Systematic Review of Plasmid AmpC Type Resistances in Escherichia coli and Klebsiella pneumoniae and Preliminary Proposal of a Simplified Screening Method for ampC
Abstract
:1. Introduction
2. Material and Methods
2.1. Systematic Review
2.2. Behavior of Enterobacteriaceae with AmpC-BL in ChromID® ESBL Medium, Using the Disk Diffusion Test with CLX Disks
3. Results
3.1. Systematic Review
3.1.1. Worldwide AmpC-BL Epidemiology
| Author (Reference) | Year of Population Study | Year of Publication | Country of Target Population | Population (H/C) a | Specific Conditions | n | AmpC (%) b | Global c | Genetic Identification | Most Frequent AmpC Enzymes | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | K.pneumoniae | ||||||||||
| Jørgensen et al. [31] | 2006 | 2010 | Denmark | H | ECI | 74 | 0.06 | - | - | PCR/WGS | CMY-2 |
| Courpon-Claudinon et al. [35] | 2005 | 2010 | France | H | 3GCR | 1051 | 0.46 | - | - | PCR/WGS | CMY-2 |
| Illiaquer et al. [42] | 2007–2009 | 2012 | France | H | KPI | 1505 | - | 0.50 | - | PCR/WGS | DHA-1 |
| Voets et al. [33] | 2009 | 2012 | Holland | C | ESBL | 636 | 3.93 | 0.47 | 5.03 | PCR/WGS | CMY-2 |
| Miró et al. [43] | 2009 | 2013 | Spain | H | EI | 100,132 | 0.69 | 1.02 | 0.64 | PCR/WGS | CMY-2 |
| Seiffert et al. [41] | 2011 | 2013 | Switzerland | H/C | ECI | 611 | 12.50 | - | - | PCR/WGS | CMY-2 |
| Gude et al. [44] | 2008–2010 | 2013 | Spain | H | EI | - | - | - | 0.56 | PCR/WGS | CMY-2 |
| Galán-Sánchez et al. [45] | 2011–2012 | 2014 | Spain | H/C | ECI | - | 0.78 | - | - | PCR/WGS | CMY-2 |
| Reuland et al. [30] | 2007 | 2014 | Holland | H | 3GCR | 503 | - | - | 2.60 | PCR | CMY-2 |
| Jones-Dias et al. [46] | 2004–2008 | 2014 | Portugal | H | 3GCR | 124 | - | - | 0.80 | PCR/WGS | CMY-2 |
| Reuland et al. [47] | 2011 | 2015 | Holland | C | EI | 550 | 1.30 | - | - | PCR | CMY-2 |
| Ibrahimagić et al. [48] | 2009–2010 | 2015 | Bosnia and Herzegovina | H/C | ESBL | 85 | - | - | 8.23 | PCR | CMY-2 |
| Alonso et al. [49] | 2010–2011 | 2016 | Spain | H/C | ECI | 21,563 | 1.10 | - | - | PCR/WGS | CMY-2 |
| Li et al. [40] | 2011–2012 | 2015 | Ireland | H | 3GCR | 95 | 19 | - | - | PCR/WGS | CIT group |
| Pascual et al. [50] | 2010–2011 | 2016 | Spain | H/C | 3GCR | 841 | 2.02 | - | - | PCR/WGS | CMY-2 |
| Zhou et al. [36] | 2012–2013 | 2017 | Holland/Germany | H/C | EI | 1087 | 0.73 | - | - | PCR/WGS | CMY-2 |
| Gómara et al. [34] | 2013–2014 | 2018 | Spain | H | CR | 63 | - | - | 14.2 | PCR | CIT group |
| Den Drijver et al. [38] | 2013–2016 | 2018 | Holland | H | EI | 2126 | 2.40 | - | - | PCR | CMY-2 |
| Ribeiro et al. [37] | 2010–2016 | 2019 | Portugal | H | 3GCR | 1246 | 1.28 | 1.04 | 2.60 | PCR/WGS | DHA-1 |
| Findlay et al. [39] | 2017–2018 | 2020 | England | C | 3GCR | 225 | 7.55 | - | - | PCR/WGS | DHA-1 |
| Rohde et al. [51] | 2014–2015 | 2020 | Germany | C | 3GCR | 828 | - | - | 11.90 | PCR/WGS | CMY-2 |
| Author (Reference) | Year of Population Study | Year of Publication | Country of Target Population | Population (H/C) a | Specific Conditions | n | AmpC (%) b | Global c | Genetic Identification | Most Frequent AmpC Enzymes | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | K.pneumoniae | ||||||||||
| Park et al. [53] | 2008–2012 | 2012 | USA | H | 3GCR | 300 | 16.33 | - | - | PCR/WGS | CMY-2 |
| Suwantarat et al. [32] | 2014–2015 | 2016 | USA | H | EI | 854 | - | - | 1.30 | PCR/WGS | CMY-2 |
| Logan et al. [55] | 2011–2015 | 2016 | USA | H | MDR | 225 | 14.22 | - | - | PCR/WGS | CMY-2 |
| Paniagua-Contreras et al. [54] | Data not available | 2018 | Mexico | C | ECI | 194 | 23.70 | - | - | PCR | CIT group |
| Tamma et al. [52] | 2014–2015 | 2019 | USA | H | EI | 1,929 | 2.23 | 0.88 | 3.42 | PCR | CMY-2 |
| Author (Reference) | Year of Population Study | Year of Publication | Country of Target Population | Population (H/C) a | Specific Conditions | n | AmpC (%) b | Global c | Genetic Identification | Most Frequent AmpC Enzymes | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | K. pneumoniae | ||||||||||
| Ogbolu et al. [62] | 2005–2007 | 2011 | Nigeria | H | EI | 134 | - | - | 4.50 | PCR/WGS | DHA-1 |
| Barguigua et al. [63] | 2010 | 2013 | Morocco | C | ECI | 1,174 | 0.59 | - | - | PCR/WGS | CIT group |
| Barguigua et al. [57] | 2010–2011 | 2013 | Morocco | C | KPI | 453 | - | 0.88 | - | PCR/WGS | EBC group |
| Yusuf et al. [59] | Data not available | 2014 | Nigeria | H/C | EI | 543 | 4.23 | 3.50 | 11.23 | - | - |
| Helmy et al. [58] | 2011–2012 | 2014 | Egypt | H | EI | 143 | 14.68 | 2.09 | 18.18 | CIT group | |
| Nakaye et al. [64] | 2013 | 2014 | Uganda | H | 3GCR | 293 | - | - | 39.60 | PCR | EBC group |
| Gharout-Said et al. [65] | 2005–2010 | 2015 | Algeria | H | EI | 922 | - | - | 1.60 | PCR/WGS | CMY-4 |
| Chérif et al. [66] | 2006–2009 | 2015 | Tunisia | H | EI | 11,393 | - | - | 0.59 | PCR/WGS | CMY-2 |
| Tellevik et al. [56] | 2010–2011 | 2016 | Tanzania | H/C | EI | 603 | 0.50 | - | - | PCR/WGS | CMY-2 |
| Zorgani et al. [61] | 2013–2014 | 2017 | Libya | H | EI | 151 | 1.98 | 3.97 | 5.96 | PCR | CIT group |
| Tanfous et al. [67] | 2002–2011 | 2018 | Tunisia | H | KPI | 128 | - | 2.30 | - | PCR/WGS | CMY-4 |
| Tanfous et al. [68] | 2002–2013 | 2018 | Tunisia | H | ESBL | 128 | - | 2.34 | - | PCR/WGS | CMY-4 |
| Rensing et al. [69] | 2013 | 2019 | Egypt | H/C | EI | 225 | 1.45 | 0.97 | 2.91 | PCR | CIT group |
| Mohamed et al. [70] | 2018 | 2020 | Egypt | C | EI | 440 | 2.04 | 2.04 | 4.09 | PCR/WGS | DHA-1 |
