Three Preceding Crops Increased the Yield of and Inhibited Clubroot Disease in Continuously Monocropped Chinese Cabbage by Regulating the Soil Properties and Rhizosphere Microbial Community
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil and Plant Preparation
2.2. Experimental Design
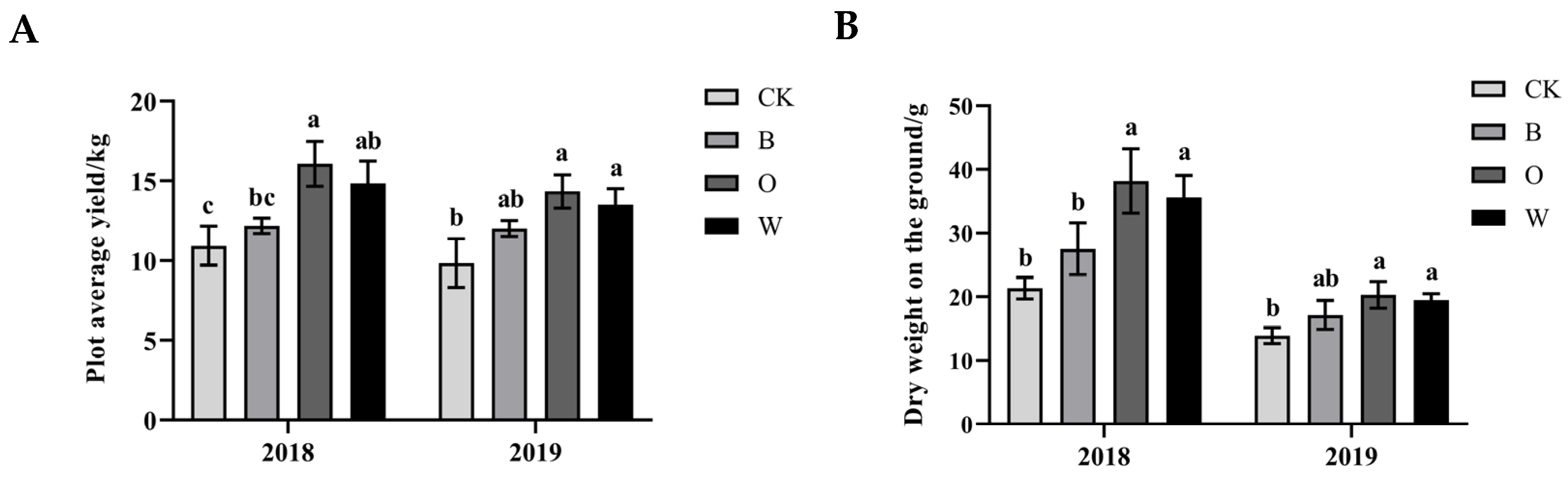
2.3. Determination of Growth and Disease
2.4. Soil Collection
2.5. Determination of Soil Chemical Properties
2.6. DNA Extraction and Quantification PCR
2.7. Analysis of Soil Microbial Diversity
3. Results
3.1. Effects of Different Preceding Crops on Growth of Chinese Cabbage
3.2. Effects of Different Preceding Crops on Incidence of Chinese Cabbage Clubroot Disease
3.3. Effects of Different Preceding Crops on Soil Chemical Properties
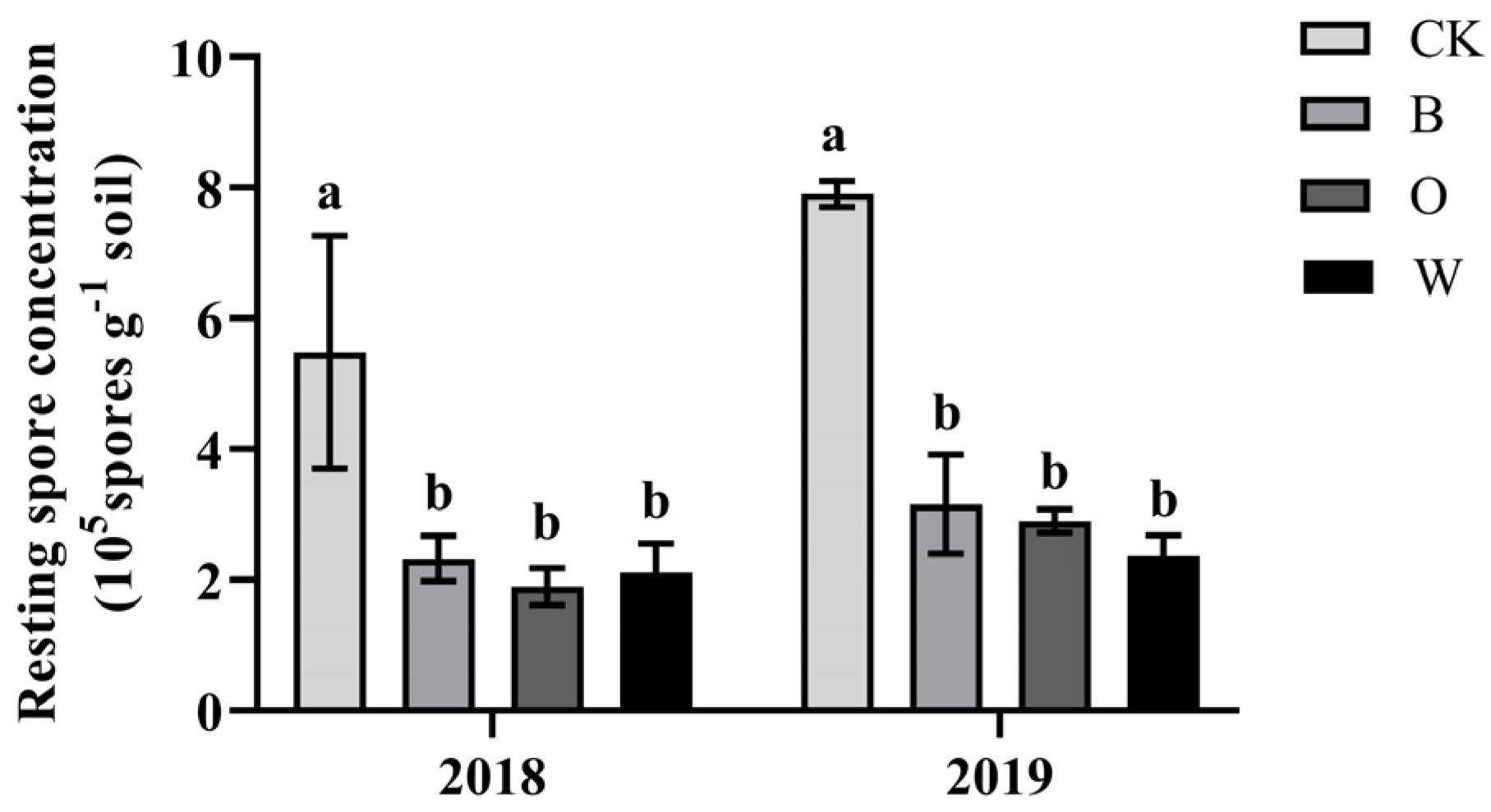
3.4. Effects of Different Preceding Crops on Resting Spores of P. brassicae
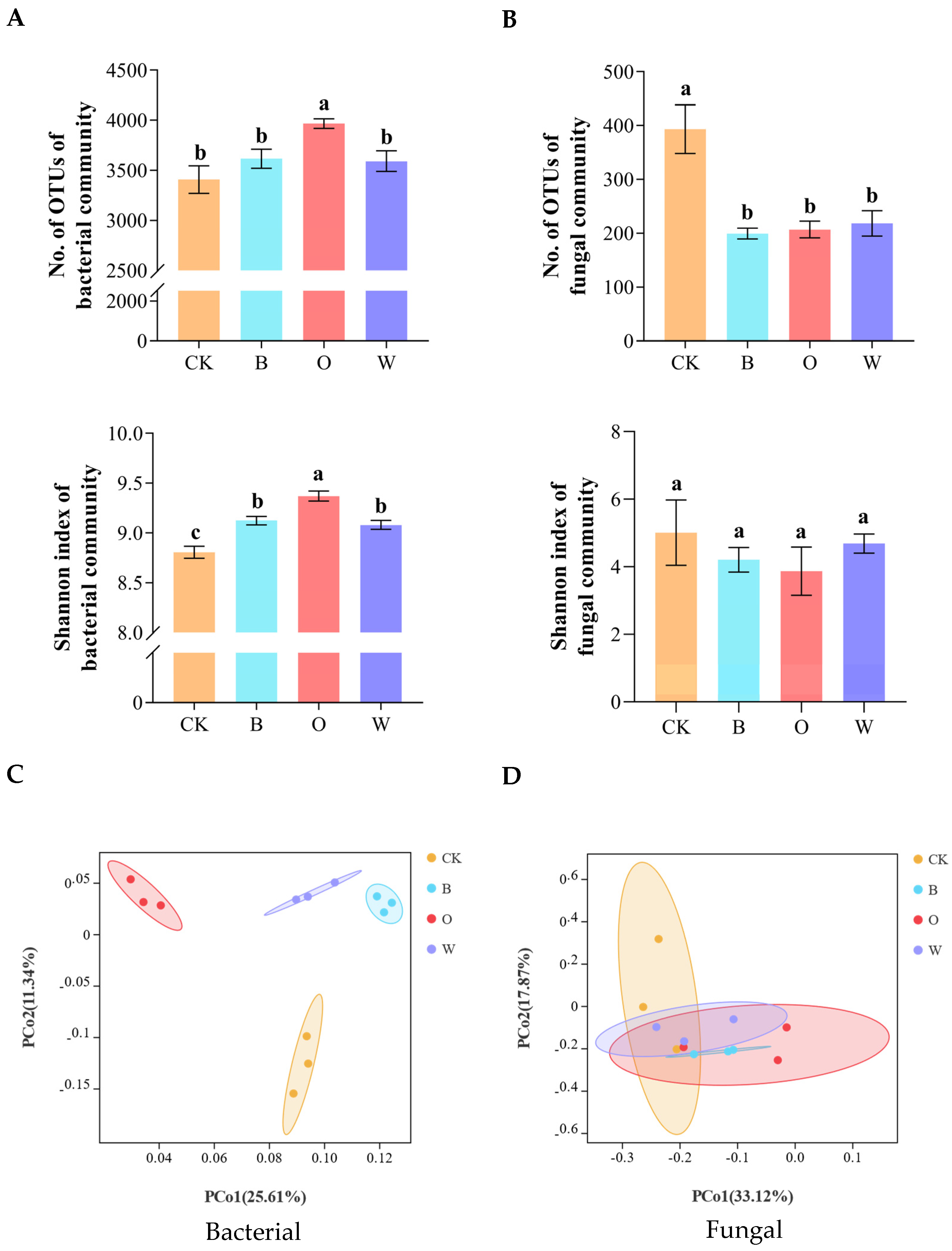
3.5. Alpha and Beta Diversities of Bacterial and Fungal Communities in Soil
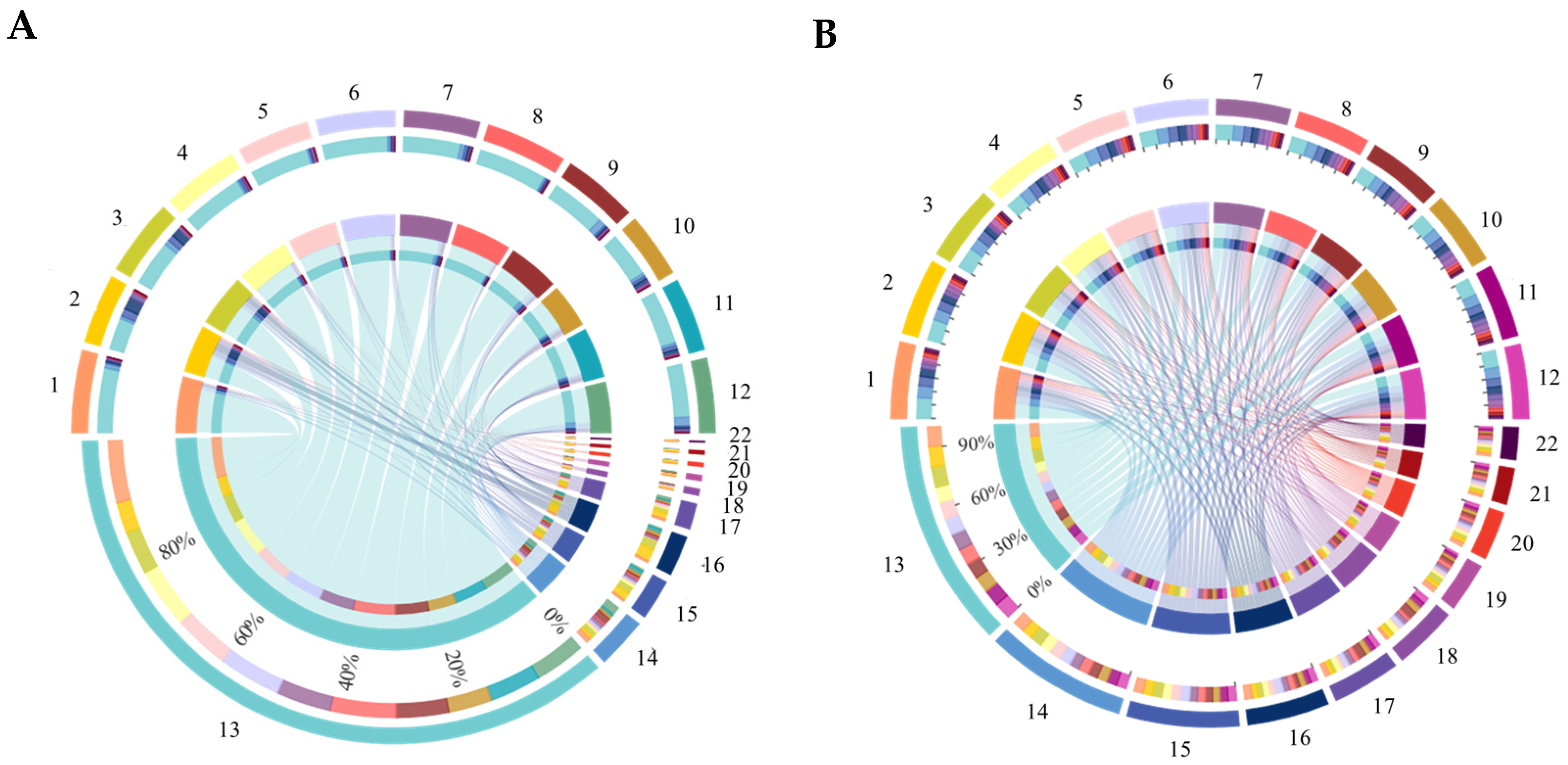
3.6. Compositions of Bacterial and Fungal Communities in Soil
3.7. Relationships between Soil Microbial Communities and Soil Chemical Properties
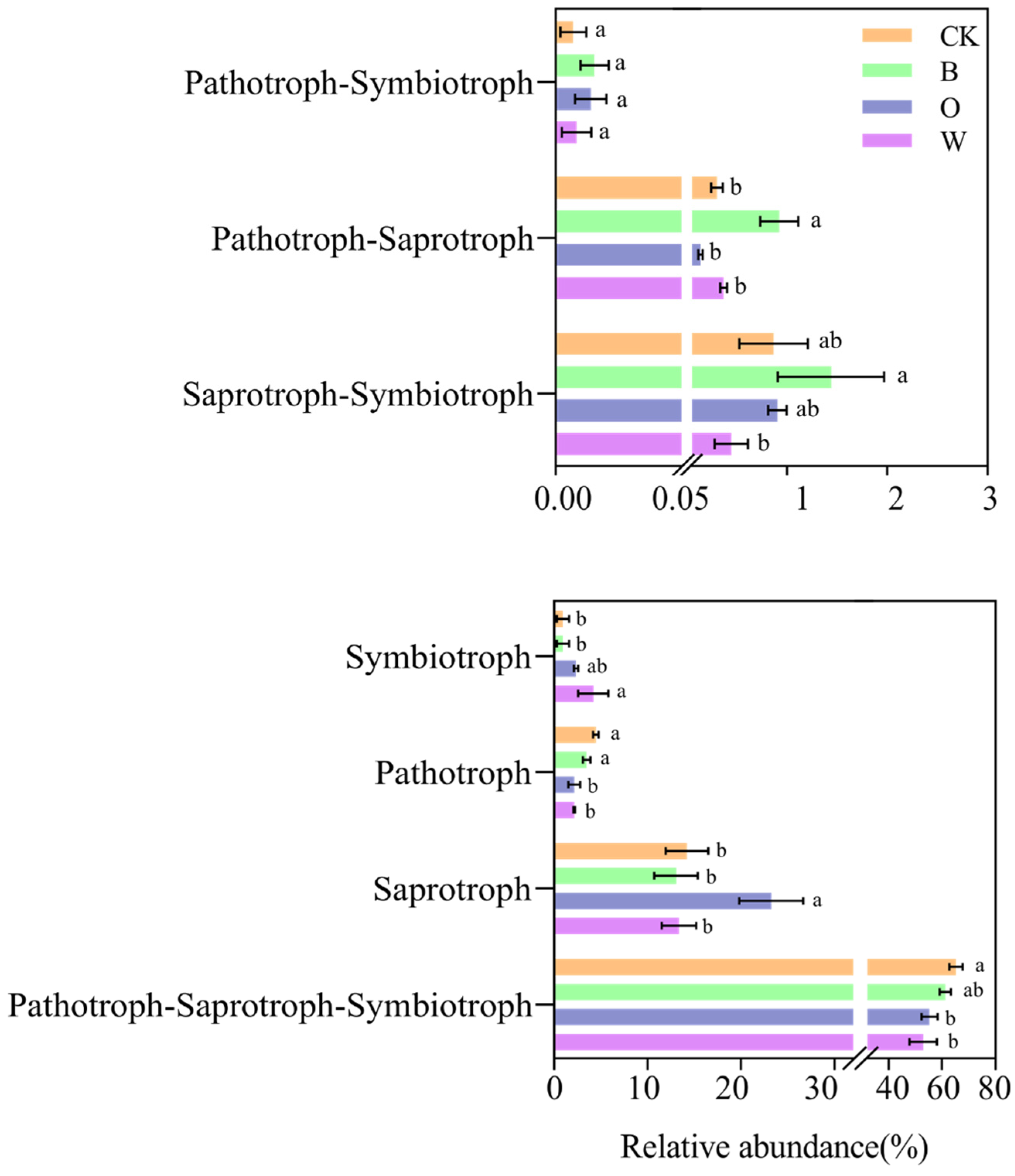
