Dihydrolipoamide Acetyltransferase AceF Influences the Type III Secretion System and Resistance to Oxidative Stresses through RsmY/Z in Pseudomonas aeruginosa
Abstract
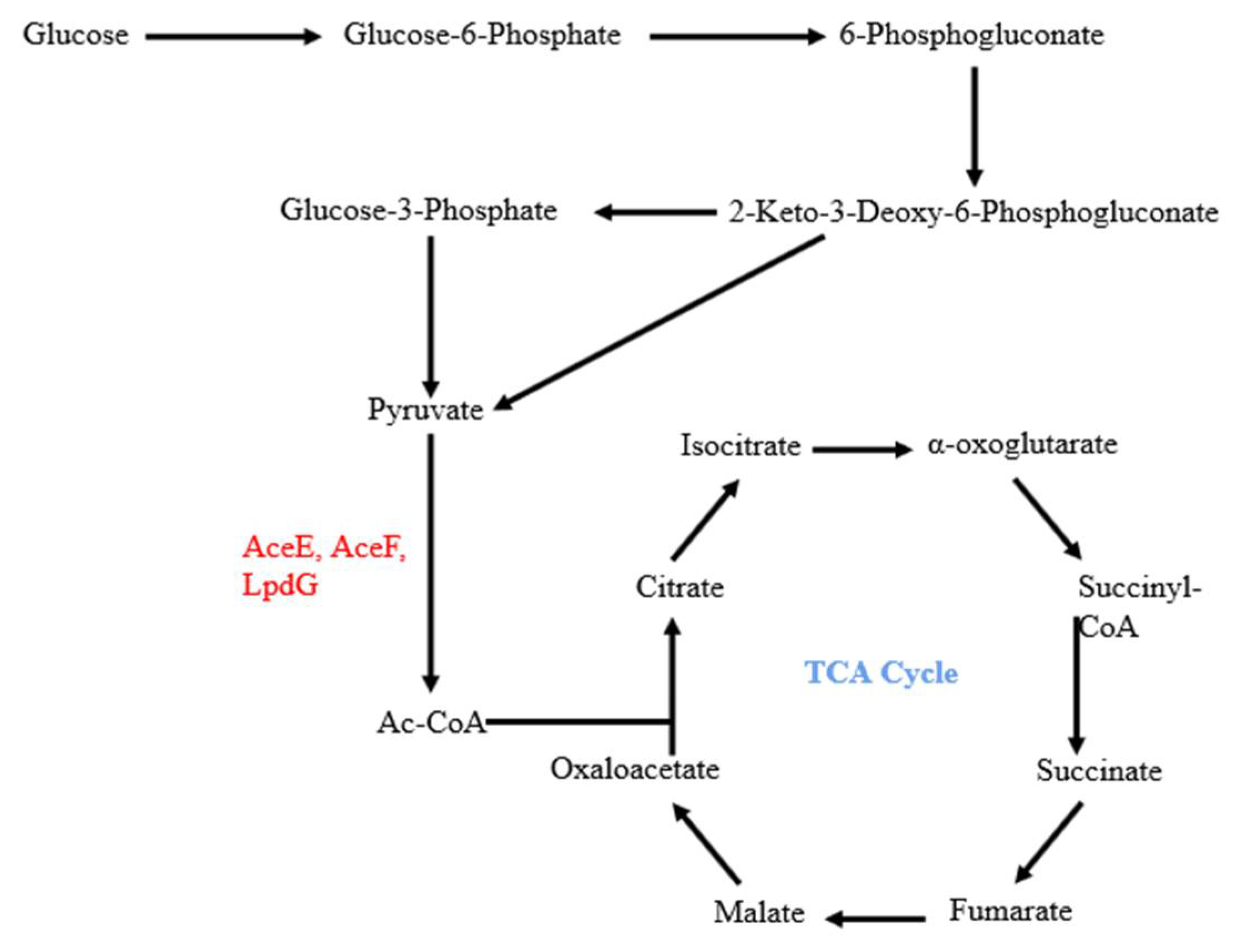
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
2.2. Cytotoxicity Assay
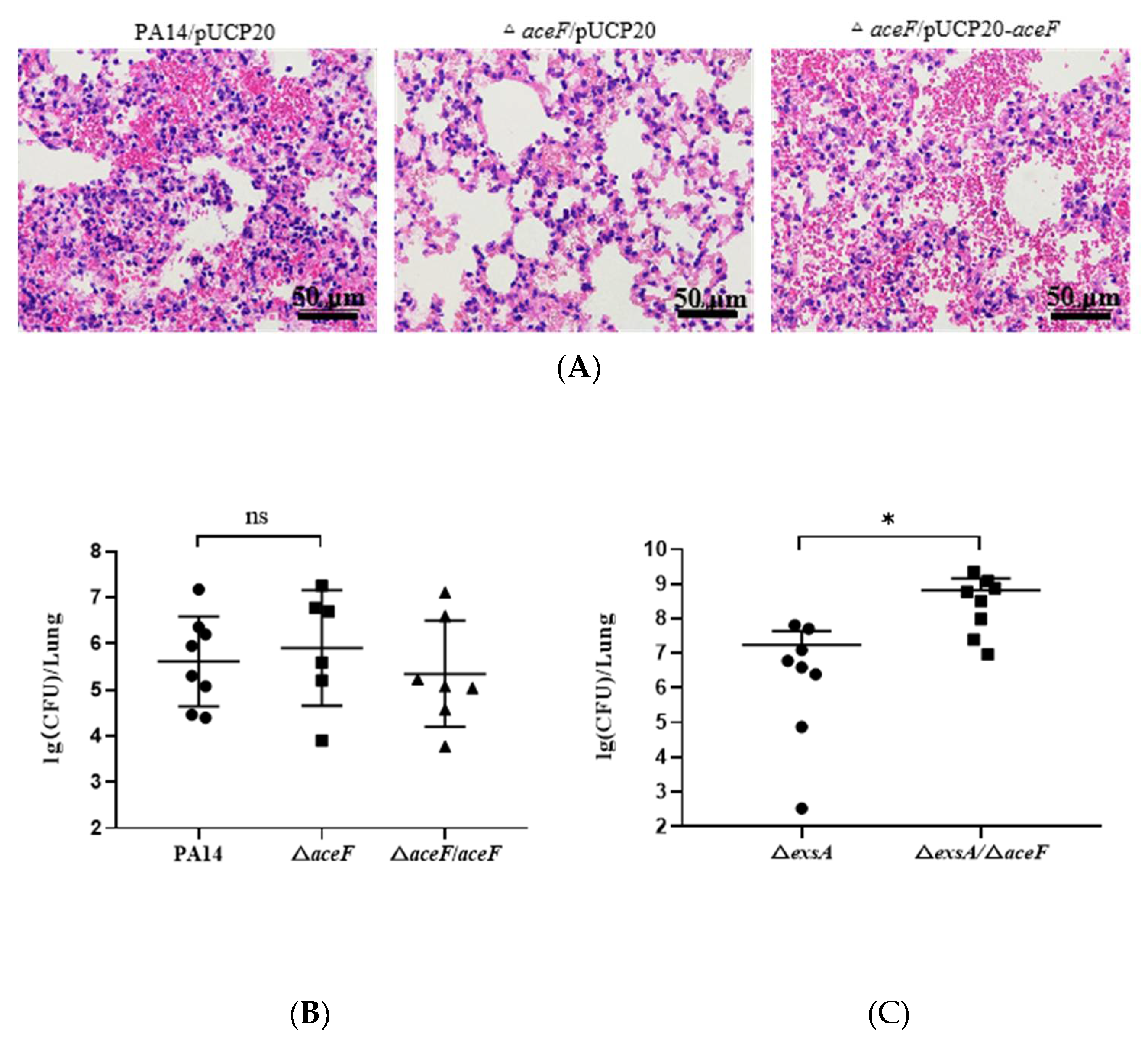
2.3. Murine Acute Pneumonia Model
2.4. Histology
2.5. Real-Time qPCR
2.6. β-Galactosidase Assay
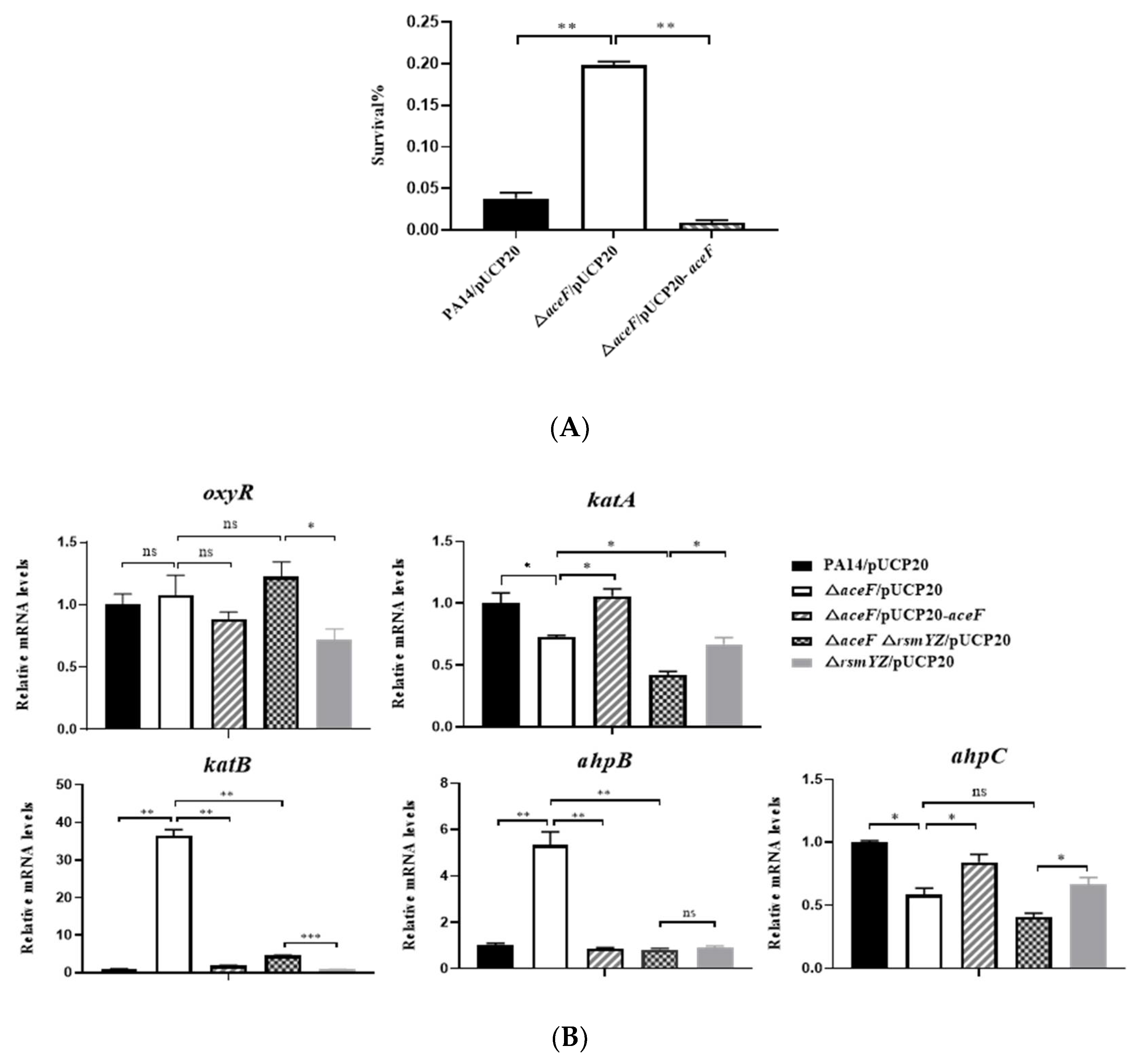
2.7. H2O2 Susceptibility Assay
3. Results
3.1. Mutation in the aceF Gene Reduces the Expression of Type III Secretion System Genes
3.2. AceF Affects the Expression of the T3SS Genes through RsmY and RsmZ
3.3. Role of AceF in Bacterial Virulence in an Acute Pneumonia Model
3.4. Mutation of the aceF gene Increases Bacterial Tolerance to Hydrogen Peroxide
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Driscoll, J.A.; Brody, S.L.; Kollef, M.H. The Epidemiology, Pathogenesis and Treatment of Pseudomonas aeruginosa Infections. Drugs 2007, 67, 351–368. [Google Scholar] [CrossRef] [PubMed]
- El-Solh, A.A.; Hattemer, A.; Hauser, A.R.; Alhajhusain, A.; Vora, H. Clinical outcomes of type III Pseudomonas aeruginosa bacteremia. Crit. Care Med. 2012, 40, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.R. The type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nat. Rev. Genet. 2009, 7, 654–665. [Google Scholar] [CrossRef] [Green Version]
- Bagayoko, S.; Leon-Icaza, S.A.; Pinilla, M.; Hessel, A.; Meunier, E. Phospholipid peroxidation fuels ExoU phospholipase-dependent cell necrosis and supports Pseudomonas aeruginosa-driven pathology. bioRxiv 2021, 17, e1009927. [Google Scholar]
- Hauser, A.R.; Cobb, E.; Bodí, M.; Mariscal, D.; Vallés, J.; Engel, J.N.; Rello, J. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 2002, 30, 521–528. [Google Scholar] [CrossRef]
- Brutinel, E.D.; Yahr, T.L. Control of gene expression by type III secretory activity. Curr. Opin. Microbiol. 2008, 11, 128–133. [Google Scholar] [CrossRef] [Green Version]
- Marsden, A.E.; Intile, P.J.; Schulmeyer, K.H.; Simmons-Patterson, E.R.; Urbanowski, M.L.; Wolfgang, M.C.; Yahr, T.L. Vfr Directly Activates exsA Transcription To Regulate Expression of the Pseudomonas aeruginosa Type III Secretion System. J. Bacteriol. 2016, 198, 1442–1450. [Google Scholar] [CrossRef] [Green Version]
