UME6 Is Involved in the Suppression of Basal Transcription of ABC Transporters and Drug Resistance in the ρ+ Cells of Saccharomyces cerevisiae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Media
2.2. Spot Dilution Assay
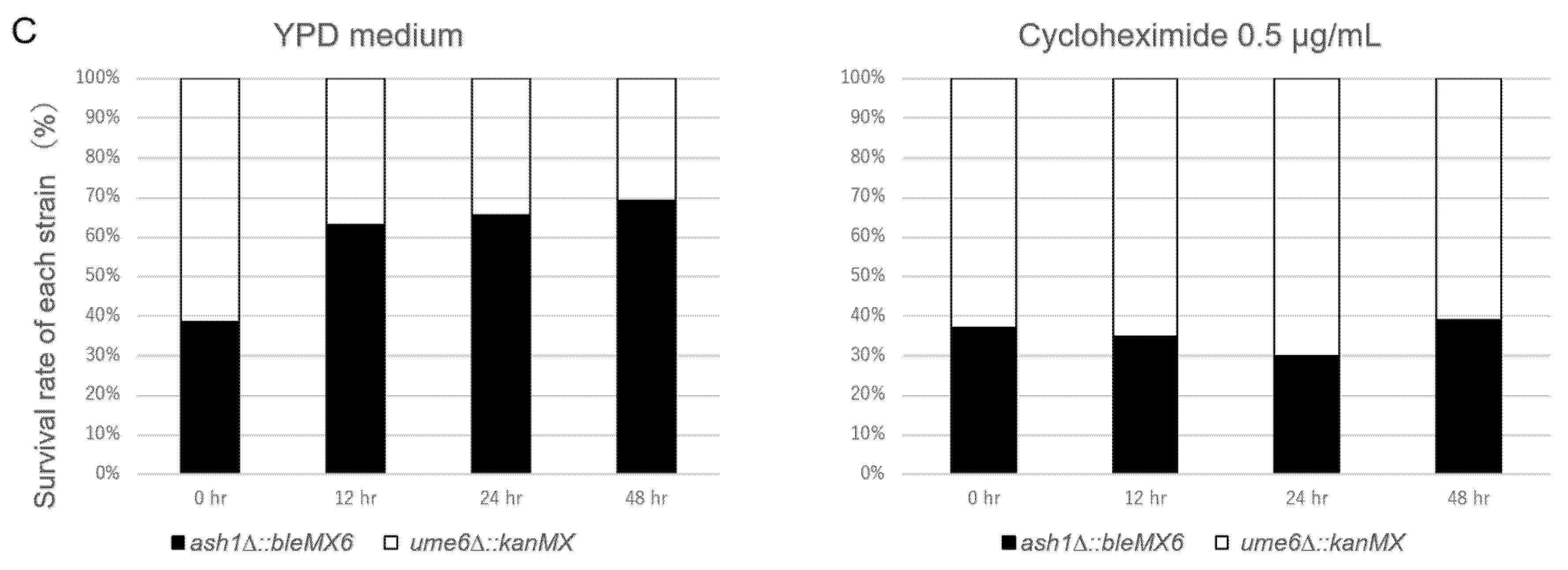
2.3. Cocultivation of Two Gene Deletion Mutants Replaced with KanMX or bleMX6 Gene Cassettes
2.4. RNA Extraction from the ρ+ Cells in Each Strain Grown to the Logarithmic Phase
2.5. RNA Extraction from ρ+ Cells before and after Cycloheximide Exposure
2.6. Real-Time RT–PCR
2.7. Statistical Analysis
3. Results
3.1. UME6 Acts as a Negative Regulator to Fluconazole- and Cycloheximide-Resistance in ρ+ Cells, While RPD3 Acts as a Positive Regulator
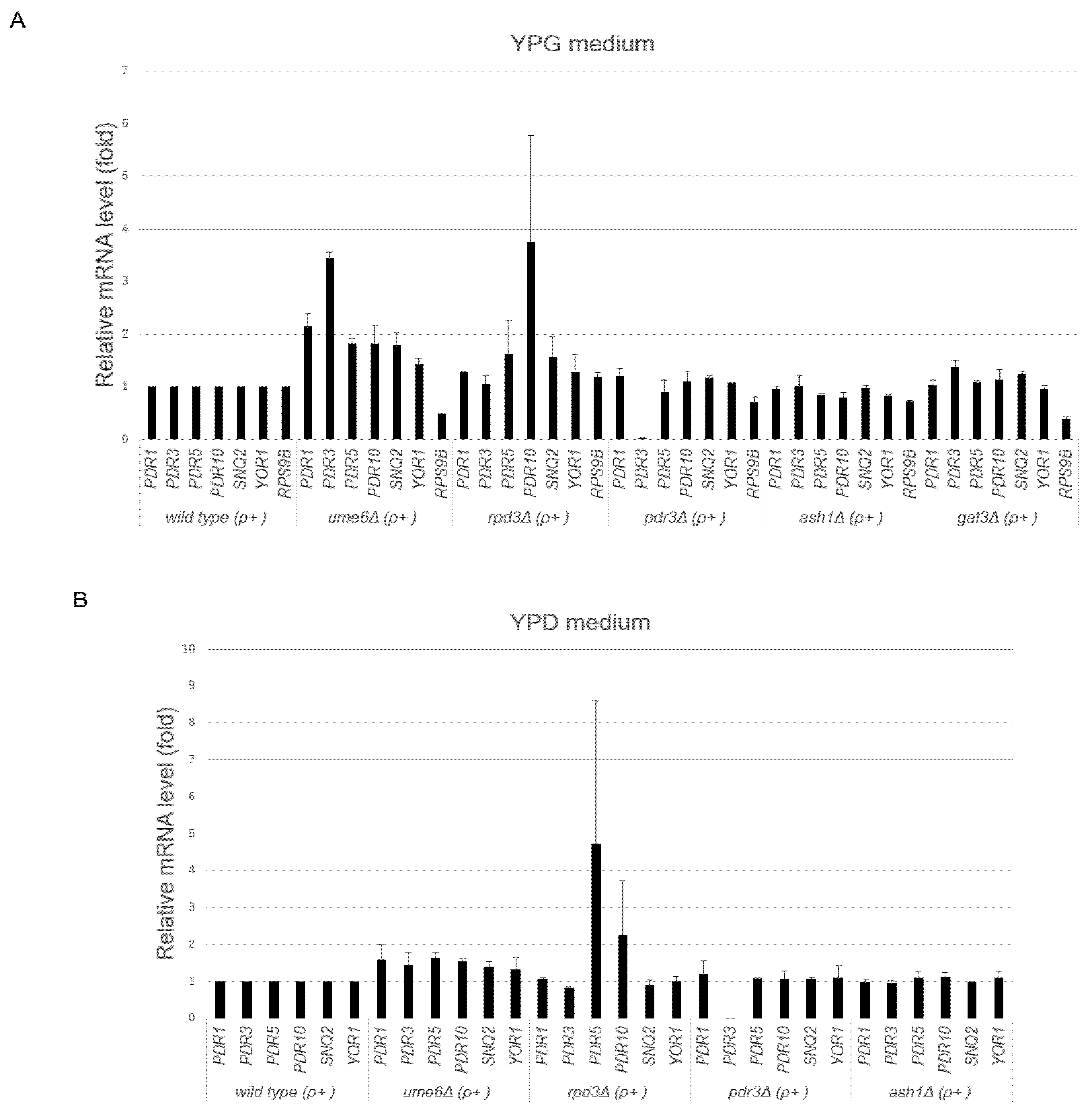
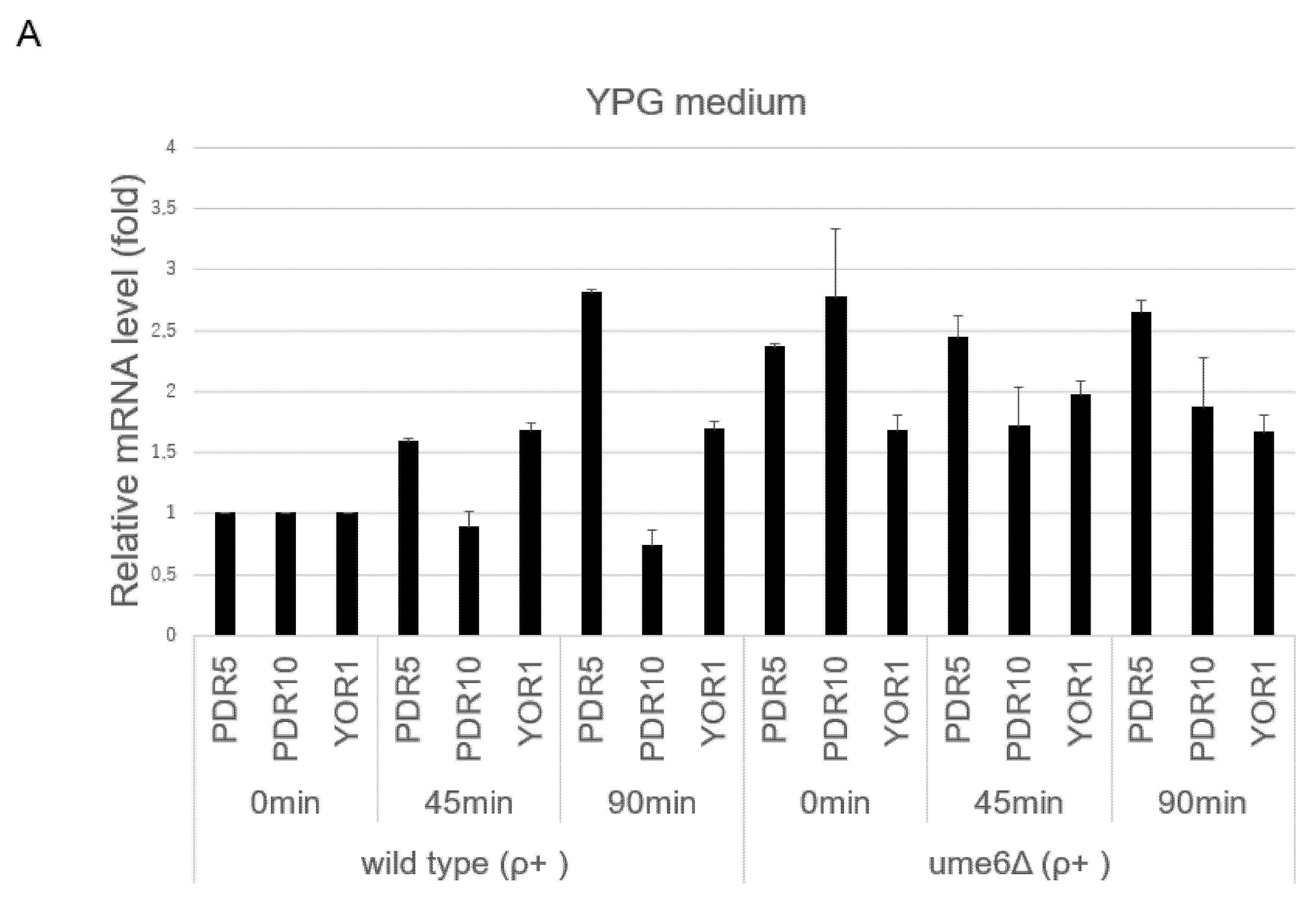
3.2. UME6 Is Required for the Suppression of Basal ABC Transporter mRNA Levels
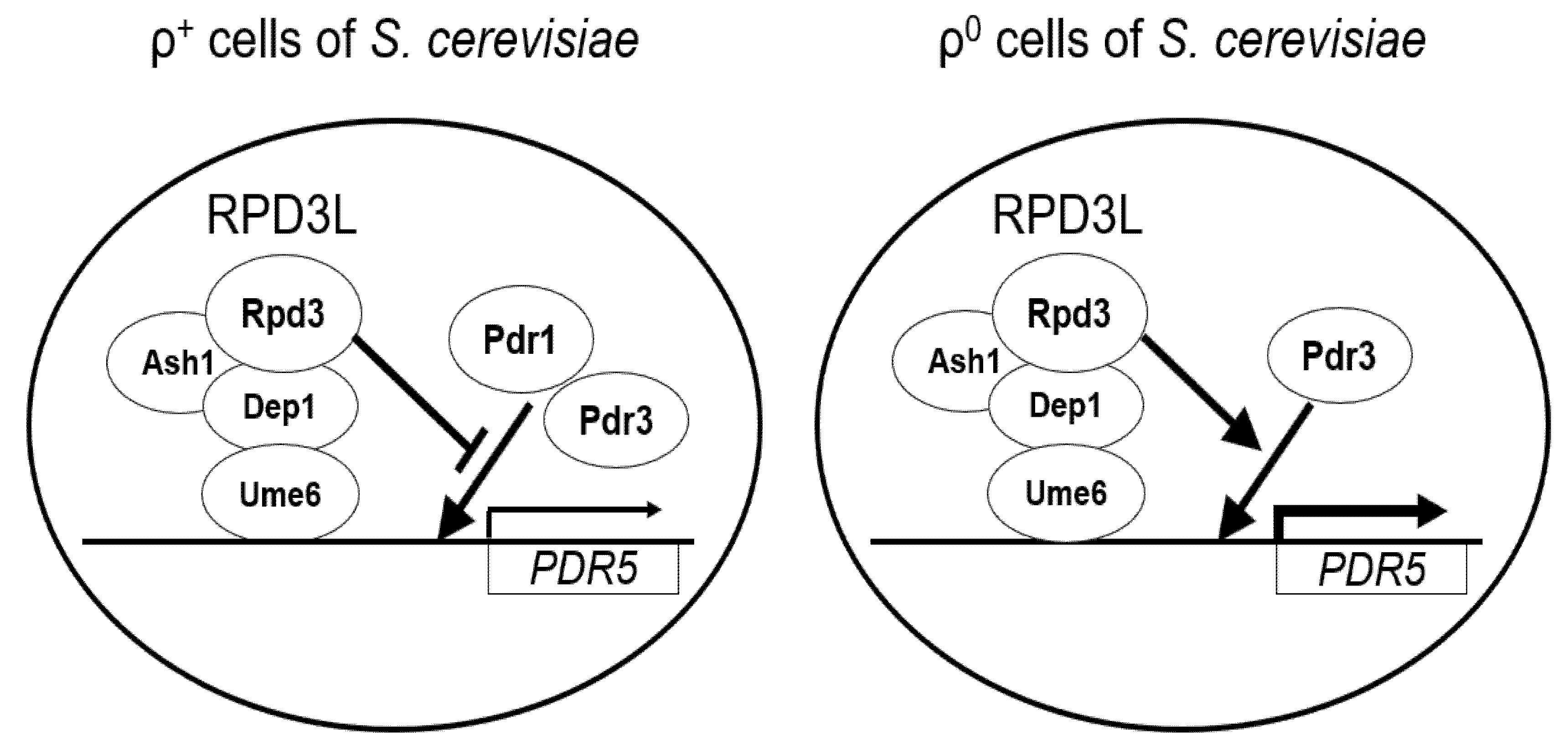
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Decottignies, A.; Grant, A.M.; Nichols, J.W.; de Wet, H.; McIntosh, D.B.; Goffeau, A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 1998, 273, 12612–12622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, A.; Wagner, M.; Du, D.; Raschka, S.; Nentwig, L.M.; Gohlke, H.; Smits, S.H.J.; Luisi, B.F.; Schmitt, L. Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5. Nat. Commun. 2021, 12, 5254. [Google Scholar] [CrossRef] [PubMed]
- Katzmann, D.J.; Hallstrom, T.C.; Voet, M.; Wysock, W.; Golin, J.; Volckaert, G.; Moye-Rowley, W.S. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995, 15, 6875–6883. [Google Scholar] [CrossRef] [Green Version]
- Kumari, S.; Kumar, M.; Gaur, N.A.; Prasad, R. Multiple roles of ABC transporters in yeast. Fungal Genet. Biol. 2021, 150, 103550. [Google Scholar] [CrossRef] [PubMed]