3.8. Microbial Ecological Guilds in Soil
3.9. Correlation Analysis between Resting Spores and Soil Microorganisms
4. Discussion
4.1. Effects of Rotation on Growth of Chinese Cabbage
4.2. Effects of Rotation on the Soil’s Chemical Properties
4.3. Effects of Rotation on Soil Microbial Communities
4.4. Effects of Rotation on the Clubroot of Chinese Cabbage
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hwang, S.F.; Strelkov, S.E.; Feng, J.; Gossen, B.D.; Howard, R.J. Plasmodiophora brassicae: A review of an emerging pathogen of the Canadian canola (Brassica napus) crop. Mol. Plant Pathol. 2012, 132, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Chai, A.L.; Xie, X.W.; Shi, Y.X.; Li, B.J. Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Can. J. Plant Pathol. 2014, 36, 142–153. [Google Scholar] [CrossRef]
- Dixon, G.R.; Tilston, E.L. Soil-Borne Pathogens and Their Interactions with the Soil Environment. In Soil Microbiology and Sustainable Crop Production; Springer Science and Business Media LLC: Berlin, Germany, 2010; pp. 197–271. [Google Scholar]
- Wang, B.; Li, R.; Ruan, Y.; Ou, Y.; Zhao, Y.; Shen, Q. Pineapple–banana rotation reduced the amount of Fusarium oxysporum more than maize–banana rotation mainly through modulating fungal communities. Soil Biol. Biochem. 2015, 86, 77–86. [Google Scholar] [CrossRef]