| Estaleva et al. [60] | 2015 | 2021 | Mozambique | H/C | ECI | 230 | 10.86 | - | - | PCR/WGS | FOX/MOX |
| Author (Reference) | Year of Population Study | Year of Publication | Country of Target Population | Population (H/C) a | Specific Conditions | n | AmpC (%) b | Global c | Genetic Identification PCR/WGS | Most Frequent AmpC Enzymes | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | K. pneumoniae | ||||||||||
| Yoo et al. [76] | 2008–2009 | 2010 | South Korea | H | EI | 276 | 1,81 | 16.66 | - | PCR | DHA-1 |
| Yamasaki et al. [21] | 2002–2008 | 2010 | Japan | H/C | EI | 22,869 | 0.07 | 0.01 | 0.13 | PCR/WGS | CMY-2 |
| Singtohin et al. [77] | 2005–2006 | 2010 | Thailand | H | EI | 2,712 | 1.62 | 0.29 | 1.91 | PCR | CMY-2 |
| Mohamudha et al. [72] | 2008 | 2010 | India | H | EI | 175 | 24.57 | 13.14 | 44.57 | - | - |
| Mohamudha et al. [73] | 2009–2010 | 2012 | India | H | EI | 241 | 24.89 | 13.27 | 38.17 | PCR | DHA-1 |
| Manoharan et al. [78] | 2007–2008 | 2012 | India | H | 3GCR | 312 | - | - | 15.38 | PCR | CIT group |
| Matsumura et al. [79] | 2010 | 2012 | Japan | H | ECI | 1,327 | 1.73 | - | - | PCR/WGS | CMY-2 |
| Gupta et al. [80] | 2008–2009 | 2012 | India | H | KPI | 100 | - | 32 | - | PCR | CMY-2 |
| Sasirekha et al. [81] | 2008 | 2012 | India | H | EI | 90 | 4.44 | 3.33 | 7.77 | - | - |
| Shafiq et al. [82] | 2008 | 2013 | Pakistan | H/C | ESBL | 511 | 7.97 | 12.37 | - | - | - |
| Azimi et al. [83] | 2013 | 2015 | Iran | H | KPI | 303 | - | 1.60 | - | PCR/WGS | CMY- |
| Hou et al. [75] | 2011 | 2015 | China | H | KPI- MDR | - | 31.50 | - | PCR | DHA- | |
| Liu et al. [84] | 2012 | 2016 | China | H | ECI | 96 | 12.50 | - | - | PCR | DHA-1 |
| Liu et al. [85] | 2012 | 2016 | China | H | KPI | 130 | - | 10.80 | - | PCR/WGS | DHA-1 |
| Ghosh et al. [86] | Data not available | 2016 | India | H | EI | 148 | 16.89 | - | - | PCR/WGS | CMY-2 |
| Luk et al. [71] | 2004–2008 | 2016 | Hong Kong | H | KPI | 109 | - | 44.95 | - | PCR | DHA-1 |
| Sadeghi et al. [87] | 2014 | 2016 | Iran | H | EI | 307 | - | - | 21.50 | PCR/WGS | CMY-2 |
| Baljin et al. [88] | 2014 | 2016 | Mongolia | H | EI | 478 | 0.41 | - | - | PCR/WGS | CMY-2 |
| Noguchi et al. [89] | 2011–2012 | 2017 | Japan | H | EI | 316 | 0.63 | 0.95 | - | PCR/WGS | DHA-1 |
| Khurana et al. [29] | 2013–2015 | 2017 | India | H | GNB | 761 | 0.52 | - | - | PCR | FOX-1/FOX-5b |
| Abdalhamid et al. [90] | 2015 | 2017 | Saudi Arabia | H | EI | 3,625 | - | - | 1 | PCR/WGS | CMY-2 |
| Harris et al. [22] | 2014–2015 | 2018 | Australia, New Zealand, Singapore | H | 3GCR | 30 | 17.10 | - | - | PCR/WGS | CMY-2 |
| Nishimura et al. [91] | 2005–2011 | 2018 | Japan | H | EI | 8,299 | 0.54 | - | 1.75 | PCR/WGS | CIT group |
| Kim et al. [92] | 2007–2016 | 2019 | South Korea | H | ECI | 1,047 | 1.52 | - | - | PCR/WGS | DHA-1 |
| Rizi et al. [93] | 2018 | 2020 | Iran | H | EI | 602 | - | - | 9.30 | PCR | CMY-2 |
| Shrestha et al. [94] | 2013–2014 | 2020 | Nepal | H/C | ECI | 2,661 | 9.86 | - | - | - | - |
| Aryal et al. [95] | 2017–2018 | 2020 | Nepal | H | GNB | 226 | - | - | 40.26 | PCR | CIT group |
| Bala et al. [96] | 2018 | 2020 | India | H | ECI | 470 | 11.10 | - | - | PCR | CIT group |
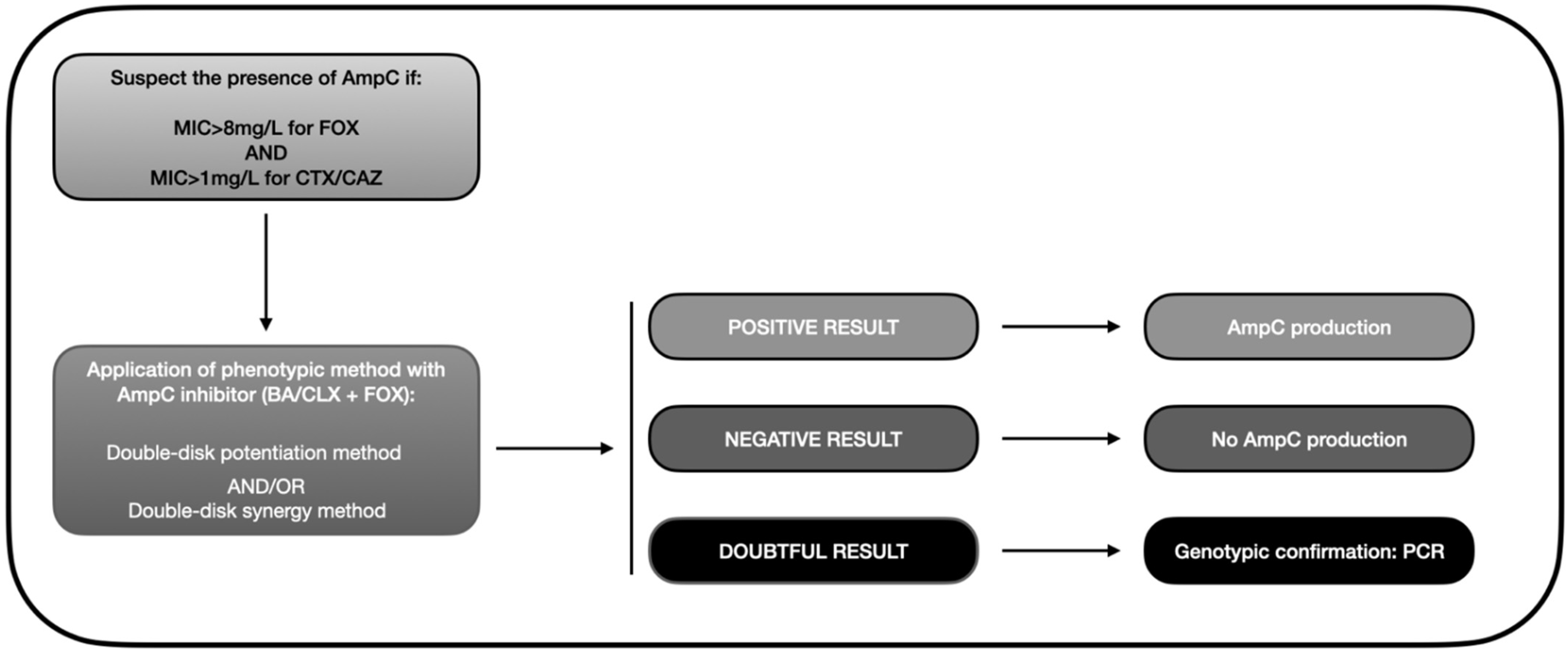
3.1.2. Phenotypic Detection Methods
- -
- Disk approximation method. This technique is employed to detect inducible AmpC-BLs. In the case of pAmpC-BLs, it would be valid for the AmpCs of DHA-1, DHA-2, ACT-1, and CMY-13 families. Two disks are used, one with a substrate antibiotic such as an oxyimino-cephalosporin (e.g., CAZ) or piperacillin/tazobactam, and the other with an inducer antibiotic (e.g., FOX, CLAV, or imipenem, etc.). The microorganism produces an inducible BL if the substrate antibiotic inhibition halo is reduced in the area close to FOX [113].
- -
- Methods with AmpC-specific inhibitors. CLX and boronic acid (BA), and their derivatives, have proven to be the most active and effective commercially available inhibitors to detect AmpC-BLs [105,113,114,115], with CLX being more specific [114,116]. The combination of CTT with other inhibitors, such as Ro48-1220 and LN-2-128 [117,118] or Syn2190 [101,118], especially Syn2190, have demonstrated high sensitivity and specificity to detect AmpC-producing microorganisms; however, they are not commercially available. The main methods include:
- (a)
- (b)
- (c)
- AmpC detection disks. This technique, described by Black et al. (2005) [122], uses Tris-EDTA to permeabilize the bacterial membrane and release beta-lactamases. The bacterium suspected of producing pAmpC-BL (study bacterium) is added to the AmpC disks, which contain Tris-EDTA. The medium is inoculated with an isolate known to be susceptible to FOX (control bacterium), and a FOX disk with AmpC disks (containing the studied bacterium) is placed on both sides. A flattening of the FOX inhibition halo indicates antibiotic inactivation (i.e., the presence of AmpC enzyme released into the medium from the studied bacterium) and therefore a positive result for the presence of pAmpC-BL [28,113,122].
- (d)
- Three-dimensional method. A FOX disk is placed in an agar plate inoculated with a strain susceptible to this antibiotic. An incision is made in the agar near the disk for inoculation with the microorganism under study. The result is positive when the inhibition halo is flattened, which is caused by the growth of the AmpC-producing microorganism [73,113,121].
- (e)
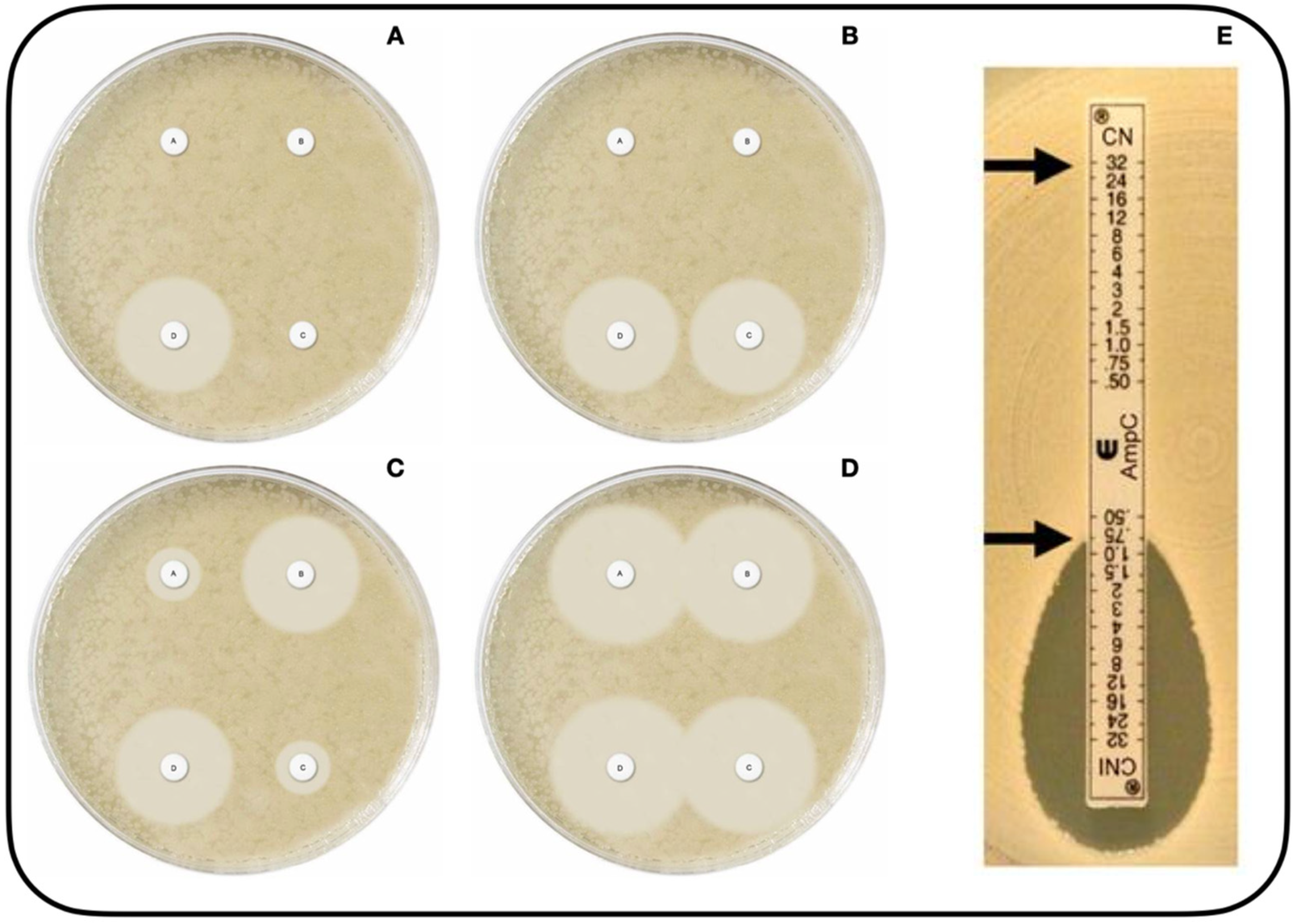
- Mast disks (MastDics® Combi AmpC and ESBL Detection Set, Merseyside, UK) (Figure 1A–D) [123]. This technique, which can be used to detect both AmpC and ESBL, utilizes cefpodoxime disks, alone and combined with AmpC inhibitor and/or ESBL inhibitor. The result is positive when the difference in halo diameter between disks with versus without inhibitor is >5 mm [112].
- (f)
- E-test® AmpC (Biomérieux SA, 69280, Marcy-l´Etoile, France) (Figure 1E) [124]. This test utilizes strips impregnated with CTT at increasing concentrations on both sides, with the presence of CLX on only one side. The result is positive if there is a CTT MIC reduction of at least three dilutions or deformation of the ellipse in the presence of CLX [52,112,121].
- (g)
3.1.3. Genotypic Detection Methods
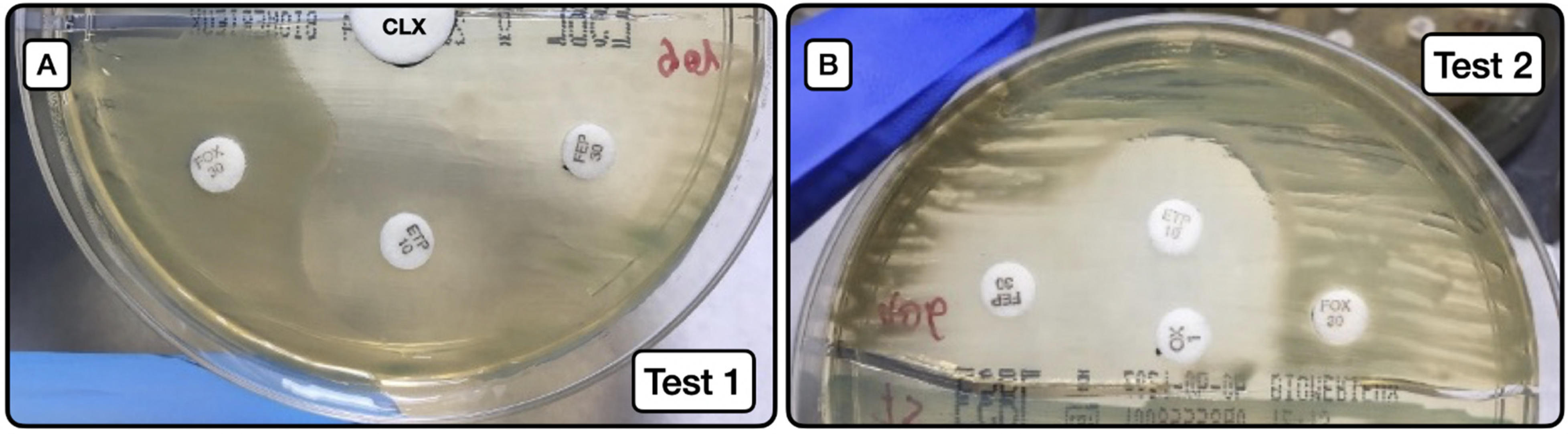
3.1.4. Behavior of Enterobacteriaceae with AmpC-BL in ChromID® ESBL Medium, Using Disk Diffusion Test with CLX Disks
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Livermore, D.M. Antibiotic resistance during and beyond COVID-19. JAC-Antimicrob Resist 2021, 3 (Suppl. 1), i5–i16. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Valverde, M.; Sojo-Dorado, J.; Pascual, A.; Rodríguez-Baño, J. Clinical management of infections caused by multidrug-resistant Enterobacteriaceae. Ther. Adv. Infect. Dis. 2013, 1, 49–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacoby, G.A. AmpC beta-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philippon, A.; Arlet, G.; Jacoby, G.A. Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother. 2002, 46, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walther-Rasmussen, J.; Høiby, N. Plasmid-borne AmpC beta-lactamases. Can. J. Microbiol. 2002, 48, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Soria-Segarra, C.; Delgado-Valverde, M.; Serrano-García, M.L.; López-Hernández, I.; Navarro-Marí, J.M.; Gutiérrez-Fernández, J. Infections in patients colonized with carbapenem-resistant Gram-negative bacteria in a medium Spanish city. Rev. Esp. Quimioter. Publ. Of. Soc. Esp. Quimioter. 2021, 34, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Cano-Martín, E.; Portillo-Calderón, I.; Pérez-Palacios, P.; Navarro-Marí, J.M.; Fernández-Sierra, M.A.; Gutiérrez-Fernández, J. A Study in a Regional Hospital of a Mid-Sized Spanish City Indicates a Major Increase in Infection/Colonization by Carbapenem-Resistant Bacteria, Coinciding with the COVID-19 Pandemic. Antibiotics 2021, 10, 1127. [Google Scholar] [CrossRef] [PubMed]
- del Castillo, M.C.; López-Cerezo, L.; Casal, M.; Pascual, A. Evaluation of chromID ESBL medium for detecting carriers of extended-spectrum beta-lactamase-producing enterobacteriaceae. Enferm. Infecc. Microbiol. Clin. 2011, 29, 471–472. [Google Scholar] [CrossRef]
- Montiel-Riquelme, F.; Calatrava-Hernández, E.; Gutiérrez-Soto, M.; Expósito-Ruiz, M.; Navarro-Marí, J.M.; Gutiérrez-Fernández, J. Clinical Relevance of Antibiotic Susceptibility Profiles for Screening Gram-negative Microorganisms Resistant to Beta-Lactam Antibiotics. Microorganisms 2020, 8, 1555. [Google Scholar] [CrossRef]
- De Oliveira, D.V.; Van Der Sand, S.T. Phenotypic Tests for the Detection of β-Lactamase-Producing Enterobacteriaceae Isolated from Different Environments. Curr. Microbiol. 2016, 73, 132–138. [Google Scholar] [CrossRef]
- EUCAST: EUCAST. Available online: https://www.eucast.org/ (accessed on 2 November 2021).
- Bush, K.; Bradford, P.A. Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfeifer, Y.; Cullik, A.; Witte, W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. IJMM 2010, 300, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Bauernfeind, A.; Chong, Y.; Lee, K. Plasmid-encoded AmpC beta-lactamases: How far have we gone 10 years after the discovery? Yonsei Med. J. 1998, 39, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meini, S.; Tascini, C.; Cei, M.; Sozio, E.; Rossolini, G.M. AmpC β-lactamase-producing Enterobacterales: What a clinician should know. Infection 2019, 47, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Su, L.-H.; Chu, C.; Cloeckaert, A.; Chiu, C.-H. An epidemic of plasmids? Dissemination of extended-spectrum cephalosporinases among Salmonella and other Enterobacteriaceae. FEMS Immunol. Med. Microbiol. 2008, 52, 155–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, T.Y.; Ng, S.Y.; Teo, L.; Koh, Y.; Teok, C.H. Detection of plasmid-mediated AmpC in Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis. J. Clin. Pathol. 2008, 61, 642–644. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.Y.; Ng, L.S.Y.; He, J.; Koh, T.H.; Hsu, L.Y. Evaluation of screening methods to detect plasmid-mediated AmpC in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis. Antimicrob. Agents. Chemother. 2009, 53, 146–149. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, K.; Komatsu, M.; Abe, N.; Fukuda, S.; Miyamoto, Y.; Higuchi, T.; Ono, T.; Nishio, H.; Sueyoshi, N.; Kida, K.; et al. Laboratory surveillance for prospective plasmid-mediated AmpC beta-lactamases in the Kinki region of Japan. J. Clin. Microbiol. 2010, 48, 3267–3273. [Google Scholar] [CrossRef] [Green Version]
- Harris, P.N.A.; Ben Zakour, N.L.; Roberts, L.W.; Wailan, A.M.; Zowawi, H.M.; Tambyah, P.A.; Lye, D.; Jureen, R.; Lee, T.H.; Yin, M.; et al. Whole genome analysis of cephalosporin-resistant Escherichia coli from bloodstream infections in Australia, New Zealand and Singapore: High prevalence of CMY-2 producers and ST131 carrying blaCTX-M-15 and blaCTX-M-27. J. Antimicrob. Chemother. 2018, 73, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Med. 2006, 119 (Suppl. 1), S20–S28, discussion S62–S70. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.-I.; Pai, H.; Kim, S.-H.; Kim, H.-B.; Kim, E.-C.; Oh, M.; Choe, K. Cefepime and the inoculum effect in tests with Klebsiella pneumoniae producing plasmid-mediated AmpC-type beta-lactamase. J. Antimicrob. Chemother. 2004, 54, 1130–1133. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Baño, J.; Miró, E.; Villar, M.; Coelho, A.; Gozalo, M.; Borrell, N.; Bou, G.; Conejo, M.C.; Pomar, V.; Aracil, B.; et al. Colonisation and infection due to Enterobacteriaceae producing plasmid-mediated AmpC β-lactamases. J. Infect. 2012, 64, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-H.; Lee, Y.-T.; Kung, C.-H.; Ku, W.-W.; Kuo, S.-C.; Chen, T.-L.; Fung, C.-P. Risk factors of community-onset urinary tract infections caused by plasmid-mediated AmpC β-lactamase-producing Enterobacteriaceae. J. Microbiol. Immunol. Infect. 2015, 48, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Pascual, V.; Ortiz, G.; Simó, M.; Alonso, N.; Garcia, M.C.; Xercavins, M.; Rivera, A.; Morera, M.A.; Miró, E.; Espejo, E.; et al. Epidemiology and risk factors for infections due to AmpC β-lactamase-producing Escherichia coli. J. Antimicrob. Chemother. 2015, 70, 899–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gude, M.J.; Seral, C.; Saenz, Y.; González-Domínguez, M.; Torres, C.; Castillo, F.J. Evaluation of four phenotypic methods to detect plasmid-mediated AmpC β-lactamases in clinical isolates. Eur. J. Clin. Microbiol. 2012, 31, 2037–2043. [Google Scholar] [CrossRef] [PubMed]
- Pai, H.; Kang, C.-I.; Byeon, J.-H.; Lee, K.-D.; Park, W.B.; Kim, H.-B.; Kim, E.-C.; Oh, M.-D.; Choe, K.-W. Epidemiology and clinical features of bloodstream infections caused by AmpC-type-beta-lactamase-producing Klebsiella pneumoniae. Antimicrob. Agents. Chemother. 2004, 48, 3720–3728. [Google Scholar] [CrossRef] [Green Version]
- Reuland, E.A.; Hays, J.P.; de Jongh, D.M.C.; Abdelrehim, E.; Willemsen, I.; Kluytmans, J.A.J.W.; Savelkoul, P.H.M.; Vandenbroucke-Grauls, C.M.J.E.; al Naiemi, N. Detection and Occurrence of Plasmid-Mediated AmpC in Highly Resistant Gram-Negative Rods. PLoS ONE 2014, 9, e91396. [Google Scholar] [CrossRef]
- Jørgensen, R.L.; Nielsen, J.B.; Friis-Møller, A.; Fjeldsøe-Nielsen, H.; Schønning, K. Prevalence and molecular characterization of clinical isolates of Escherichia coli expressing an AmpC phenotype. J. Antimicrob. Chemother. 2010, 65, 460–464. [Google Scholar] [CrossRef] [Green Version]
- Suwantarat, N.; Logan, L.K.; Carroll, K.C.; Bonomo, R.A.; Simner, P.J.; Rudin, S.D.; Milstone, A.M.; Tekle, T.; Ross, T.; Tamma, P.D. The Prevalence and Molecular Epidemiology of Multidrug-Resistant Enterobacteriaceae Colonization in a Pediatric Intensive Care Unit. Infect. Control. Hosp. Epidemiol. 2016, 37, 535–543. [Google Scholar] [CrossRef] [Green Version]
- Voets, G.M.; Platteel, T.N.; Fluit, A.C.; Scharringa, J.; Schapendonk, C.M.; Stuart, J.C.; Bonten, M.J.M.; Leverstein-van Hall, M.A.; Hall, M.A.L.; National ESBL Surveillance Working Group. Population distribution of Beta-lactamase conferring resistance to third-generation cephalosporins in human clinical Enterobacteriaceae in the Netherlands. PLoS ONE 2012, 7, e52102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómara, M.; López-Calleja, A.I.; Iglesia, B.M.P.V.; Cerón, I.F.; López, A.R.; Pinilla, M.J.R. Detection of carbapenemases and other mechanisms of enzymatic resistance to β-lactams in Enterobacteriaceae with diminished susceptibility to carbapenems in a tertiary care hospital. Enfermedades Infecc. Microbiol. Clin. Engl. Ed. 2018, 36, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Courpon-Claudinon, A.; Lefort, A.; Panhard, X.; Clermont, O.; Dornic, Q.; Fantin, B.; Mentré, F.; Wolff, M.; Denamur, E.; Branger, C.; et al. Bacteraemia caused by third-generation cephalosporin-resistant Escherichia coli in France: Prevalence, molecular epidemiology and clinical features. Clin. Microbiol. Infect. 2011, 17, 557–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; García-Cobos, S.; Ruijs, G.J.H.M.; Kampinga, G.A.; Arends, J.P.; Borst, D.M.; Möller, L.V.; Holman, N.D.; Schuurs, T.A.; Bruijnesteijn van Coppenraet, L.E.; et al. Epidemiology of Extended-Spectrum β-Lactamase-Producing E. coli and Vancomycin-Resistant Enterococci in the Northern Dutch-German Cross-Border Region. Front. Microbiol. 2017, 8, 1914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, T.G.; Novais, Â.; Rodrigues, C.; Nascimento, R.; Freitas, F.; Machado, E.; Peixe, L. Dynamics of clonal and plasmid backgrounds of Enterobacteriaceae producing acquired AmpC in Portuguese clinical settings over time. Int. J. Antimicrob. Agents. 2019, 53, 650–656. [Google Scholar] [CrossRef] [PubMed]
- den Drijver, E.; Verweij, J.J.; Verhulst, C.; Oome, S.; Soer, J.; Willemsen, I.; Schrauwen, E.J.A.; Kluytmans-van den Bergh, M.F.Q.; Kluytmans, J.A.J.W. Decline in AmpC β-lactamase-producing Escherichia coli in a Dutch teaching hospital (2013–2016). PLoS ONE 2018, 13, e0204864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Findlay, J.; Gould, V.C.; North, P.; Bowker, K.E.; Williams, M.O.; MacGowan, A.P.; Avison, M.B. Characterization of cefotaxime-resistant urinary Escherichia coli from primary care in South-West England 2017–18. J. Antimicrob. Chemother. 2019, 75, 65–71. [Google Scholar] [CrossRef]
- Li, Y.; Cassidy, F.; Salmon, A.; Keating, D.; Herra, C.; Schaffer, K. Detection and epidemiology of plasmid-mediated AmpC β-lactamase producing Escherichia coli in two Irish tertiary care hospitals. J. Glob. Antimicrob. Resist. 2015, 3, 242–246. [Google Scholar] [CrossRef] [Green Version]
- Seiffert, S.N.; Hilty, M.; Kronenberg, A.; Droz, S.; Perreten, V.; Endimiani, A. Extended-spectrum cephalosporin-resistant Escherichia coli in community, specialized outpatient clinic and hospital settings in Switzerland. J. Antimicrob. Chemother. 2013, 68, 2249–2254. [Google Scholar] [CrossRef] [Green Version]
- Illiaquer, M.; Caroff, N.; Bémer, P.; Aubin, G.G.; Juvin, M.-E.; Lepelletier, D.; Reynaud, A.; Corvec, S. Occurrence and molecular characterization of Klebsiella pneumoniae ST37 clinical isolates producing plasmid-mediated AmpC recovered over a 3-year period. Diagn. Microbiol. Infect. Dis. 2012, 74, 95–97. [Google Scholar] [CrossRef] [PubMed]
- Miró, E.; Agüero, J.; Larrosa, M.N.; Fernández, A.; Conejo, M.C.; Bou, G.; González-López, J.J.; Lara, N.; Martínez-Martínez, L.; Oliver, A.; et al. Prevalence and molecular epidemiology of acquired AmpC β-lactamases and carbapenemases in Enterobacteriaceae isolates from 35 hospitals in Spain. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Gude, M.J.; Seral, C.; Sáenz, Y.; Cebollada, R.; González-Domínguez, M.; Torres, C.; Castillo, F.J. Molecular epidemiology, resistance profiles and clinical features in clinical plasmid-mediated AmpC-producing Enterobacteriaceae. Int. J. Med. Microbiol. IJMM 2013, 303, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Galán-Sánchez, F.; Aznar-Marín, P.; Marín-Casanova, P.; Rodríguez-Iglesias, M. Diversity of bla genes and low incidence of CTX-M in plasmid-mediated AmpC-producing Escherichia coli clinical isolates. APMIS Acta. Pathol. Microbiol. Immunol. Scand. 2014, 122, 796–799. [Google Scholar] [CrossRef] [PubMed]
- Jones-Dias, D.; Manageiro, V.; Ferreira, E.; Louro, D.; Antibiotic Resistance Surveillance Program in Portugal (ARSIP) participants; Caniça, M. Diversity of extended-spectrum and plasmid-mediated AmpC β-lactamases in Enterobacteriaceae isolates from portuguese health care facilities. J. Microbiol. 2014, 52, 496–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reuland, E.A.; Halaby, T.; Hays, J.P.; de Jongh, D.M.C.; Snetselaar, H.D.R.; van Keulen, M.; Elders, P.J.M.; Savelkoul, P.H.M.; Vandenbroucke-Grauls, C.M.J.E.; Al Naiemi, N. Plasmid-mediated AmpC: Prevalence in community-acquired isolates in Amsterdam, the Netherlands, and risk factors for carriage. PLoS ONE 2015, 10, e0113033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibrahimagić, A.; Bedenić, B.; Kamberović, F.; Uzunović, S. High prevalence of CTX-M-15 and first report of CTX-M-3, CTX-M-22, CTX-M-28 and plasmid-mediated AmpC beta-lactamase producing Enterobacteriaceae causing urinary tract infections in Bosnia and Herzegovina in hospital and community settings. J. Infect. Chemother. 2015, 21, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Alonso, N.A.; Miro, E.; Pascual, V.; Rivera, A.; Simó, M.; Garcia, M.C.; Xercavins, M.; Morera, M.A.; Espejo, E.; Gurguí, M.; et al. Molecular characterisation of acquired and overproduced chromosomal blaAmpC in Escherichia coli clinical isolates. Int. J. Antimicrob. Agents 2015, 47, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Pascual, V.; Alonso, N.; Simó, M.; Ortiz, G.; Garcia, M.C.; Xercavins, M.; Rivera, A.; Morera, M.A.; Miró, E.; Espejo, E.; et al. Bloodstream infections caused by Escherichia coli producing AmpC β-lactamases: Epidemiology and clinical features. Eur. J. Clin. Microbiol. 2016, 35, 1997–2003. [Google Scholar] [CrossRef]
- Rohde, A.M.; Zweigner, J.; Wiese-Posselt, M.; Schwab, F.; Behnke, M.; Kola, A.; Schröder, W.; Peter, S.; Tacconelli, E.; Wille, T.; et al. Prevalence of third-generation cephalosporin-resistant Enterobacterales colonization on hospital admission and ESBL genotype-specific risk factors: A cross-sectional study in six German university hospitals. J. Antimicrob. Chemother. 2020, 75, 1631–1638. [Google Scholar] [CrossRef]
- Tamma, P.D.; Sharara, S.L.; Pana, Z.D.; Amoah, J.; Fisher, S.L.; Tekle, T.; Doi, Y.; Simner, P.J. Molecular Epidemiology of Ceftriaxone Non-Susceptible Enterobacterales Isolates in an Academic Medical Center in the United States. Open Forum. Infect. Dis. 2019, 6, ofz353. [Google Scholar] [CrossRef]
- Park, Y.S.; Adams-Haduch, J.M.; Shutt, K.; Iii, D.M.Y.; Johnson, L.E.; Hingwe, A.; Lewis, J.S.; Jorgensen, J.H.; Doi, Y. Clinical and Microbiologic Characteristics of Cephalosporin-Resistant Escherichia coli at Three Centers in the United States. Antimicrob. Agents Chemother. 2012, 56, 1870–1876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paniagua-Contreras, G.L.; Monroy-Pérez, E.; Bautista, A.; Reyes, R.; Vicente, A.; Vaca-Paniagua, F.; Díaz, C.E.; Martínez, S.; Domínguez, P.; García, L.R.; et al. Multiple antibiotic resistances and virulence markers of uropathogenic Escherichia coli from Mexico. Pathog. Glob. Health 2018, 112, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Logan, L.K.; Hujer, A.M.; Marshall, S.H.; Domitrovic, T.N.; Rudin, S.D.; Zheng, X.; Qureshi, N.K.; Hayden, M.K.; Scaggs, F.A.; Karadkhele, A.; et al. Analysis of β-Lactamase Resistance Determinants in Enterobacteriaceae from Chicago Children: A Multicenter Survey. Antimicrob. Agents Chemother. 2016, 60, 3462–3469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tellevik, M.G.; Blomberg, B.; Kommedal, Ø.; Maselle, S.Y.; Langeland, N.; Moyo, S.J. High Prevalence of Faecal Carriage of ESBL-Producing Enterobacteriaceae among Children in Dar es Salaam, Tanzania. PLoS ONE 2016, 11, e0168024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barguigua, A.; El Otmani, F.; Talmi, M.; Reguig, A.; Jamali, L.; Zerouali, K.; Timinouni, M. Prevalence and genotypic analysis of plasmid-mediated β-lactamases among urinary Klebsiella pneumoniae isolates in Moroccan community. J. Antibiot. 2012, 66, 11–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helmy, M.M.; Wasfi, R. Phenotypic and molecular characterization of plasmid mediated AmpC β-lactamases among Escherichia coli, Klebsiella spp., and Proteus mirabilis isolated from urinary tract infections in Egyptian hospitals. BioMed Res. Int. 2014, 2014, 171548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yusuf, I.; Arzai, A.; Haruna, M.; Sharif, A.; Getso, M. Detection of multi drug resistant bacteria in major hospitals in Kano, North-West, Nigeria. Braz. J. Microbiol. 2014, 45, 791–798. [Google Scholar] [CrossRef] [Green Version]
- Estaleva, C.E.L.; Zimba, T.F.; Sekyere, J.O.; Govinden, U.; Chenia, H.Y.; Simonsen, G.S.; Haldorsen, B.; Essack, S.Y.; Sundsfjord, A. High prevalence of multidrug resistant ESBL- and plasmid mediated AmpC-producing clinical isolates of Escherichia coli at Maputo Central Hospital, Mozambique. BMC Infect. Dis. 2021, 21, 16. [Google Scholar] [CrossRef] [PubMed]
- Zorgani, A.; Daw, H.; Sufya, N.; Bashein, A.; Elahmer, O.; Chouchani, C. Co-Occurrence of Plasmid-Mediated AmpC β-Lactamase Activity among Klebsiella pneumoniae and Escherichia Coli. Open Microbiol. J. 2017, 11, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Ogbolu, D.; Daini, O.; Ogunledun, A.; Alli, A.; Webber, M. High levels of multidrug resistance in clinical isolates of Gram-negative pathogens from Nigeria. Int. J. Antimicrob. Agents 2011, 37, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Barguigua, A.; El Otmani, F.; Talmi, M.; Zerouali, K.; Timinouni, M. Prevalence and types of extended spectrum β-lactamases among urinary Escherichia coli isolates in Moroccan community. Microb. Pathog. 2013, 61–62, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Nakaye, M.; Bwanga, F.; Itabangi, H.; Stanley, I.J.; Bashir, M.; Bazira, J. AmpC-BETA Lactamases among Enterobacteriaceae Isolated at a Tertiary Hospital, South Western Uganda. Br. Biotechnol. J. 2014, 4, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Gharout-Sait, A.; Touati, A.; Guillard, T.; Brasme, L.; De Champs, C. Molecular characterization and epidemiology of cefoxitin resistance among Enterobacteriaceae lacking inducible chromosomal ampC genes from hospitalized and non-hospitalized patients in Algeria: Description of new sequence type in Klebsiella pneumoniae isolates. Braz. J. Infect. Dis. 2015, 19, 187–195. [Google Scholar] [PubMed] [Green Version]
- Chérif, T.; Saidani, M.; Decré, D.; Boutiba-Ben Boubaker, I.; Arlet, G. Cooccurrence of Multiple AmpC β-Lactamases in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis in Tunisia. Antimicrob. Agents. Chemother. 2016, 60, 4–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Tanfous, F.; Achour, W.; Raddaoui, A.; Ben Hassen, A. Molecular characterisation and epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates from immunocompromised patients in Tunisia. J. Glob. Antimicrob. Resist. 2018, 13, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Ben Tanfous, F.; Raddaoui, A.; Chebbi, Y.; Achour, W. Epidemiology and molecular characterisation of colistin-resistant Klebsiella pneumoniae isolates from immunocompromised patients in Tunisia. Int. J. Antimicrob. Agents 2018, 52, 861–865. [Google Scholar] [CrossRef]
- Rensing, K.L.; Abdallah, H.M.; Koek, A.; Elmowalid, G.A.; Vandenbroucke-Grauls, C.M.J.E.; Al Naiemi, N.; Van Dijk, K. Prevalence of plasmid-mediated AmpC in Enterobacteriaceae isolated from humans and from retail meat in Zagazig, Egypt. Antimicrob. Resist. Infect. Control 2019, 8, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, E.S.; Khairy, R.M.M.; Abdelrahim, S.S. Prevalence and molecular characteristics of ESBL and AmpC β -lactamase producing Enterobacteriaceae strains isolated from UTIs in Egypt. Antimicrob. Resist. Infect. Control 2020, 9, 198. [Google Scholar] [CrossRef]
- Luk, S.; Wong, W.-K.; Ho, A.Y.-M.; Yu, K.C.-H.; To, W.-K.; Ng, T.-K. Clinical features and molecular epidemiology of plasmid-mediated DHA-type AmpC β-lactamase-producing Klebsiella pneumoniae blood culture isolates, Hong Kong. J. Glob. Antimicrob. Resist. 2016, 7, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Mohamudha Parveen, R.; Harish, B.N.; Parija, S.C. Ampc Beta lactamases among gram negative clinical isolates from a tertiary hospital, South India. Braz. J. Microbiol. Publ. Braz. Soc. Microbiol. 2010, 41, 596–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harish, B.; Mohamudha, P.R.; Parija, S.C. Molecular description of plasmid-mediated AmpC β-lactamases among nosocomial isolates of Escherichia coli & Klebsiella pneumoniae from six different hospitals in India. Indian J. Med Res. 2012, 135, 114–119. [Google Scholar]
- Habeeb, M.A.; Haque, A.; Nematzadeh, S.; Iversen, A.; Giske, C.G. High prevalence of 16S rRNA methylase RmtB among CTX-M extended-spectrum β-lactamase-producing Klebsiella pneumoniae from Islamabad, Pakistan. Int. J. Antimicrob. Agents 2013, 41, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.-H.; Song, X.-Y.; Ma, X.-B.; Zhang, S.-Y.; Zhang, J.-Q. Molecular characterization of multidrug-resistant Klebsiella pneumoniae isolates. Braz. J. Microbiol. 2015, 46, 759–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, J.S.; Byeon, J.; Yang, J.; Yoo, J.I.; Chung, G.T.; Lee, Y.S. High prevalence of extended-spectrum beta-lactamases and plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae isolated from long-term care facilities in Korea. Diagn. Microbiol. Infect. Dis. 2010, 67, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Singtohin, S.; Chanawong, A.; Lulitanond, A.; Sribenjalux, P.; Auncharoen, A.; Kaewkes, W.; Songsri, J.; Pienthaweechai, K. CMY-2, CMY-8b, and DHA-1 plasmid-mediated AmpC β-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae from a university hospital, Thailand. Diagn. Microbiol. Infect. Dis. 2010, 68, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, A.; Sugumar, M.; Kumar, A.; Jose, H.; Mathai, D.; Khilnani, G.C.; Kapil, A.; Francis, G.; Radhakrishnan, K.; Dutta, T.K.; et al. Phenotypic & molecular characterization of AmpC β-lactamases among Escherichia coli, Klebsiella spp. & Enterobacter spp. from five Indian Medical Centers. Indian J. Med. Res. 2012, 135, 359–364. [Google Scholar] [PubMed]
- Matsumura, Y.; Yamamoto, M.; Higuchi, T.; Komori, T.; Tsuboi, F.; Hayashi, A.; Sugimoto, Y.; Hotta, G.; Matsushima, A.; Ngao, M.; et al. Prevalence of plasmid-mediated AmpC β-lactamase-producing Escherichia coli and spread of the ST131 clone among extended-spectrum β-lactamase-producing E. coli in Japan. Int. J. Antimicrob. Agents 2012, 40, 158–162. [Google Scholar] [CrossRef] [Green Version]
- Gupta, V.; Kumarasamy, K.; Gulati, N.; Garg, R.; Krishnan, P.; Chander, J. AmpC β-lactamases in nosocomial isolates of Klebsiella pneumoniae from India. Indian J. Med Res. 2012, 136, 237–241. [Google Scholar]
- Sasirekha, B.; Shivakumar, S. Occurrence of Plasmid-Mediated AmpC β-Lactamases Among Escherichia coli and Klebsiella pneumoniae Clinical Isolates in a Tertiary Care Hospital in Bangalore. Indian J. Microbiol. 2011, 52, 174–179. [Google Scholar] [CrossRef] [Green Version]
- Shafiq, M.; Rahman, H.; Qasim, M.; Ayub, N.; Hussain, S.; Khan, J.; Naeem, M. Prevalence of plasmid-mediated AmpC β-lactamases in Escherichia coli and Klebsiella pneumonia at tertiary care hospital of Islamabad, Pakistan. Eur. J. Microbiol. Immunol. 2013, 3, 267–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azimi, L.; Erajiyan, G.; Talebi, M.; Owlia, P.; Bina, M.; Shojaie, A.; Lari, A.R. Phenotypic and Molecular Characterization of Plasmid Mediated AmpC among Clinical Isolates of Klebsiella pneumoniae Isolated from Different Hospitals in Tehran. J. Clin. Diagn. Res. 2015, 9, DC01–DC03. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, Y. Detection of plasmid-mediated AmpC β-lactamase in Escherichia coli. Biomed. Rep. 2016, 4, 687–690. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-Q.; Liu, Y.-R. Detection and genotype analysis of AmpC β-lactamase in Klebsiella pneumoniae from tertiary hospitals. Exp. Ther. Med. 2016, 12, 480–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, B.; Mukherjee, M. Emergence of co-production of plasmid-mediated AmpC beta-lactamase and ESBL in cefoxitin-resistant uropathogenic Escherichia coli. Eur. J. Clin. Microbiol. 2016, 35, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, M.R.; Ghotaslou, R.; Akhi, M.T.; Asgharzadeh, M.; Hasani, A. Molecular characterization of extended-spectrum β-lactamase, plasmid-mediated AmpC cephalosporinase and carbapenemase genes among Enterobacteriaceae isolates in five medical centres of East and West Azerbaijan, Iran. J. Med Microbiol. 2016, 65, 1322–1331. [Google Scholar] [CrossRef] [PubMed]
- Baljin, B.; Baldan, G.; Chimeddorj, B.; Tulgaa, K.; Gunchin, B.; Sandag, T.; Pfeffer, K.; MacKenzie, C.R.; Wendel, A.F. Faecal Carriage of Gram-Negative Multidrug-Resistant Bacteria among Patients Hospitalized in Two Centres in Ulaanbaatar, Mongolia. PLoS ONE 2016, 11, e0168146. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, T.; Matsumura, Y.; Yamamoto, M.; Nagao, M.; Takakura, S.; Ichiyama, S. Clinical and microbiologic characteristics of cefotaxime-non-susceptible Enterobacteriaceae bacteremia: A case control study. BMC Infect. Dis. 2017, 17, 44. [Google Scholar] [CrossRef] [Green Version]
- Abdalhamid, B.; Albunayan, S.; Shaikh, A.; Elhadi, N.; Aljindan, R. Prevalence study of plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking inducible ampC from Saudi hospitals. J. Med Microbiol. 2017, 66, 1286–1290. [Google Scholar] [CrossRef]
- Nishimura, F.; Morinaga, Y.; Akamatsu, N.; Matsuda, J.; Kaku, N.; Takeda, K.; Uno, N.; Kosai, K.; Hasegawa, H.; Yanagihara, K. Plasmid-Mediated AmpC β-Lactamase and Underestimation of Extended-Spectrum β-Lactamase in Cefepime-Susceptible Elevated-Ceftazidime-MIC Enterobacteriaceae Isolates. Jpn. J. Infect. Dis. 2018, 71, 281–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.G.; Jeong, J.; Kim, M.J.; Park, D.W.; Shin, J.H.; Park, H.J.; Chung, J.K.; Kee, H.Y. Prevalence and molecular epidemiology of ESBLs, plasmid-determined AmpC-type β-lactamases and carbapenemases among diarrhoeagenic Escherichia coli isolates from children in Gwangju, Korea: 2007-16. J. Antimicrob. Chemother. 2019, 74, 2181–2187. [Google Scholar] [CrossRef] [PubMed]
- Rizi, K.S.; Mosavat, A.; Youssefi, M.; Jamehdar, S.A.; Ghazvini, K.; Safdari, H.; Amini, Y.; Farsiani, H. High prevalence of blaCMY AmpC beta-lactamase in ESBL co-producing Escherichia coli and Klebsiella spp. clinical isolates in the northeast of Iran. J. Glob. Antimicrob. Resist. 2020, 22, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Thapa Shrestha, U.; Shrestha, S.; Adhikari, N.; Rijal, K.R.; Shrestha, B.; Adhikari, B.; Banjara, M.R.; Ghimire, P. Plasmid Profiling and Occurrence of β-Lactamase Enzymes in Multidrug-Resistant Uropathogenic Escherichia coli in Kathmandu, Nepal. Infect. Drug Resist. 2020, 13, 1905–1917. [Google Scholar] [CrossRef] [PubMed]
- Aryal, S.C.; Upreti, M.K.; Sah, A.K.; Ansari, M.; Nepal, K.; Dhungel, B.; Adhikari, N.; Lekhak, B.; Rijal, K.R. Plasmid-Mediated AmpC β-Lactamase CITM and DHAM Genes Among Gram-Negative Clinical Isolates. Infect. Drug Resist. 2020, 13, 4249–4261. [Google Scholar] [CrossRef] [PubMed]
- Bala, R.; Singh, V.A.; Gupta, N.; Rakshit, P. Prevalence, multidrug-resistance and risk factors for AmpC β-lactamases producing Escherichia coli from hospitalized patients. J. Infect. Dev. Ctries. 2020, 14, 1466–1469. [Google Scholar] [CrossRef] [PubMed]
- Conejo, M.C.; Mata, C.; Navarro, F.; Pascual, A.; GEMARA Collaborative Group. Detection and reporting beta-lactam resistance phenotypes in Escherichia coli and Klebsiella pneumoniae: A multicenter proficiency study in Spain. Diagn. Microbiol. Infect. Dis. 2008, 62, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Coolen, J.P.M.; Drijver, E.D.; Kluytmans, J.A.J.W.; Verweij, J.J.; Lamberts, B.A.; Soer, J.A.C.J.; Verhulst, C.; Wertheim, H.F.L.; Kolwijck, E. Development of an algorithm to discriminate between plasmid- and chromosomal-mediated AmpC β-lactamase production in Escherichia coli by elaborate phenotypic and genotypic characterization. J. Antimicrob. Chemother. 2019, 74, 3481–3488. [Google Scholar] [CrossRef] [PubMed]
- Thomson, K. Controversies about Extended-Spectrum and AmpC Beta-Lactamases. Emerg. Infect. Dis. 2001, 7, 333–334. [Google Scholar] [CrossRef]
- Oteo, J.; Bou, G.; Chaves, F.; Oliver, A. Microbiological methods for surveillance of carrier status of multiresistant bacteria. Enfermedades Infecc. Y Microbiol. Clin. 2017, 35, 667–675. [Google Scholar] [CrossRef]
- Jacoby, G.A. Extended-spectrum beta-lactamases and other enzymes providing resistance to oxyimino-beta-lactams. Infect. Dis. Clin. North Am. 1997, 11, 875–887. [Google Scholar] [CrossRef]
- Coudron, P.E. Inhibitor-based methods for detection of plasmid-mediated AmpC beta-lactamases in Klebsiella spp., Escherichia coli, and Proteus mirabilis. J. Clin. Microbiol. 2005, 3, 4163–4167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peter-Getzlaff, S.; Polsfuss, S.; Poledica, M.; Hombach, M.; Giger, J.; Böttger, E.C.; Zbinden, R.; Bloemberg, G.V. Detection of AmpC beta-lactamase in Escherichia coli: Comparison of three phenotypic confirmation assays and genetic analysis. J. Clin. Microbiol. 2011, 49, 2924–2932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohner, P.C.; Robberts, F.J.L.; Cockerill, F.R.; Patel, R. Cephalosporin MIC distribution of extended-spectrum-{beta}-lactamase- and pAmpC-producing Escherichia coli and Klebsiella species. J. Clin. Microbiol. 2009, 47, 2419–2425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robberts, F.J.L.; Kohner, P.C.; Patel, R. Unreliable extended-spectrum beta-lactamase detection in the presence of plasmid-mediated AmpC in Escherichia coli clinical isolates. J. Clin. Microbiol. 2009, 47, 358–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doi, Y.; Paterson, D.L. Detection of plasmid-mediated class C beta-lactamases. Int. J. Infect. Dis. IJID 2007, 11, 191–197. [Google Scholar]
- Yang, K.; Guglielmo, B.J. Diagnosis and treatment of extended-spectrum and AmpC beta-lactamase-producing organisms. Ann. Pharmacother. 2007, 41, 1427–1435. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Segreti, J. Overview of the epidemiological profile and laboratory detection of extended-spectrum beta-lactamases. Clin. Infect. Dis. 2006, 42 (Suppl. 4), S153–S163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingram, P.R.; Inglis, T.J.J.; Vanzetti, T.R.; Henderson, B.A.; Harnett, G.B.; Murray, R.J. Comparison of methods for AmpC β-lactamase detection in Enterobacteriaceae. J. Med Microbiol. 2011, 60, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Châtre, P.; Madec, J.-Y. Emergence of Escherichia coli producing extended-spectrum AmpC β-lactamases (ESAC) in animals. Front. Microbiol. 2014, 5, 53. [Google Scholar] [CrossRef]
- Calvo, J.; Cantón, R.; Fernández Cuenca, F.; Mirelis, B.; Navarro, F. Available online: https://www.seimc.org/contenidos/documentoscientificos/procedimientosmicrobiologia/seimc-procedimientomicrobiologia39.pdf (accessed on 31 October 2021).
- Pitout, J.D.D. Extraintestinal pathogenic Escherichia coli: An update on antimicrobial resistance, laboratory diagnosis and treatment. Expert. Rev. Anti. Infect. Ther. 2012, 10, 1165–1176. [Google Scholar] [CrossRef]
- Martínez Rojas, D.D.V. Betalactamasas tipo AmpC: Generalidades y métodos para detección fenotípica. Rev. Soc. Venez. Microbiol. 2009, 29, 78–83. [Google Scholar]
- Polsfuss, S.; Bloemberg, G.V.; Giger, J.; Meyer, V.; Böttger, E.C.; Hombach, M. Practical approach for reliable detection of AmpC beta-lactamase-producing Enterobacteriaceae. J. Clin. Microbiol. 2011, 49, 2798–2803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamma, P.D.; Doi, Y.; Bonomo, R.A.; Johnson, J.K.; Simner, P.J.; Antibacterial Resistance Leadership Group. A Primer on AmpC β-Lactamases: Necessary Knowledge for an Increasingly Multidrug-resistant World. Clin. Infect. Dis. 2019, 69, 1446–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edquist, P.; Ringman, M.; Liljequist, B.O.; Wisell, K.T.; Giske, C.G. Phenotypic detection of plasmid-acquired AmpC in Escherichia coli—evaluation of screening criteria and performance of two commercial methods for the phenotypic confirmation of AmpC production. Eur. J. Clin. Microbiol. 2013, 32, 1205–1210. [Google Scholar] [CrossRef]
- Black, J.A.; Thomson, K.S.; Pitout, J.D.D. Use of beta-lactamase inhibitors in disk tests to detect plasmid-mediated AmpC beta-lactamases. J. Clin. Microbiol. 2004, 42, 2203–2206. [Google Scholar] [CrossRef] [Green Version]
- Black, J.A.; Thomson, K.S.; Buynak, J.D.; Pitout, J.D.D. Evaluation of beta-lactamase inhibitors in disk tests for detection of plasmid-mediated AmpC beta-lactamases in well-characterized clinical strains of Klebsiella spp. J. Clin. Microbiol. 2005, 43, 4168–4171. [Google Scholar] [CrossRef] [Green Version]
- Willems, E.; Verhaegen, J.; Magerman, K.; Nys, S.; Cartuyvels, R. Towards a phenotypic screening strategy for emerging β-lactamases in Gram-negative bacilli. Int. J. Antimicrob. Agents 2013, 41, 99–109. [Google Scholar] [CrossRef]
- Navarro, F.; Calvo, J.; Cantón, R.; Fernández-Cuenca, F.; Mirelis, B. Detection of resistance phenotypes in gram-negative bacteria. Enferm. Infecc. Microbiol. Clin. 2011, 29, 524–534. [Google Scholar] [CrossRef]
- Seral, C.; Gude, M.J.; Castillo, F.J. Emergence of plasmid mediated AmpC β-lactamasas: Origin, importance, detection and therapeutical options. Rev. Esp. Quimioter. Publ. Of. Soc. Esp. Quimioter. 2012, 25, 89–99. [Google Scholar]
- Black, J.A.; Moland, E.S.; Thomson, K.S. AmpC disk test for detection of plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae lacking chromosomal AmpC beta-lactamases. J. Clin. Microbiol. 2005, 43, 3110–3113. [Google Scholar] [CrossRef] [Green Version]
- Ampc Esbl Detection Set. Available online: https://mast-group.com/uk/products/amr/antibiotic-resistance-detection-sets/d68c/ (accessed on 25 October 2021).
- Etest® Para la Detección de Resistencia Antimicrobiana (ARD). bioMérieux España. Available online: https://www.biomerieux.es/diagnostico-clinico/productos/etestr-para-la-deteccion-de-resistencia-antimicrobiana-ard (accessed on 25 October 2021).
- Scapaticci, M.; Fossen, G.; Ius, V. Epidemiology of extended spectrum β-lactamase, AmpC and class A carbapenemases-producing organisms isolated at San Camillo Hospital of Treviso (Italy) between April 2012 and March 2014. Microbiol. Medica 2016, 31. Available online: https://www.pagepressjournals.org/index.php/mm/article/view/4622 (accessed on 10 February 2022). [CrossRef] [Green Version]
- Fröding, I.; Vondracek, M.; Giske, C.G. Rapid EUCAST disc diffusion testing of MDR Escherichia coli and Klebsiella pneumoniae: Inhibition zones for extended-spectrum cephalosporins can be reliably read after 6 h of incubation. J. Antimicrob. Chemother. 2017, 72, 1094–1102. [Google Scholar] [PubMed] [Green Version]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brolund, A.; Wisell, K.T.; Edquist, P.J.; Elfström, L.; Walder, M.; Giske, C.G. Development of a real-time SYBRGreen PCR assay for rapid detection of acquired AmpC in Enterobacteriaceae. J. Microbiol. Methods 2010, 82, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Chavda, K.D.; Satlin, M.J.; Chen, L.; Manca, C.; Jenkins, S.G.; Walsh, T.J.; Kreiswirth, B.N. Evaluation of a Multiplex PCR Assay To Rapidly Detect Enterobacteriaceae with a Broad Range of β-Lactamases Directly from Perianal Swabs. Antimicrob. Agents Chemother. 2016, 60, 6957–6961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voets, G.M.; Fluit, A.C.; Scharringa, J.; Cohen Stuart, J.; Leverstein-van Hall, M.A. A set of multiplex PCRs for genotypic detection of extended-spectrum β-lactamases, carbapenemases, plasmid-mediated AmpC β-lactamases and OXA β-lactamases. Int. J. Antimicrob. Agents. 2011, 37, 356–359. [Google Scholar] [CrossRef] [Green Version]
- Geyer, C.N.; Reisbig, M.D.; Hanson, N.D. Development of a TaqMan Multiplex PCR Assay for Detection of Plasmid-Mediated AmpC β-Lactamase Genes. J. Clin. Microbiol. 2012, 50, 3722–3725. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Zhang, J.; Rao, S.; Sun, L.; Zhang, J.; Liu, R.; Zheng, G.; Ma, X.; Hou, S.; Zhuang, X.; et al. Heptaplex PCR melting curve analysis for rapid detection of plasmid-mediated AmpC β-lactamase genes. J. Microbiol. Methods 2015, 110, 1–6. [Google Scholar] [CrossRef]
- Caliskan, E.; Coskun, U.S.S.; Dulger, G.; Kilincel, O.; Ankarali, H.; Sahin, I. Investigation of plasmid mediated AmpC beta-lactamases in Escherichia coli and Klebsiella pneumoniae isolates by phenotypic and genotypic. J. Pak. Med Assoc. 2019, 69, 834–839. [Google Scholar]
- Kis, Z.; Tóth, Á.; Jánvári, L.; Damjanova, I. Countrywide dissemination of a DHA-1-type plasmid-mediated AmpC β-lactamase-producing Klebsiella pneumoniae ST11 international high-risk clone in Hungary, 2009–2013. J. Med. Microbiol. 2016, 65, 1020–1027. [Google Scholar] [CrossRef]
- Tenover, F.C.; Emery, S.L.; Spiegel, C.A.; Bradford, P.; Eells, S.; Endimiani, A.; Bonomo, R.A.; McGowan, J.E. Identification of Plasmid-Mediated AmpC β-Lactamases in Escherichia coli, Klebsiella spp., and Proteus Species Can Potentially Improve Reporting of Cephalosporin Susceptibility Testing Results. J. Clin. Microbiol. 2009, 47, 294–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agyekum, A.; Fajardo-Lubián, A.; Ai, X.; Ginn, A.N.; Zong, Z.; Guo, X.; Turnidge, J.; Partridge, S.R.; Iredell, J.R. Predictability of Phenotype in Relation to Common β-Lactam Resistance Mechanisms in Escherichia coli and Klebsiella pneumoniae. J. Clin. Microbiol. 2016, 54, 1243–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Hong, S.G.; Park, Y.J.; Lee, H.S.; Song, W.; Jeong, J.; Yong, D.; Chong, Y. Evaluation of phenotypic screening methods for detecting plasmid-mediated AmpC beta-lactamases-producing isolates of Escherichia coli and Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 2005, 53, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Park, Y.-J.; Yu, J.; Lee, J.; Ahn, D.-R.; Min, S.-J. Performance of a novel fluorogenic probe assay for the detection of extended-spectrum-β-lactamase or plasmid AmpC β-lactamase–producing Enterobacterales directly from simulated blood culture bottles. J. Microbiol. Methods 2020, 175, 105988. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Bae, I.K.; Lee, Y.-N.; Lee, C.-H.; Lee, S.H.; Jeong, S.H. Detection of extended-spectrum beta-lactamases by using boronic acid as an AmpC beta-lactamase inhibitor in clinical isolates of Klebsiella spp. and Escherichia coli. J. Clin. Microbiol. 2007, 45, 1180–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, W.; Jeong, S.H.; Kim, J.-S.; Kim, H.-S.; Shin, D.H.; Roh, K.H.; Lee, K.M. Use of boronic acid disk methods to detect the combined expression of plasmid-mediated AmpC beta-lactamases and extended-spectrum beta-lactamases in clinical isolates of Klebsiella spp., Salmonella spp., and Proteus mirabilis. Diagn. Microbiol. Infect. Dis. 2007, 57, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Le, P.G.; Moore, K.L.; Church, D.L.; Gregson, D.B. Detection of AmpC β-lactamases in Escherichia coli, Klebsiella spp., Salmonella spp. and Proteus mirabilis in a regional clinical microbiology laboratory. Clin. Microbiol. Infect. 2010, 16, 165–170. [Google Scholar] [CrossRef] [Green Version]
| pAMPC-BL Enzyme | Country of Discovery | Year of Isolation | First Species in Which It Was Isolated | Chromosomal Origin Species | % Similarity (with Respect to the Chromosomal Gene) |
|---|---|---|---|---|---|
| CMY-1 | South Korea | 1989 | K. pneumoniae | A. hydrophila | 82 |
| CMY-2 | Greece | 1996 | K. pneumoniae | C. freundii | 96 |
| MIR-1 | USA | 1990 | K. pneumoniae | E. cloacae | 99 |
| MOX-1 | Japan | 1993 | K. pneumoniae | A. hydrophila | 80 |
| LAT-1 | Greece | 1993 | K. pneumoniae | C. freundii | 95 |
| FOX-1 | Argentina | 1994 | K. pneumoniae | A. caviae | 99 |
| DHA-1 | Saudi Arabia | 1997 | S. enteriditis | M. morganii | 99 |
| ACT-1 | USA | 1997 | K. pneumoniae | E. asburiae | 98 |
| ACC-1 | Germany | 1999 | K. pneumoniae | H. alvei | 99 |
| CFE-1 | Japan | 2004 | E. coli | C. freundii | 99 |
| Test 1 Negative | Test 1 Positive | Total | |
| No AmpC | 86 | 0 | 86 |
| AmpC ± ESBL | 11 | 5 | 16 |
| 97 | 5 | 102 | |
| Test 1 Negative | Test 1 Positive | Total | |
| No AmpC | 86 | 0 | 86 |
| AmpC alone | 4 | 5 | 9 |
| 90 | 5 | 95 |
| Test 2 Negative | Test 2 Positive | Total | |
| No AmpC | 86 | 0 | 86 |
| AmpC ± ESBL | 6 | 10 | 16 |
| 92 | 10 | 102 | |
| Test 2 Negative | Test 2 Positive | Total | |
| No AmpC | 86 | 0 | 86 |
| AmpC alone | 1 | 8 | 9 |
| 87 | 8 | 95 |
| (CI 95%) | Test 1 | Test 2 | ||
|---|---|---|---|---|
| Indicators | AmpC ± ESBL | AmpC Alone | AmpC ± ESBL | AmpC Alone |
| Prevalence | 15.7% | 9.5% | 15.7% | 9.5% |
| Sensitivity | 31.3% (14.2–55.6) | 55.6% (26.7–81.1) | 62.5% (38.6–81.5) | 88.9% (56.5–98) |
| Specificity | 100% (95.7–100) | 100% (95.7–100) | 100% (95.7–100) | 100% (95.7–100) |
| PPV | 100% (56.6–100) | 100 (55.6–100) | 100% (72.2–100) | 100% (67.6–100) |
| NPV | 88.7% (80.8–93.5) | 95.6% (89.1–98.3) | 93.5% (86.5–97) | 98.9% (93.8–99.8) |
| Validity Index | 89.2% (81.7–93.9) | 95.8% (89.7–98.4) | 94.1% (87.8–97.3) | 98.9% (94.3–99.8) |
| Youden’s Index | 0.313 | 0.556 | 0.625 | 0.889 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Guerrero, E.; Callejas-Rodelas, J.C.; Navarro-Marí, J.M.; Gutiérrez-Fernández, J. Systematic Review of Plasmid AmpC Type Resistances in Escherichia coli and Klebsiella pneumoniae and Preliminary Proposal of a Simplified Screening Method for ampC. Microorganisms 2022, 10, 611. https://doi.org/10.3390/microorganisms10030611
Rodríguez-Guerrero E, Callejas-Rodelas JC, Navarro-Marí JM, Gutiérrez-Fernández J. Systematic Review of Plasmid AmpC Type Resistances in Escherichia coli and Klebsiella pneumoniae and Preliminary Proposal of a Simplified Screening Method for ampC. Microorganisms. 2022; 10(3):611. https://doi.org/10.3390/microorganisms10030611
Chicago/Turabian StyleRodríguez-Guerrero, Enrique, Juan Carlos Callejas-Rodelas, José María Navarro-Marí, and José Gutiérrez-Fernández. 2022. "Systematic Review of Plasmid AmpC Type Resistances in Escherichia coli and Klebsiella pneumoniae and Preliminary Proposal of a Simplified Screening Method for ampC" Microorganisms 10, no. 3: 611. https://doi.org/10.3390/microorganisms10030611
APA StyleRodríguez-Guerrero, E., Callejas-Rodelas, J. C., Navarro-Marí, J. M., & Gutiérrez-Fernández, J. (2022). Systematic Review of Plasmid AmpC Type Resistances in Escherichia coli and Klebsiella pneumoniae and Preliminary Proposal of a Simplified Screening Method for ampC. Microorganisms, 10(3), 611. https://doi.org/10.3390/microorganisms10030611