- Godinho, C.P.; Dias, P.J.; Ponçot, E.; Sá-Correia, I. The Paralogous Genes PDR18 and SNQ2, Encoding Multidrug Resistance ABC Transporters, Derive From a Recent Duplication Event, PDR18 Being Specific to the Saccharomyces Genus. Front. Genet. 2018, 9, 476. [Google Scholar] [CrossRef]
- Carvajal, E.; Van Den Hazel, H.B.; Cybularz-Kolaczkowska, A.; Balzi, E.; Goffeau, A. Molecular and phenotypic: Characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol. Gen. Genet. 1997, 256, 406–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buechel, E.R.; Pinkett, H.W. Transcription factors and ABC transporters: From pleiotropic drug resistance to cellular signaling in yeast. FEBS Lett. 2020, 594, 3943–3964. [Google Scholar] [CrossRef]
- Mamnun, Y.M.; Schüller, C.; Kuchler, K. Expression regulation of the yeast PDR5 ATP-binding cassette (ABC) transporter suggests a role in cellular detoxification during the exponential growth phase. FEBS Lett. 2004, 559, 111–117. [Google Scholar] [CrossRef] [Green Version]
- Wolfger, H.; Mahé, Y.; Parle-McDermott, A.; Delahodde, A.; Kuchler, K. The yeast ATP binding cassette (ABC) protein genes PDR10 and PDR15 are novel targets for the Pdr1 and Pdr3 transcriptional regulators. FEBS Lett. 1997, 418, 269–274. [Google Scholar] [CrossRef] [Green Version]
- Katzmann, D.J.; Burnett, P.E.; Golin, J.; Mahé, Y.; Moye-Rowley, W.S. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol. Cell. Biol. 1994, 14, 4653–4661. [Google Scholar]
- Katzmann, D.J.; Hallstrom, T.C.; Mahé, Y.; Moye-Rowley, W.S. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J. Biol. Chem. 1996, 271, 23049–23054. [Google Scholar] [CrossRef] [Green Version]