- Thibault, J.; Faudry, E.; Ebel, C.; Attree, I.; Elsen, S. Anti-activator ExsD Forms a 1:1 Complex with ExsA to Inhibit Transcription of Type III Secretion Operons. J. Biol. Chem. 2009, 284, 15762–15770. [Google Scholar] [CrossRef] [Green Version]
- McMackin, E.A.W.; Djapgne, L.; Corley, J.M.; Yahr, T.L. Fitting Pieces into the Puzzle of Pseudomonas aeruginosa Type III Secretion System Gene Expression. J. Bacteriol. 2019, 201, e00209-19. [Google Scholar] [CrossRef] [Green Version]
- Janssen, K.H.; Corley, J.M.; Djapgne, L.; Cribbs, J.T.; Voelker, D.; Slusher, Z.; Nordell, R.; Regulski, E.E.; Kazmierczak, B.I.; McMackin, E.W.; et al. Hfq and sRNA 179 Inhibit Expression of the Pseudomonas aeruginosa cAMP-Vfr and Type III Secretion Regulons. MBio 2020, 11, e00363-20. [Google Scholar] [CrossRef]
- Janssen, K.H.; Diaz, M.R.; Golden, M.; Graham, J.W.; Yahr, T.L. Functional analyses of the RsmY and RsmZ small non-coding regulatory RNAs in Pseudomonas aeruginosa. J. Bacteriol. 2018, 200, e00736-17. [Google Scholar] [PubMed] [Green Version]
- Allsopp, L.P.; Wood, T.E.; Howard, S.A.; Maggiorelli, F.; Nolan, L.M.; Wettstadt, S.; Filloux, A. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2017, 114, 7707–7712. [Google Scholar] [CrossRef] [Green Version]
- Irie, Y.; Starkey, M.; Edwards, A.N.; Wozniak, D.J.; Romeo, T.; Parsek, M.R. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 2010, 78, 158–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapouge, K.; Schubert, M.; Allain, F.H.-T.; Haas, D. Gac/Rsm signal transduction pathway of γ-proteobacteria: From RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008, 67, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Chambonnier, G.; Roux, L.; Redelberger, D.; Fadel, F.; Bordi, C. The Hybrid Histidine Kinase LadS Forms a Multicomponent Signal Transduction System with the GacS/GacA Two-Component System in Pseudomonas aeruginosa. PLoS Genet. 2016, 12, e1006032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, A.L.; Merighi, M.; Hyodo, M.; Ventre, I.; Filloux, A.; Lory, S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009, 23, 249–259. [Google Scholar] [CrossRef] [Green Version]