- Howard, R.J.; Strelkov, S.E.; Harding, M.W. Clubroot of cruciferous crops—New perspectives on an old disease. Can. J. Plant Pathol. 2010, 321, 43–57. [Google Scholar] [CrossRef]
- Han, Z.; Di, C.; Khashi, U.; Rahman, M.; Gao, D.; Wu, F.; Pan, K. Repeated Application of Rice Straw Stabilizes Soil Bacterial Community Composition and Inhibits Clubroot Disease. Agriculture 2021, 11, 108. [Google Scholar] [CrossRef]
- Tang, L.; Nie, S.; Li, W.; Fan, C.; Wang, S.; Wu, F.; Pan, K. Wheat straw increases the defense response and resistance of watermelon monoculture to Fusarium wilt. BMC Plant Biol. 2019, 191, 551. [Google Scholar] [CrossRef]
- Xu, W.; Liu, D.; Wu, F.; Liu, S. Root exudates of wheat are involved in suppression of Fusarium wilt in watermelon in watermelon-wheat companion cropping. Eur. J. Plant Pathol. 2014, 1411, 209–216. [Google Scholar] [CrossRef]
- Chen, T.; Lin, S.; Wu, L.; Lin, W.; Sampietro, D. Soil Sickness: Current Status and Future Perspectives. Allelopath. J. 2015, 36, 167–195. [Google Scholar]