- Mahé, Y.; Parle-McDermott, A.; Nourani, A.; Delahodde, A.; Lamprecht, A.; Kuchler, K. The ATP-binding cassette multidrug transporter Snq2 of Saccharomyces cerevisiae: A novel target for the transcription factors Pdr1 and Pdr3. Mol. Microbiol. 1996, 20, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Delahodde, A.; Delaveau, T.; Jacq, C. Positive autoregulation of the yeast transcription factor Pdr3p, which is involved in control of drug resistance. Mol. Cell. Biol. 1995, 15, 4043–4051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeRisi, J.; van den Hazel, B.; Marc, P.; Balzi, E.; Brown, P.; Jacq, C.; Goffeau, A. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 2000, 470, 156–160. [Google Scholar] [CrossRef] [Green Version]
- Devaux, F.; Carvajal, E.; Moye-Rowley, S.; Jacq, C. Genome-wide studies on the nuclear PDR3-controlled response to mitochondrial dysfunction in yeast. FEBS Lett. 2002, 515, 25–28. [Google Scholar] [CrossRef] [Green Version]
- Nourani, A.; Papajova, D.; Delahodde, A.; Jacq, C.; Subik, J. Clustered amino acid substitutions in the yeast transcription regulator Pdr3p increase pleiotropic drug resistance and identify a new central regulatory domain. Mol. Gen. Genet. 1997, 256, 397–405. [Google Scholar] [CrossRef]
- Thakur, J.K.; Arthanari, H.; Yang, F.; Pan, S.J.; Fan, X.; Breger, J.; Frueh, D.P.; Gulshan, K.; Li, D.K.; Mylonakis, E.; et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 2008, 452, 604–609. [Google Scholar] [CrossRef]
- Fardeau, V.; Lelandais, G.; Oldfield, A.; Salin, H.N.; Lemoine, S.; Garcia, M.; Tanty, V.; Le Crom, S.; Jacq, C.; Devaux, F. The central role of PDR1 in the foundation of yeast drug resistance. J. Biol. Chem. 2007, 282, 5063–5074. [Google Scholar] [CrossRef] [Green Version]