- Ryan Kaler, K.M.; Nix, J.C.; Schubot, F.D. RetS inhibits Pseudomonas aeruginosa biofilm formation by disrupting the canonical histidine kinase dimerization interface of GacS. J. Biol. Chem. 2021, 297, 101193. [Google Scholar] [CrossRef]
- Francis, V.I.; Waters, E.; Finton-James, S.E.; Gori, A.; Kadioglu, A.; Brown, A.R.; Porter, S.L. Multiple communication mechanisms between sensor kinases are crucial for virulence in Pseudomonas aeruginosa. Nat. Commun. 2018, 9, 2219. [Google Scholar] [CrossRef]
- Laskowski, M.A.; Kazmierczak, B.I. Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infect. Immun. 2006, 74, 4462–4473. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.X.; Wheeler, K.M.; Cady, K.C.; Lehoux, S.; Cummings, R.D.; Laub, M.T.; Ribbeck, K. Mucin Glycans Signal through the Sensor Kinase RetS to Inhibit Virulence-Associated Traits in Pseudomonas aeruginosa. Curr. Biol. 2021, 31, 90–102. [Google Scholar] [CrossRef]
- Kong, W.; Lin, C.; Zhao, J.; Shen, T.; Surette, M.G.; Shen, L.; Duan, K.J. Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol. 2013, 88, 784–797. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Wang, D.; Pan, X.; Xia, B.; Weng, Y.; Long, Y.; Ren, H.; Zhou, J.; Jin, Y.; Bai, F.; et al. TpiA is a Key Metabolic Enzyme That Affects Virulence and Resistance to Aminoglycoside Antibiotics through CrcZ in Pseudomonas aeruginosa. MBio 2020, 11, e02079-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, D.-K.; Filopon, D.; Chaker, H.; Boullanger, S.; Derouazi, M.; Polack, B.; Toussaint, B. High-cell-density regulation of the Pseudomonas aeruginosa type III secretion system: Implications for tryptophan catabolites. Microbiology 2008, 154, 2195–2208. [Google Scholar] [CrossRef] [Green Version]
- Rietsch, A.; Wolfgang, M.C.; Mekalanos, J.J. Effect of Metabolic Imbalance on Expression of Type III Secretion Genes in Pseudomonas aeruginosa. Infect. Immun. 2004, 72, 1383–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, J.C.S.; Rzhepishevska, O.; Ramstedt, M.; Welch, M. Type III secretion system expression in oxygen-limited Pseudomonas aeruginosa cultures is stimulated by isocitrate lyase activity. Open Biol. 2013, 3, 120131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonnleitner, E.; Abdou, L.; Haas, D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2009, 106, 21866–21871. [Google Scholar] [CrossRef] [PubMed]