- Li, Y.; Chi, J.; Ao, J.; Gao, X.; Liu, X.; Sun, Y.; Zhu, W. Effects of Different Continuous Cropping Years on Bacterial Community and Diversity of Cucumber Rhizosphere Soil in Solar-Greenhouse. Curr. Microbiol. 2021, 78, 2380–2390. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, F. Changes in soil chemical characters and enzyme activities during continuous monocropping of cucumber (Cucumis sativus). Pak. J. Bot. 2015, 47, 691–697. [Google Scholar]
- Larkin, R.P.; Fravel, D.R. Effects of Varying Environmental Conditions on Biological Control of Fusarium Wilt of Tomato by Nonpathogenic Fusarium spp. Phytopathology 2002, 9211, 1160–1166. [Google Scholar] [CrossRef]
- Huang, L.-F.; Song, L.-X.; Xia, X.-J.; Mao, W.-H.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q. Plant-Soil Feedbacks and Soil Sickness: From Mechanisms to Application in Agriculture. J. Chem. Ecol. 2013, 392, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, X.; Kong, W.; Wu, Y.; Wang, J. Effect of monoculture soybean on soil microbial community in the Northeast China. Plant Soil 2010, 3301, 423–433. [Google Scholar] [CrossRef]
- Jin, X.; Zhang, J.; Shi, Y.; Wu, F.; Zhou, X. Green manures of Indian mustard and wild rocket enhance cucumber resistance to Fusarium wilt through modulating rhizosphere bacterial community composition. Plant Soil 2019, 441, 283–300. [Google Scholar] [CrossRef]