- Miranda, M.N.; Masuda, C.A.; Ferreira-Pereira, A.; Carvajal, E.; Ghislain, M.; Montero-Lomelí, M. The serine/threonine protein phosphatase Sit4p activates multidrug resistance in Saccharomyces cerevisiae. FEMS Yeast Res. 2010, 10, 674–686. [Google Scholar] [CrossRef] [Green Version]
- Swagatika, S.; Tomar, R.S. ABC transporter Pdr5 is required for cantharidin resistance in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2021, 553, 141–147. [Google Scholar] [CrossRef]
- Cui, Z.; Shiraki, T.; Hirata, D.; Miyakawa, T. Yeast gene YRR1, which is required for resistance to 4-nitroquinoline N-oxide, mediates transcriptional activation of the multidrug resistance transporter gene SNQ2. Mol. Microbiol. 1998, 29, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- Schüller, C.; Mamnun, Y.M.; Wolfger, H.; Rockwell, N.; Thorner, J.; Kuchler, K. Membrane-active compounds activate the transcription factors Pdr1 and Pdr3 connecting pleiotropic drug resistance and membrane lipid homeostasis in saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 4932–4944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, M.C.; Sá-Correia, I. Saccharomyces cerevisiae resistance to chlorinated phenoxyacetic acid herbicides involves Pdr1p-mediated transcriptional activation of TPO1 and PDR5 genes. Biochem. Biophys. Res. Commun. 2002, 292, 530–537. [Google Scholar] [CrossRef]
- Hallstrom, T.C.; Moye-Rowley, W.S. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 37347–37356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrozza, M.J.; Florens, L.; Swanson, S.K.; Shia, W.J.; Anderson, S.; Yates, J.; Washburn, M.P.; Workman, J.L. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 2005, 1731, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Rundlett, S.E.; Carmen, A.A.; Suka, N.; Turner, B.M.; Grunstein, M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 1998, 392, 831–835. [Google Scholar] [CrossRef]