- Naftalin, R.J.; Neale, H.; Simmons, N.L. Proceedings: Methods used to estimate the unidirectional fluxes of sodium simultaneously across the mucosal and serosal borders of rabbit ileum. J. Physiol. 1975, 252, 7P–8P. [Google Scholar] [PubMed]
- Rietsch, A.; Mekalanos, J.J. Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 2005, 59, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.T.; Karkhoff-Schweizer, R.R.; Kutchma, A.J.; Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998, 212, 77–86. [Google Scholar] [CrossRef]
- Wu, W.; Huang, J.; Duan, B.; Traficante, D.C.; Hong, H.; Risech, M.; Lory, S.; Priebe, G.P. Th17-stimulating protein vaccines confer protection against Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 2012, 186, 420–427. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.; Pan, X.; Xia, B.; Fei, C.; Jin, Y.; Fang, B.; Gregory, P.; Cheng, Z.; Jin, S.; Wu, W. Construction of a Protective Vaccine Against Lipopolysaccharide-Heterologous Pseudomonas aeruginosa Strains Based on Expression Profiling of Outer Membrane Proteins During Infection. Front. Immunol. 2018, 9, 1737. [Google Scholar]
- Weng, Y.; Chen, F.; Liu, Y.; Zhao, Q.; Chen, R.; Pan, X.; Liu, C.; Cheng, Z.; Jin, S.; Jin, Y.; et al. Pseudomonas aeruginosa Enolase Influences Bacterial Tolerance to Oxidative Stresses and Virulence. Front. Microbiol. 2016, 7, 1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savli, H.; Karadenizli, A.; Kolayli, F.; Gundes, S.; Ozbek, U.; Vahaboglu, H. Expression stability of six housekeeping genes: A proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J. Med. Microbiol. 2003, 52, 403–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, M.S.; Matthews, W.J., Jr.; Kang, Y.; Nguyen, D.T.; Hoang, T.T. In Vivo Evidence of Pseudomonas aeruginosa Nutrient Acquisition and Pathogenesis in the Lungs of Cystic Fibrosis Patients. Infect. Immun. 2007, 75, 5313–5324. [Google Scholar] [CrossRef] [Green Version]
- Miller, J.H. Assay of β-Galactosidase. In Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]
- Diaz, M.H.; Hauser, A.R. Pseudomonas aeruginosa Cytotoxin ExoU Is Injected into Phagocytic Cells during Acute Pneumonia. Infect. Immun. 2010, 78, 1447–1456. [Google Scholar] [CrossRef] [Green Version]