- Tian, X.; Wang, D.; Mao, Z.; Pan, L.; Liao, J.; Cai, Z. Infection of Plasmodiophora brassicae changes the fungal endophyte community of tumourous stem mustard roots as revealed by high-throughput sequencing and culture-dependent methods. PLoS ONE 2019, 146, e0214975. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, L.; Guillerm-Erckelboudt, A.Y.; Gazengel, K.; Linglin, J.; Ourry, M.; Glory, P.; Sarniguet, A.; Daval, S.; Manzanares-Dauleux, M.J.; Mougel, C. Temporal dynamics of bacterial and fungal communities during the infection of Brassica rapa roots by the protist Plasmodiophora brassicae. PLoS ONE 2019, 142, e0204195. [Google Scholar] [CrossRef]
- Sharma, K.; Gossen, B.; Greenshields, D.; Selvaraj, G.; Strelkov, S.; McDonald, M.R. Reaction of Lines of the Rapid Cycling Brassica Collection and Arabidopsis thaliana to Four Pathotypes of Plasmodiophora brassicae. Plant Dis. 2013, 97, 720–727. [Google Scholar] [CrossRef][Green Version]
- Liu, R.; Pan, Y.; Bao, H.; Liang, S.; Jiang, Y.; Tu, H.; Nong, J.; Huang, W. Variations in Soil Physico-Chemical Properties along Slope Position Gradient in Secondary Vegetation of the Hilly Region, Guilin, Southwest China. Sustainability 2020, 12, 1303. [Google Scholar] [CrossRef]