- Goldmark, J.P.; Fazzio, T.G.; Estep, P.W.; Church, G.M.; Tsukiyama, T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 2000, 103, 423–433. [Google Scholar] [CrossRef] [Green Version]
- David, K.; Kevin, S. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 1997, 89, 365–371. [Google Scholar]
- Fazzio, T.G.; Kooperberg, C.; Goldmark, J.P.; Neal, C.; Basom, R.; Delrow, J.; Tsukiyama, T. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 2001, 21, 6450–6460. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Martínez, M.E.; Benet, M.; Alepuz, P.; Tordera, V. Nut1/Hos1 and Sas2/Rpd3 control the H3 acetylation of two different sets of osmotic stress-induced genes. Epigenetics 2020, 15, 251–271. [Google Scholar] [CrossRef]
- Sharma, V.M.; Tomar, R.S.; Dempsey, A.E.; Reese, J.C. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol. Cell. Biol. 2007, 27, 3199–3210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, Y. RPD3 and UME6 are involved in the activation of PDR5 transcription and pleiotropic drug resistance in ρ0 cells of Saccharomyces cerevisiae. BMC Microbiol. 2021, 21, 311. [Google Scholar] [CrossRef] [PubMed]
- Borecka-Melkusova, S.; Kozovska, Z.; Hikkel, I.; Dzugasova, V.; Subik, J. RPD3 and ROM2 are required for multidrug resistance in Saccharomyces cerevisiae. FEMS Yeast Res. 2008, 8, 414–424. [Google Scholar] [CrossRef] [Green Version]
- Robbins, N.; Leach, M.D.; Cowen, L.E. Lysine Deacetylases Hda1 and Rpd3 Regulate Hsp90 Function thereby Governing Fungal Drug Resistance. Cell Rep. 2012, 2, 878–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yibmantasiri, P.; Bircham, P.W.; Maass, D.R.; Bellows, D.S.; Atkinson, P.H. Networks of genes modulating the pleiotropic drug response in Saccharomyces cerevisiae. Mol. Biosyst. 2014, 10, 128–137. [Google Scholar] [CrossRef]
- Knorre, D.A.; Azbarova, A.V.; Galkina, K.V.; Feniouk, B.A.; Severin, F.F. Replicative aging as a source of cell heterogeneity in budding yeast. Mech. Ageing Dev. 2018, 176, 24–31. [Google Scholar] [CrossRef]
- Kochmak, S.A.; Knorre, D.A.; Sokolov, S.S.; Severin, F.F. Physiological scenarios of programmed loss of mitochondrial DNA function and death of yeast. Biochemistry 2011, 76, 167–171. [Google Scholar] [CrossRef]
- Berman, J.; Krysan, D.J. Drug resistance and tolerance in fungi. Nat. Rev. Microbiol. 2020, 18, 319–331. [Google Scholar] [CrossRef]
- Hentges, P.; Van Driessche, B.; Tafforeau, L.; Vandenhaute, J.; Carr, A.M. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 2005, 22, 1013–1019. [Google Scholar] [CrossRef]
- Garcia, J.M.; Schwabe, M.J.; Voelker, D.R.; Riekhof, W.R. A functional genomic screen in Saccharomyces cerevisiae reveals divergent mechanisms of resistance to different alkylphosphocholine chemotherapeutic agents. G3 2021, 11, jkab233. [Google Scholar] [CrossRef]
- Huang, Z.; Dai, H.; Zhang, X.; Wang, Q.; Sun, J.; Deng, Y.; Shi, P. BSC2 induces multidrug resistance via contributing to the formation of biofilm in Saccharomyces cerevisiae. Cell. Microbiol. 2021, 23, e13391. [Google Scholar] [CrossRef] [PubMed]
- Onda, M.; Ota, K.; Chiba, T.; Sakaki, Y.; Ito, T. Analysis of gene network regulating yeast multidrug resistance by artificial activation of transcription factors: Involvement of Pdr3 in salt tolerance. Gene 2004, 332, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Elsztein, C.; de Lucena, R.M.; de Morais, M.A. The resistance of the yeast Saccharomyces cerevisiae to the biocide polyhexamethylene biguanide: Involvement of cell wall integrity pathway and emerging role for YAP1. BMC Mol. Biol. 2011, 12, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Yue, Z.; Xu, F.; Wang, S.; Hu, X.; Dai, J.; Zhao, G. Coevolution of ribosomal RNA expansion segment 7L and assembly factor Noc2p specializes the ribosome biogenesis pathway between Saccharomyces cerevisiae and Candida albicans. Nucleic Acids Res. 2021, 49, 4655–4667. [Google Scholar] [CrossRef]
- Jensen, A.N.; Chindaudomsate, W.; Thitiananpakorn, K.; Mongkolsuk, S.; Jensen, L.T. Improper protein trafficking contributes to artemisinin sensitivity in cells lacking the KDAC Rpd3p. FEBS Lett. 2014, 588, 4018–4025. [Google Scholar] [CrossRef] [Green Version]
- Hallstrom, T.C.; Katzmann, D.J.; Torres, R.J.; Sharp, W.J.; Moye-Rowley, W.S. Regulation of transcription factor Pdr1p function by an Hsp70 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998, 18, 1147–1155. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, B.E.; Tong, J.K.; Schreiber, S.L. Genome wide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 13708–13713. [Google Scholar] [CrossRef] [Green Version]
- Venters, B.J.; Wachi, S.; Mavrich, T.N.; Andersen, B.E.; Jena, P.; Sinnamon, A.J.; Jain, P.; Rolleri, N.S.; Jiang, C.; Hemeryck-Walsh, C.; et al. A Comprehensive Genomic Binding Map of Gene and Chromatin Regulatory Proteins in Saccharomyces. Mol. Cell 2011, 41, 480–492. [Google Scholar] [CrossRef] [Green Version]
- Horvath, R.; Hawe, N.; Lam, C.; Mestnikov, K.; Eji-Lasisi, M.; Rohde, J.; Sadowski, I. TORC1 signaling modulates Cdk8-dependent GAL gene expression in Saccharomyces cerevisiae. Genetics 2021, 219, iyab168. [Google Scholar] [CrossRef]
- Gonzalez, D.; Hamidi, N.; Del Sol, R.; Benschop, J.J.; Nancy, T.; Li, C.; Francis, L.; Tzouros, M.; Krijgsveld, J.; Holstege, F.C.; et al. Suppression of Mediator is regulated by Cdk8-dependent Grr1 turnover of the Med3 coactivator. Proc. Natl. Acad. Sci. USA 2014, 111, 2500–2505. [Google Scholar] [CrossRef] [Green Version]
- Osman, S.; Mohammad, E.; Lidschreiber, M.; Stuetzer, A.; Bazsó, F.L.; Maier, K.C.; Urlaub, H.; Cramer, P. The Cdk8 kinase module regulates interaction of the mediator complex with RNA polymerase II. J. Biol. Chem. 2021, 296, 100734. [Google Scholar] [CrossRef] [PubMed]
- Shahi, P.; Gulshan, K.; Näär, A.M.; Moye-Rowley, W.S. Differential Roles of Transcriptional Mediator Subunits in Regulation of Multidrug Resistance Gene Expression in Saccharomyces cerevisiae. Mol. Biol. Cell 2010, 21, 2469–2482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, D.A.; Shi, L.; Tu, B.P.; Weissman, J.S. Cycloheximide can distort measurements of mRNA levels and translation efficiency. Nucleic Acids Res. 2019, 47, 4974–4985. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamada, Y. UME6 Is Involved in the Suppression of Basal Transcription of ABC Transporters and Drug Resistance in the ρ+ Cells of Saccharomyces cerevisiae. Microorganisms 2022, 10, 601. https://doi.org/10.3390/microorganisms10030601
Yamada Y. UME6 Is Involved in the Suppression of Basal Transcription of ABC Transporters and Drug Resistance in the ρ+ Cells of Saccharomyces cerevisiae. Microorganisms. 2022; 10(3):601. https://doi.org/10.3390/microorganisms10030601
Chicago/Turabian StyleYamada, Yoichi. 2022. "UME6 Is Involved in the Suppression of Basal Transcription of ABC Transporters and Drug Resistance in the ρ+ Cells of Saccharomyces cerevisiae" Microorganisms 10, no. 3: 601. https://doi.org/10.3390/microorganisms10030601