- Howell, H.A.; Logan, L.K.; Hauser, A.R. Type III Secretion of ExoU Is Critical during Early Pseudomonas aeruginosa Pneumonia. MBio 2013, 4, e00032-13. [Google Scholar] [CrossRef] [Green Version]
- Rangel, S.M.; Diaz, M.H.; Knoten, C.A.; Zhang, A.; Hauser, A.R. The Role of ExoS in Dissemination of Pseudomonas aeruginosa during Pneumonia. PLoS Pathog. 2015, 11, e1004945. [Google Scholar] [CrossRef] [Green Version]
- Arai, T.; Kamiya, J.; Nagino, M.; Uesaka, K.; Yuasa, N.-H.; Oda, K.; Sano, T.; Yoshikai, Y.; Nimura, Y. Bilirubin impairs neutrophil bactericidal activity through the antioxidant mechanism in vitro. Gastroenterology 2000, 118, A1332. [Google Scholar] [CrossRef]
- Ziltener, P.; Reinheckel, T.; Oxenius, A. Neutrophil and Alveolar Macrophage-Mediated Innate Immune Control of Legionella pneumophila Lung Infection via TNF and ROS. PLoS Pathog. 2016, 12, e1005591. [Google Scholar] [CrossRef]
- Vogt, S.L.; Green, C.; Stevens, K.M.; Day, B.; Erickson, D.L.; Woods, D.E.; Storey, D.G. The Stringent Response Is Essential for Pseudomonas aeruginosa Virulence in the Rat Lung Agar Bead and Drosophila melanogaster Feeding Models of Infection. Infect. Immun. 2011, 79, 4094–4104. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, F.A.; Shaker, G.H.; Askoura, M.M. Oxidative Stress Influences Pseudomonas aeruginosa Susceptibility to Antibiotics and Reduces Its Pathogenesis in Host. Curr. Microbiol. 2020, 77, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Heo, Y.J.; Lee, J.K.; Cho, Y.H. KatA, the major catalase, is critical for osmoprotection and virulence in Pseudomonas aeruginosa PA14. Infect. Immun. 2005, 73, 4399–4403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochsner, U.A.; Vasil, M.L.; Alsabbagh, E.; Parvatiyar, K.; Hassett, D.J. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 2000, 182, 4533–4544. [Google Scholar] [CrossRef] [Green Version]
- Jo, I.; Chung, I.Y.; Bae, H.W.; Kim, J.S.; Song, S.; Cho, Y.H.; Ha, N.C. Structural details of the OxyR peroxide-sensing mechanism. Proc. Natl. Acad. Sci. USA 2015, 112, 6443–6448. [Google Scholar] [CrossRef] [Green Version]
- Wei, Q.; Le, M.; Andreas, D.; Falk, H.; Warunya, P.; Ameer, E.; Sebastian, S.; Stéphane, P.; Daniel, C.; Daniel, H. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 2012, 40, 4320–4333. [Google Scholar] [CrossRef] [PubMed]