- Wallenhammar, A.C.; Almquist, C.; Söderström, M.; Jonsson, A. In-field distribution of Plasmodiophora brassicae measured using quantitative real-time PCR. Plant Pathol. 2012, 611, 16–28. [Google Scholar] [CrossRef]
- Ibañez, F.; Arroyo, M.E.; Angelini, J.; Tonelli, M.L.; Muñoz, V.; Ludueña, L.; Valetti, L.; Fabra, A. Non-rhizobial peanut nodule bacteria promote maize (Zea mays L.) and peanut (Arachis hypogaea L.) growth in a simulated crop rotation system. Appl. Soil Ecol. 2014, 84, 208–212. [Google Scholar] [CrossRef]
- Jimenez-Osornio, J.J.; Gliessman, S.R. Allelopathic interference in a wild mustard (Brassica campestris L.) and broccoli (Brassica oleracea L. var. italica) Intercrop agroecosystem. In Allelochemicals: Role in Agriculture and Forestry. ACS Symposium Series; ACS Publications: Washington, DC, USA, 1987; Volume 330, pp. 262–274. [Google Scholar]
- Li, N.; Gao, D.; Zhou, X.; Chen, S.; Li, C.; Wu, F. Intercropping with Potato-Onion Enhanced the Soil Microbial Diversity of Tomato. Microorganisms 2020, 8, 834. [Google Scholar] [CrossRef]
- Ye, S.F.; Yu, J.Q.; Peng, Y.H.; