1. Introduction
Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) is a widely used explosive in the military and construction industries [
1]. RDX is characterized by relatively high thermal stability, high density, and high detonation velocity, all of which promote its extensive use [
2,
3]. Over the past century, RDX has become a significant global pollutant, and there are mounting concerns over its toxicity to biological and environmental systems. The manufacturing, loading, assembly, and packaging of RDX into munitions for use in frontline research, at training sites, and on the battlefield have resulted in terrestrial and aquatic pollution [
4]. Consequently, some sites have been reported to contain explosives and associated compounds at concentrations that threaten the environment and human health [
5]. One such site is Israel’s coastal aquifer below manufacturing facilities, which is contaminated with concentrations of up to 2 mg/L RDX [
6], 1200 mg/L perchlorate, 268 mg/L chlorate, 204 mg/L, and nitrate [
7,
8]. Groundwater supply to surrounding municipalities has been halted because of the pollution. Thus, there is an urgent need to remove the hazardous RDX to ensure public safety and protect natural resources, especially in a semiarid environment such as Israel.
RDX can be biodegraded under both anaerobic and aerobic conditions through various pathways [
9]: reductive, anaerobic, and aerobic ring cleavage leading to mineralization. Under anaerobic conditions, subsequent reduction involves two-electron transfer of the -N-NO
2 functional groups, giving the corresponding nitroso derivatives, such as MNX, DNX, and TNX, (mono-, di-, and tri-nitroso derivatives, respectively), which are more toxic [
10]. On the other hand, aerobic denitration involves cleaving the -N-NO
2 bonds, leading to the formation of 4-nitro-2,4-diazabutanal (NDAB), nitrite, ammonia, formaldehyde, and formic acid [
11]. Several strains of the genus
Rhodococcus, such as DN22 from Australia [
12], 11Y from England [
13], and YH1 [
14], T7, and T9N from Israel [
15], have been identified as RDX degraders via these steps. The denitration mechanism was found to be catalyzed by a unique form of the enzyme cytochrome P450, encoded by the gene
xplA, with NADPH as an electron donor and resulting in NDAB as a stable, non-toxic product [
16]. Thus, the biodegradation capacity of RDX by different bacteria can be used to treat polluted groundwater.
Because RDX is used as a nitrogen source, it is expected that external inorganic nitrogen compounds will affect
xplA expression. For instance, in
Gordonia sp. strain KTR9, nitrogen availability, as ammonium or nitrate, is a significant determinant of RDX degradation and
xplA expression [
17]. Although deletion of
glnR (encoding a regulatory protein affecting nitrogen assimilation in diverse
Actinobacteria) also abolished the inhibition of
xplA expression by nitrite, there was no evidence for a direct role of
glnR in regulating
xplA expression. Instead, the general availability of nitrogen repressed
xplA expression [
18]. Similarly, in
Rhodococcus rhodochrous 11Y,
xplA expression was induced under nitrogen-limiting conditions and further enhanced by the presence of RDX [
19].
Groundwater contaminated with explosives and propellants (perchlorate, for example) is often treated in an aboveground bioreactor, especially when pumping can control fast-migrating pollutants. The advantage of this method is the ability to precisely control biodegradation by maintaining physical and hydrochemical conditions, and the production of high-quality effluents. However, when inorganic co-contaminants are present together with RDX in the bioreactor, they can interfere with each other’s removal, as demonstrated in anaerobic fluidized bed reactors [
20]. In another example, nitrate, RDX, and perchlorate were effectively reduced in microcosms and flow-through columns using a vegetable oil substrate. Nitrate and perchlorate were rapidly biodegraded, followed by RDX and HMX, implying their inhibitory effect [
21]. A later field experiment showed that a bio-barrier containing vegetable oil substrate in a polluted shallow aquifer removed both explosives and perchlorate. However, the toxic intermediate(s) of anaerobic degradation of RDX and metals such as arsenic were observed in the downgradient treated water, emphasizing the limitation of the anaerobic remediation approach [
22].
The potential effect of inorganic co-contaminants in the groundwater on RDX degradation depends on the redox conditions—aerobic or anaerobic. The co-contaminants can compete with RDX as substrate and may have a toxic effect on the degrading organisms. Under aerobic conditions, nitrate can serve as an alternative nitrogen source for the RDX-degrading bacteria [
23,
24]; however, the effects of perchlorate and chlorate are not known. Under anaerobic conditions, perchlorate, chlorate, or nitrate can serve as favored electron acceptors [
22]. Nitrate can also serve as an alternative to nitrogen. Perchlorate or chlorate (strong oxidizers) can induce oxidative stress in the cells and thus slow down degradation [
25]. It has been shown that in the genomes of perchlorate-reducing bacteria, the perchlorate reduction genomic island always includes oxidative stress genes, such as
rpoS, which indicates the oxidative damage caused by perchlorate [
26].
Because the aerobic degradation of RDX does not produce toxic intermediates, the use of RDX-degrading strains, such as
Gordonia KTR9, has been tested for the bioremediation of polluted water [
27,
28,
29]. Those studies showed that in situ bioaugmentation and biostimulation of aerobic bacteria lead to rapid degradation of RDX in a polluted aquifer over a long period [
30]. In Israel, in contrast, the planned remediation of the coastal aquifer is via the “pump and treat” method to control the migration of perchlorate and chlorate [
31], where six production wells are expected to supply 650 m
3/h each to a biological treatment plant. This provides an opportunity to design an aerobic–anaerobic system to treat RDX first aerobically, and then the rest of the contaminants in an anaerobic stage. However, the fundamental question of how aerobic RDX degradation is affected by co-contaminants has never been addressed.
We hypothesized that during ex situ (pump and treat) bioremediation of polluted groundwater, the occurrence of toxic co-contaminants will affect the efficiency of aerobic RDX degradation by
Rhodococcus strains in bioreactors treating contaminated groundwater. Our research objectives were first, to test and compare the effects of the co-contaminants on the RDX degraders’ activity and growth. These degraders had been isolated from different environments, such as the Israeli coastal aquifer, and included
Rhodococcus strains YH1 [
14] and T7 [
15], and
Gordonia strain YY1 [
32]. Then, we examined the effects of the co-contaminants (individually or as a mixture) on the intrinsic degradation activity of strain YH1. Finally, we tested whether the co-contaminants affect
xplA expression in strain YH1. Our results are expected to contribute to the better design of an ex-situ bioreactor for treating groundwater containing RDX and co-contaminants in Israel using the effective RDX degraders—
Rhodococcus strains.
3. Results
The study’s basic assumption was that perchlorate, chlorate, and nitrate, individually and as a mixture, would have either an adverse effect or no effect on RDX degradation by the three tested bacterial strains. The initial examination involved analyzing these strains’ growth and degradation activity in the presence of these environmentally relevant co-contaminants, found in polluted groundwater in Israel. In the presence of perchlorate, chlorate, nitrate, and their mixture, biomass yield after 96 h depended on the strain and treatment. Two-way RM (repeated measures) ANOVA revealed a significant effect of strain type on the biomass yield for all treatments (p < 0.0001). Pairwise comparison of biomass yield after 96 h of control treatment showed significantly higher yield for YH1 vs. T7 (p < 0.05) but not as compared to strain YY1. In the presence of perchlorate and chlorate, the biomass yield of YH1 (96 h) was significantly higher than those of the other two strains. This was not the case with the nitrate treatment. With the mixture of co-contaminants, the biomass yield of strain YY1 was higher than that of strain T7 (p < 0.001).
3.1. Growth Rates in the Presence of Environmentally Relevant Co-Contaminants
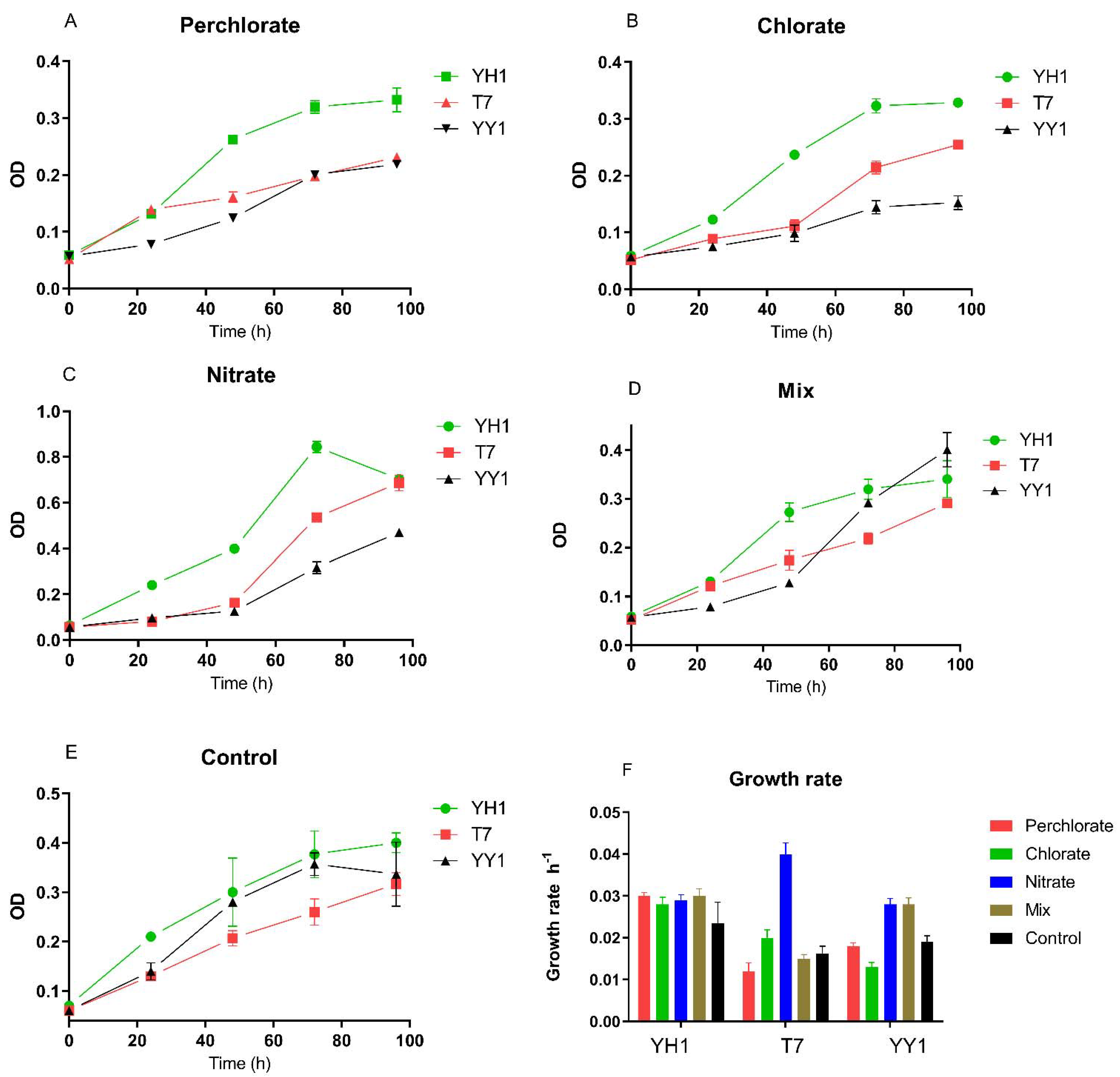
Growth rates and doubling times were extracted from the growth curves by nonlinear regression of the log phase (
Figure 1). We observed varied effects of perchlorate and chlorate on the growth of the strains when RDX was the sole nitrogen source. Perchlorate increased the growth rate of YH1 compared to the control, but this increase was not significant. For strain T7, the growth rate on nitrate was significantly higher than that for the control and the rest of the treatments. For strain YY1, the growth rate on nitrate and mixed contaminants was not significantly different but was higher than for the other treatments. Neither perchlorate nor chlorate were transformed in any of the experiments.
3.2. RDX Degradation Rates in the Presence of Environmentally Relevant Co-Contaminants
A surprising observation was the acceleration of RDX degradation in the presence of co-contaminants (
Figure 2). The effect was obvious in the presence of perchlorate, chlorate, and the contaminant mixture for strain YH1. Two-way RM ANOVA of the data revealed a significant effect of strain type on degradation constant for all treatments (
p < 0.0001). For strain YH1, degradation constants were significantly higher for the perchlorate, chlorate, and mixed treatments than for the nitrate and control treatments. For strain T7, chlorate significantly decreased the degradation constant compared to the other treatments, but the degradation constants in the other treatments also differed. For YY1, only nitrate significantly increased degradation constants, whereas the other treatments were not significantly different.
3.3. Response of the Different Strains to Increasing Co-Contaminant Concentration
The dose response of the different strains was tested with a series of increasing concentrations of contaminants. It was hypothesized that the effect of increasing nitrate concentration on RDX degradation would be competitive; the strains would utilize nitrate and glucose, resulting in carbon limitation in the medium, and impairing RDX degradation. Furthermore, increasing concentrations of perchlorate and chlorate might affect bacterial growth and activity either via toxicity, or by affecting the salinity of the medium. To distinguish between these mechanisms, we included NaCl as a control.
Nitrate did not influence the extent of RDX degradation by strains YH1 and YY1, even at 50 mM after 96 h incubation. Strain T7 was affected by 20 and 50 mM nitrate, with 71.5 ± 2.8 and 61.2 ± 6.7% degradation (of the degradation in controls) after 96 h, respectively. Increasing nitrate concentration to 1 mM resulted in a 2-fold increase in biomass yield after 96 h compared to controls; however, further increases in nitrate concentration did not increase the yield for any of the strains (
Figure S1). When evaluating the total organic carbon and total nitrogen remaining after 96 h (
Figure 3), it appeared that even at 1 mM nitrate, 90% of the carbon was consumed. This suggests that the carbon in the medium becomes limiting, which may prevent further RDX degradation. However, this suggestion is probably only valid for strain T7, which did not degrade RDX completely. At the same time, concurrent RDX and nitrate utilization was apparent for all strains, but the results imply that from 5 to 50 mM nitrate, RDX is preferentially utilized. Overall, RDX degradation was not sensitive to nitrate competition, even under extreme concentrations.
Increasing perchlorate and chlorate concentrations did not affect the extent of RDX degradation by strains YH1 or YY1. For strain T7, 50 mM chlorate reduced RDX degradation to 60% (data not shown) at 96 h. The average control OD
600 after 96 h was 0.3, 0.28, and 0.28 for strains YH1, T7, and YY1, respectively (
Figure 4). For strain YH1, neither perchlorate nor NaCl influenced degradation or yield; a 33% decrease in yield was observed under 50 mM chlorate with no impact on degradation (
Figure 4). Although neither perchlorate nor chlorate affected RDX degradation by strain YY1, they both reduced the biomass yield of this strain. At the same time, increased salinity did not affect either degradation or yield, implying that perchlorate and chlorate have a toxic effect on strain YY1. For strain T7, NaCl had no effect, and increasing concentrations of perchlorate did not change the culture OD. Increasing chlorate reduced biomass yield by up to 50% compared to the control at 50 mM (
Figure 4).
3.4. Effect of Co-Contaminants on the Intrinsic Activity of XplA in Strains YH1, T7, and YY1
The effects of co-contaminants perchlorate and chlorate were also tested in a short-term assay. In the current study, chlorate and perchlorate unexpectedly enhanced the RDX degradation. Cells were grown in minimal medium containing RDX and perchlorate (800 mg/L) or chlorate (200 mg/L), and glucose as a carbon source. The cells were harvested and washed twice in minimal medium and transferred to fresh medium containing different concentrations of RDX (1–20 mg/L), and co-contaminants perchlorate (800 mg/L) or chlorate (200 mg/L), without carbon. The initial OD600 was 0.05 and the reaction time was 3 h.
The resting cell experiment aims to test the direct effect of the perchlorate and chlorate on the intrinsic activity of cytochrome P450, whereas in
Figure 2, the effect was tested on growing cultures, so different degradation rates are to be expected. Nonlinear curve fitting produced the kinetic parameters of the Michaelis–Menten equation. As seen in
Figure 5, there was no clear trend for the effects of perchlorate and chlorate on Vmax and Km. For example, Vmax in the presence of chlorate was significantly higher than in the presence of perchlorate for YH1, but not for T7 or YY1. For these latter two strains, Vmax of the control was considerably higher than with the chlorate and perchlorate treatments, suggesting their sensitivity to these oxyanions. In strain YH1, only chlorate significantly reduced Km relative to the control.
3.5. Expression of xplA in Co-Contaminant-Exposed Culture of Strain YH1
An additional effect of co-contaminants on RDX degradation might be related to interference with
xplA expression. Thus, we determined
xplA expression in the presence of perchlorate, the main co-contaminant in the groundwater because no previous studies have reported RDX degradation and
xplA expression in the presence of perchlorate under aerobic conditions. Strain YH1 was pre-grown in minimal medium supplemented with RDX (20 mg/L). Then, the following treatments were applied: RDX + perchlorate, perchlorate alone, perchlorate + ammonium, and perchlorate + nitrate, with glucose as the energy and carbon source. When the culture was spiked with perchlorate + RDX,
xplA expression was enhanced about 3-fold at 12 h and then declined (
Figure 6). A significant constant increase in expression level was seen between 12 and 24 h incubation with perchlorate alone; expression then declined significantly at 36 h.
The observation of peak
xplA expression for RDX + perchlorate at 12 h followed by a significant decline agreed with our observation of complete degradation of RDX in the presence of perchlorate (
Figure 2). As expected, ammonium and nitrate with perchlorate did not induce
xplA expression.
No growth occurred in the treatment with perchlorate alone and, therefore, the question of whether XplA is active was investigated (
Figure S2). XplA in the perchlorate-exposed cell suspension was active, albeit less than in the control (RDX), and at the same level as in the ammonium + perchlorate treatment (
Figure S3). The lowest degradation activity was found for the perchlorate + nitrate treatment.
To further examine the factors affecting
xplA expression, we investigated the effect of ammonium. Only 30% degradation of RDX was observed with ammonium alone (
Figure 7A), whereas complete degradation was achieved in the control after 36 h. The expression of
xplA in this culture was significantly higher (~6- to 7-fold) after 24 and 36 h incubation (
Figure 7B). However, in the treatment with both RDX and ammonium, the expression of
xplA decreased compared to ammonia alone but was still significantly higher than the control, from 2-fold to 3-fold at 24 h and 36 h. The increased expression in the ammonium + RDX treatment was not translated to degradation activity, suggesting post-transcriptional regulation. Furthermore, when 24 h culture was harvested from the ammonium treatment, washed, and suspended in medium with RDX (no carbon), the degradation rate was 0.43 mg/L per h, 50% lower than for the RDX control (0.82 mg/L per h) (
Figure S4).
4. Discussion
The polluted groundwater in the Israeli coastal aquifer contains high concentrations of perchlorate, chlorate, and nitrate at the plume center, which can affect RDX biodegradation. In contrast to anaerobic treatment, aerobic treatment of the polluted water followed by anaerobic perchlorate reduction can provide effluents without toxic reduced RDX intermediates. However, the co-contaminants’ potential to affect aerobic RDX degradation had never been evaluated. Our results reveal a significant strain-specific effect of the co-contaminants, with faster growth and faster degradation for strain YH1 compared to strains T7 and YY1, suggesting better adaptation.
In Israel’s polluted groundwater, nitrate is the primary inorganic nitrogen that can affect RDX degradation. In previous studies, the effect of nitrate on the growth and activities of the tested strains was examined. Strain YH1 degraded RDX in the presence of 1000 mg/L nitrate, albeit at a slower rate than in the control, suggesting competition with nitrogen [
24]. Strain T7, however, did not degrade RDX in the presence of nitrate, but at the same time, it proliferated. With this strain, not all of the nitrate was consumed during incubation, implying that the culture had become carbon-limited [
15]. Strain YY1 was not examined so far, and the current results suggest that its degradation activity is insensitive to nitrate (at the level tested). In comparison, strain 11Y cells grown under nitrogen-limiting conditions (450 μM NH
4Cl, 450 μM KNO
3, 450 μM KNO
2, and 150 μM RDX) or incubated with no nitrogen, degraded RDX faster than cells grown under non-limiting nitrogen conditions (5 mM NH
4Cl or KNO
3) [
19]. For this strain, external nitrogen, including nitrate, suppressed
xplA expression, and this might be the mechanism affecting RDX degradation by strain YH1.
In contrast to nitrate, the effects of perchlorate or chlorate on RDX degradation are not known. It appears that in strains T7 and YY1, RDX degradation is insensitive to perchlorate, whereas strain T7 was negatively affected by chlorate. For strain YH1, the negative effect of nitrate on RDX degradation was offset by perchlorate and chlorate in the mixed treatment. These co-contaminants can impact either xplA directly or the expression of
xplA. Our results disagreed with the observation of nitrate’s effect on RDX degradation by strain 11Y, where at 5 mM, it retarded the degradation rate [
19]. Our combined observations of the growth and degradation kinetics of the three strains (
Figure 1 and
Figure 2) and the effect of nitrate concentrations indicate that they can also be effective in a high nitrate environment.
Our results suggest that chlorate is more toxic than perchlorate to all strains. Chlorate toxicity can result from its nonspecific reduction to chlorite by respiratory nitrate reductase [
36,
37]. In the current study, however, all strains could use nitrate as nitrogen through assimilatory nitrate reduction to ammonium. None contained the membrane-bound dissimilatory nitrate reductase (
narG) [
38].
The effect of the different co-contaminants on RDX degradation kinetics can be competitive, non-competitive, or uncompetitive, reflecting other interactions with the cytochrome P450 or enzyme-substrate complex. There was no effect for perchlorate, whereas for chlorate, the significant reduction in Vmax and Km were characteristic of uncompetitive inhibition. In the mixed treatment, the increase in Km was characteristic of a competitive effect; however, the rise in Vmax did not support this possibility. Chlorate is an inhibitor of various enzymes, including nitrite oxidoreductase and nitrate reductase, and acts as a competitive substrate for ATP-sulfurylase, a highly conserved prerequisite enzyme of the sulfate-reduction pathway [
39,
40,
41,
42]. Neither chlorate nor perchlorate is known to be an inhibitor of cytochrome P450 (XplA); however, chlorate may interfere with oxygen binding to the ferrous heme center of the enzyme. The observed increase in reaction velocity with the mixed co-contaminants was unusual, implying faster turnover of RDX by XplA. The addition of nitrate was responsible for this effect, yet the mechanism remains unclear in our study.
In other studies, expression of
xplA in
Gordonia sp. strain KTR9 was strongly induced by nitrogen limitation [
18]. In experiments where ammonium (0.9 mM) in the culture medium was consumed,
xplA expression increased up to 150-fold.
R. rhodochrous 11Y expression of
xplA (1.5-fold increase) and translation of XplA were also regulated by nitrogen limitation [
19]. Resting cells of strain 11Y degraded RDX fastest after pre-growth on 250 μM RDX, and then 150 μM RDX, followed by pre-growth on 750 μM NH
4Cl and then 450 μM NH
4Cl. High nitrate and ammonium concentrations slowed down the activity of the resting cells. As for YH1, ammonium and nitrite strongly repressed cytochrome P450 (XplA) expression and RDX degradation in growing cultures. Nevertheless, ammonium itself did not suppress
xplA transcription (
Figure 7B); however, it seems that post-transcriptional regulation stops RDX degradation. Taken together,
xplA expression is controlled by nitrogen limitation; its fold increase in the presence of perchlorate in YH1 is likely due to nitrogen limitation in the culture. Nevertheless, the activity of xplA in the 24 h cell suspension was lower (0.43 mg/L per h) than in the control (0.82 mg/L per h). This lower activity is likely due to the absence of reducing power (NADPH) in the non-growing culture (
Figure S2). Our results indicate that there is no need for RDX as an inducer for active xplA, agreeing with the findings for strains 11Y and KTR9 [
18,
19].
5. Conclusions
We hypothesized that during ex-situ (pump and treat) bioremediation of RDX-contaminated groundwater, the presence of co-contaminants would hinder the aerobic degradation by Rhodococcus strains. Among the tested co-contaminants, we assumed that perchlorate and chlorate would be toxic, and nitrate would be a competitive inhibitor for degradation. The assumptions were tested for three RDX-degrading strains under different conditions. First, the effect of co-contaminants (at environmentally relevant concentrations) was found to be strain-specific, with YH1 being the least sensitive to them as its growth rate was not influenced. However, the first-order degradation constant was higher in the presence of perchlorate than in the control. Degradation and growth inhibition by chlorate were observed only for T7, raising a few questions: Is the effect concentration-dependent? Is the impact directly on cytochrome P450, or is it due to suppression of xplA expression?
With respect to nitrate, only T7 was affected by extremely high nitrate, whereas degradation by YH1 and YY1 was not affected. The amount of DOC remaining after 96 h suggests that T7 was carbon-limited. Nevertheless, most of the DN (nitrate) remained, and thus, nitrate may act directly on the enzyme or on xplA expression (in this strain). Similarly, high chlorate and perchlorate did not reduce the extent of RDX degradation. Still, chlorate reduced the biomass yield for all strains, particularly T7, indicating its toxicity, even at 1 mM. The mechanisms underlying this toxicity are not yet known. The strains were able to overcome this stress, as inferred from the absence of biomass decline with increased chlorate.
Expression of xplA increased in the presence of perchlorate alone after 24 h incubation of strain YH1, in contrast to the control treatments (perchlorate + RDX, perchlorate + nitrate, and perchlorate + ammonium). The xplA at this stage is active but at a lower rate than in the control, suggesting post-transcriptional regulation. In the treatment with perchlorate alone, the increased expression is likely due to nitrogen limitation. As expected, ammonium + nitrate repressed xplA expression. Although strong xplA expression was observed in ammonium-exposed cells, xplA activity was much lower than in the control, again indicating post-transcriptional regulation.
In conclusion, the examined strains demonstrated variable responses to the co-contaminants, with strain YH1 being the least affected and strain T7 the most. All three strains can be used for bioremediation of RDX-polluted groundwater containing an environmentally relevant mixture of co-contaminants.