- Khakimova, M.; Ahlgren, H.G.; Harrison, J.; English, A.M.; Nguyen, D. The Stringent Response Controls Catalases in Pseudomonas aeruginosa and Is Required for Hydrogen Peroxide and Antibiotic Tolerance. J. Bacteriol. 2013, 195, 2011–2020. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Weng, Y.; Xu, C.; Wang, D.; Pan, X.; Tian, Z.; Xia, B.; Li, H.; Chen, R.; Liu, C.; et al. Endoribonuclease YbeY Is Essential for RNA Processing and Virulence in Pseudomonas aeruginosa. MBio 2020, 11, e00659-20. [Google Scholar] [CrossRef]
- Stephan, H.; Claudio, V.; Cécile, G.-B.; Dieter, H. Role of the stress sigma factor RpoS in GacA/RsmA-controlled secondary metabolism and resistance to oxidative stress in Pseudomonas fluorescens CHA0. FEMS Microbiol. Lett. 2005, 243, 251–258. [Google Scholar]
- Pletzer, D.; Blimkie, T.; Wolfmeier, H.; Li, Y.; Baghela, A.; Lee, A.H.Y.; Falsafi, R.; Hancock, R.E.W. The Stringent Stress Response Controls Proteases and Global Regulators under Optimal Growth Conditions in Pseudomonas aeruginosa. MSystems 2020, 5, e00495-20. [Google Scholar] [CrossRef]
- Chowdhury, N.; Kwan, B.W.; Wood, T.K. Persistence Increases in the Absence of the Alarmone Guanosine Tetraphosphate by Reducing Cell Growth. Sci. Rep. 2016, 6, 20519. [Google Scholar] [CrossRef]
- Battesti, A.; Bouveret, E. Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J. Bacteriol. 2009, 191, 616–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battesti, A.; Bouveret, E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 2006, 62, 1048–1063. [Google Scholar] [CrossRef] [PubMed]
- Sekowska, A.; Dénervaud, V.; Ashida, H.; Michoud, K.; Haas, D.; Yokota, A.; Danchin, A. Bacterial variations on the methionine salvage pathway. BMC Microbiol. 2004, 4, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpousis, A.J. The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007, 61, 71–87. [Google Scholar] [CrossRef] [Green Version]
- Mildenhall, K.B.; Wiese, N.; Chung, D.; Maples, V.F.; Mohanty, B.K.; Kushner, S.R. RNase E-based degradosome modulates polyadenylation of mRNAs after Rho-independent transcription terminators in Escherichia coli. Mol. Microbiol. 2016, 101, 645–655. [Google Scholar] [CrossRef] [Green Version]
- Liberati, N.T.; Urbach, J.M.; Miyata, S.; Lee, D.G.; Drenkard, E.; Wu, G.; Villanueva, J.; Wei, T.; Ausubel, F.M. Anordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 2006, 103, 2833–2838. [Google Scholar] [CrossRef] [Green Version]
- Olsen, R.H.; DeBusscher, G.; McCombie, W.R. Development of broad-host-range vectors and gene banks: Self-cloning of the Pseudomonas aeruginosa PAO chromosome. J. Bacteriol. 1982, 150, 60–69. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Xu, C.; Jin, Y.; Sun, Z. SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. mBio 2013, 4, e00419-13. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Weng, Y.; Feng, Z. Polynucleotide Phosphorylase Regulates Multiple Virulence Factors and the Stabilities of Small RNAs RsmY/Z in Pseudomonas aeruginosa. Front. Microbiol. 2016, 7, 247. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Xia, Y.; Tian, Z.; Jin, Y.; Bai, F.; Cheng, Z.; Swietnicki, W.; Wu, W.; Pan, X. Dihydrolipoamide Acetyltransferase AceF Influences the Type III Secretion System and Resistance to Oxidative Stresses through RsmY/Z in Pseudomonas aeruginosa. Microorganisms 2022, 10, 666. https://doi.org/10.3390/microorganisms10030666
Li H, Xia Y, Tian Z, Jin Y, Bai F, Cheng Z, Swietnicki W, Wu W, Pan X. Dihydrolipoamide Acetyltransferase AceF Influences the Type III Secretion System and Resistance to Oxidative Stresses through RsmY/Z in Pseudomonas aeruginosa. Microorganisms. 2022; 10(3):666. https://doi.org/10.3390/microorganisms10030666
Chicago/Turabian StyleLi, Haozhou, Yushan Xia, Zhenyang Tian, Yongxin Jin, Fang Bai, Zhihui Cheng, Wieslaw Swietnicki, Weihui Wu, and Xiaolei Pan. 2022. "Dihydrolipoamide Acetyltransferase AceF Influences the Type III Secretion System and Resistance to Oxidative Stresses through RsmY/Z in Pseudomonas aeruginosa" Microorganisms 10, no. 3: 666. https://doi.org/10.3390/microorganisms10030666
APA StyleLi, H., Xia, Y., Tian, Z., Jin, Y., Bai, F., Cheng, Z., Swietnicki, W., Wu, W., & Pan, X. (2022). Dihydrolipoamide Acetyltransferase AceF Influences the Type III Secretion System and Resistance to Oxidative Stresses through RsmY/Z in Pseudomonas aeruginosa. Microorganisms, 10(3), 666. https://doi.org/10.3390/microorganisms10030666






