukhari, Z.; LeChevallier, M.; Haas, C.N.; Gurian, P.L. Risk-Based Critical Concentrations of Legionella pneumophila for Indoor Residential Water Uses. Environ. Sci. Technol. 2019, 53, 4528–4541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lampl, B.M.J.; Lang, M.; Wodnick, S. Can mandatory monitoring in rental apartments effectively prevent legionellosis? A retrospective analysis of data from Regensburg with a review of the literature. GMS Hyg. Infect. Control 2020, 15, Doc14. [Google Scholar] [PubMed]
- ECDC. European Technical Guidelines for the Prevention, Control and Investigation of Infections Caused by Legionella Species Data. Available online: https://www.ecdc.europa.eu/en/publications-data/european-technical-guidelines-prevention-control-and-investigation-infections (accessed on 22 January 2022).
- Stout, J.E.; Yu, V.L. Environmental culturing for Legionella: Can we build a better mouse trap? Am. J. Infect. Control 2010, 38, 341–343. [Google Scholar] [CrossRef]
- Allegheny County Health Department. Updated Guidelines for the Control of Legionella in Western Pennsylvania. 2014. Available online: https://www.alleghenycounty.us/uploadedFiles/Allegheny_Home/Health_Department/Resources/Data_and_Reporting/Infectious_Disease_Epidemiology/2014_FINAL_Legionella_Guidelines_for_Western_PA.pdf (accessed on 22 January 2022).
- Helbig, J.H.; Kurtz, J.B.; Pastoris, M.C.; Pelaz, C.; Lück, P.C. Antigenic lipopolysaccharide components of Legionella pneumophila recognized by monoclonal antibodies: Possibilities and limitations for division of the species into serogroups. J. Clin. Microbiol. 1997, 35, 2841–2845. [Google Scholar] [CrossRef] [Green Version]
- Krueger, F. TrimGalore. Available online: https://github.com/FelixKrueger/TrimGalore (accessed on 22 January 2022).
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Andrews, S. Github/s-andrews/FastQC. Available online: https://github.com/s-andrews/FastQC (accessed on 22 January 2022).
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Legsta. Available online: https://github.com/tseemann/legsta (accessed on 22 January 2022).
- Guppy. Basecalling with Guppy—de. NBI Nanopore Training Course Latest Documentation. Available online: https://nanoporetech.com/community (accessed on 22 January 2022).
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyres, K.L.; Wick, R.R.; Judd, L.M.; Froumine, R.; Tokolyi, A.; Gorrie, C.L.; Lam, M.M.C.; Duchêne, S.; Jenney, A.; Holt, K.E. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019, 15, e1008114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Lee, H.K.; Shim, J.I.; Kim, H.E.; Yu, J.Y.; Kang, Y.H. Distribution of Legionella Species from Environmental Water Sources of Public Facilities and Genetic Diversity of L. pneumophila Serogroup 1 in South Korea. Appl. Environ. Microbiol. 2010, 76, 6547–6554. [Google Scholar] [CrossRef] [Green Version]
- Lück, P.C. Diagnostics and Clinical Disease Treatment. In Legionella Molecular Microbiology; Hauner, K., Swansom, M., Eds.; Academic Press: Cambridge, MA, USA, 2008; Volume 15, p. 139. [Google Scholar]
- Stavanger Kommune. Velferd og Helse, Samfunnsmedisin. Legionella—Revidering av Rutiner i Kommunale Bygg. Saksdokument 18/10696-1, 12.4.18. 2018. Available online: https://www.stavanger.kommune.no/helse-og-omsorg/helsesjefen-i-stavanger/rutiner-ved-funn-av-legionella/ (accessed on 30 January 2022). (In Norwegian).
- Sciuto, E.; Laganà, P.; Filice, S.; Scalese, S.; Libertino, S.; Corso, D.; Faro, G.; Coniglio, M. Environmental Management of Legionella in Domestic Water Systems: Consolidated and Innovative Approaches for Disinfection Methods and Risk Assessment. Microorganisms 2021, 9, 577. [Google Scholar] [CrossRef]
- Whiley, H.; Bentham, R.; Brown, M. Legionella Persistence in Manufactured Water Systems: Pasteurization Potentially Selecting for Thermal Tolerance. Front. Microbiol. 2017, 8, 1330. [Google Scholar] [CrossRef]
- Girolamini, L.; Salaris, S.; Lizzadro, J.; Mazzotta, M.; Pascale, M.R.; Pellati, T.; Cristino, S. How Molecular Typing Can Support Legionella Environmental Surveillance in Hot Water Distribution Systems: A Hospital Experience. Int. J. Environ. Res. Public Health 2020, 17, 8662. [Google Scholar] [CrossRef] [PubMed]
- ECDC. Surevillance Atlas of Infectious Diseases. Available online: https://atlas.ecdc.europa.eu/public/index.aspx?Dataset=27&HealthTopic=30 (accessed on 22 January 2022).
- Technical Implementation of the Water Directive for Environmental Testing of Legionella. ELDSNet Annual Meeting. Presentation by Maria Luisa Ricci. 2021. Available online: https://www.ecdc.europa.eu/en/about-us/partnerships-and-networks/disease-and-laboratory-networks/eldsnet (accessed on 21 January 2022).
- Directive (EU) 2020/2184 of the European Parliament and of the Council of 16 December 2020 on the Quality of Water Intended for Human Consumption. Available online: https://eur-lex.europa.eu/eli/dir/2020/2184/oj (accessed on 21 January 2022).
- Harrison, T.G.; Afshar, B.; Doshi, N.; Fry, N.; Lee, J.V. Distribution of Legionella pneumophila serogroups, monoclonal antibody subgroups and DNA sequence types in recent clinical and environmental isolates from England and Wales (2000–2008). Eur. J. Clin. Microbiol. 2009, 28, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Lück, C.; Fry, N.K.; Helbig, J.H.; Jarraud, S.; Harrison, T.G. Typing Methods for Legionella. In Legionella; Humana Press: Totowa, NJ, USA, 2013; pp. 119–148. [Google Scholar] [CrossRef]
- Neoh, H.-M.; Tan, X.-E.; Sapri, H.F.; Tan, T.L. Pulsed-field gel electrophoresis (PFGE): A review of the “gold standard” for bacteria typing and current alternatives. Infect. Genet. Evol. 2019, 74, 103935. [Google Scholar] [CrossRef] [PubMed]
- Wells, M.; Lasek-Nesselquist, E.; Schoonmaker-Bopp, D.; Baker, D.; Thompson, L.; Wroblewski, D.; Nazarian, E.; Lapierre, P.; Musser, K.A. Insights into the long-term persistence of Legionella in facilities from whole-genome sequencing. Infect. Genet. Evol. 2018, 65, 200–209. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Mentasti, M.; Tewolde, R.; Aslett, M.; Harris, S.R.; Afshar, B.; Underwood, A.; Fry, N.K.; Parkhill, J.; Harrison, T.G. Evaluation of an Optimal Epidemiological Typing Scheme for Legionella pneumophila with Whole-Genome Sequence Data Using Validation Guidelines. J. Clin. Microbiol. 2016, 54, 2135–2148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivče, D.G.; Rončević, D.; Šantić, M.; Cenov, A.; Linšak, D.T.; Mićović, V.; Lušić, D.; Glad, M.; Ljubas, D.; Lušić, D.V. Is a Proactive Approach to Controlling Legionella in the Environment Justified? Food Technol. Biotechnol. 2021, 59, 314–324. [Google Scholar] [CrossRef]


| Risk Category | Category Criteria | Sample Frequency as Listed in Municipal Guidelines (2012) | Sample Frequency in Project Period (2012–2021) |
|---|---|---|---|
| 1 | Known user groups with compromised immunity and/or detected L. pneumophila sg1 in the system | 2 times a year; if consistent results, reduce to yearly. | Preventive measures and extensive sampling (minimum once a week for 4 weeks) to document the effect of the measures. |
| 2 | Open access to the public and/or detected L. pneumophila sg2–14 or Legionella spp. in the system | Yearly; if consistent results, reduce to every 2 years. | Yearly for the whole test period. Preventive measures and extensive sampling (minimum once a week for 4 weeks) to document the effect of the measures. |
| 3 | Restricted access and no Legionella spp. detected in the system | Yearly; if no Legionella detected, reduce to every 5 years. | Every 4 years for kindergartens. Every 3 years for smaller sports complexes with restricted access. Others as described in guidelines. |
| Initial Status | Current Status | ||||||
|---|---|---|---|---|---|---|---|
| Code | Sg | ST | Positive/Total | CFU/mL | Treatment Measures | Positive/Total | CFU/mL |
| H5 | 1 | 1/574 1 | 7/20 | ++ | DL, T | 1/22 | + |
| K2 | 1 | 1 | 2/4 | + | T | Not in use | |
| K3 | 1 | 1 | 3/5 | +++ | PC, T | Not in use | |
| SC4 | 1 | 1 | 5/5 | +++ 2 | DL, PC, T, F, CD, DF | 0/8 | + 3 |
| S8 * | 1 | 59 | 6/6 | +++ | DL, T, CC, F | 5/6 | + |
| H3 | 1 | 154 | 2/12 | + | DL, PC, T | 0/12 | ND |
| H4 | 1 | 154 | 6/17 | ++ | DL, PC, T | 1/17 | + |
| S3 | 1 | 154 | 6/6 | ++ | DL, PC, T, CD | 0/9 | ND |
| S4 | 1 | 154 | 5/6 | +++ | DL, PC, T, CD, F | 2/10 | + |
| S5 | 1 | 154 | 4/6 | +++ | DL, T | 4/8 | + |
| SC6 | 1 | 487 | 5/6 | ++ | DL, PC, CD | 0/6 | ND |
| K1 | 1 | 574 | 2/2 | +++ | DL, T | Not in use | |
| O1 | 1 | 574 | 1/1 | + | PC, T | Not in use | |
| SC3 | 1 | 574 | 5/11 | + | DL, PC, CD | 0/12 | ND |
| SC5 | 1 | 574 | 7/7 | ++ 2 | DL, PC, T, CD, F, CC | 8/22 | + |
| SC7 | 1 | 574 | 1/4 | ++ | PC, T | 0/15 | ND |
| S6 | 3 | 87 | 8/8 | ++ | T, F | 7/8 | ++ |
| S7 | 6 | 114 | 1/1 | + | PC, T | 2/3 | + |
| SC9 | 4 | 1334 | 4/6 | ++ | DL, T | Not in use | |
| SC2 | 3 | 1341 | 1/6 | + | DL, T | 2/6 | + 4 |
| H1 | 6 | 1341 | 6/6 | ++ 2 | DL, T | Not in use | |
| O2 | 6 | 1341 | 3/3 | ++ | None | Not in use | |
| O3 | 6 | 1341 | 1/1 | + | None | Not in use | |
| O4 | 6 | 1341 | 4/6 | ++ | T | Not in use | |
| O5 ** | 2–14 *** | 1341 | 2/2 | + | None | 2/2 | ++ |
| S2 | 6 | 1341 | 5/5 | +++ | PC, T, F | 5/5 | ++ |
| SC1 | 6 | 1341 | 6/6 | ++++ | DL, PC, T | 8/10 | ++ |
| SC8 | 6 | 1341 | 5/5 | +++ | PC, T, CD | Not in use | |
| H2 | 4 | 1361 | 3/3 | ++ 2 | DL, T | Not in use | |
| S1 | 10 | 1361 | 4/4 | +++ | DL, PC, T, CD, F | 4/4 | ++ |
| SBT | flaA | pilE | asd | mip | mompS | proA | neuA |
|---|---|---|---|---|---|---|---|
| 154 | 11 | 14 | 16 | 16 | 15 | 13 | 2 |
| 574 | 11 | 14 | 16 | 16 | 15 | 13 | 11 |
| 1334 | 11 | 14 | 16 | 25 | 7 | 13 | 206 |
| 1 | 1 | 4 | 3 | 1 | 1 | 1 | 1 |
| 59 | 7 | 6 | 17 | 3 | 13 | 11 | 11 |
| 87 | 2 | 10 | 3 | 28 | 9 | 4 | 13 |
| 114 | 3 | 6 | 1 | 6 | 14 | 11 | 9 |
| 487 | 3 | 6 | 1 | 28 | 14 | 11 | 11 |
| 1341 | 3 | 13 | 1 | 3 | 14 | 9 | 207 |
| 1361 | 3 | 13 | 1 | 1 | 14 | 9 | 207 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krøvel, A.V.; Bernhoff, E.; Austerheim, E.; Soma, M.A.; Romstad, M.R.; Löhr, I.H. Legionella pneumophila in Municipal Shower Systems in Stavanger, Norway; A Longitudinal Surveillance Study Using Whole Genome Sequencing in Risk Management. Microorganisms 2022, 10, 536. https://doi.org/10.3390/microorganisms10030536
Krøvel AV, Bernhoff E, Austerheim E, Soma MA, Romstad MR, Löhr IH. Legionella pneumophila in Municipal Shower Systems in Stavanger, Norway; A Longitudinal Surveillance Study Using Whole Genome Sequencing in Risk Management. Microorganisms. 2022; 10(3):536. https://doi.org/10.3390/microorganisms10030536
Chicago/Turabian StyleKrøvel, Anne Vatland, Eva Bernhoff, Elin Austerheim, Markus André Soma, Monica Regine Romstad, and Iren Høyland Löhr. 2022. "Legionella pneumophila in Municipal Shower Systems in Stavanger, Norway; A Longitudinal Surveillance Study Using Whole Genome Sequencing in Risk Management" Microorganisms 10, no. 3: 536. https://doi.org/10.3390/microorganisms10030536
APA StyleKrøvel, A. V., Bernhoff, E., Austerheim, E., Soma, M. A., Romstad, M. R., & Löhr, I. H. (2022). Legionella pneumophila in Municipal Shower Systems in Stavanger, Norway; A Longitudinal Surveillance Study Using Whole Genome Sequencing in Risk Management. Microorganisms, 10(3), 536. https://doi.org/10.3390/microorganisms10030536






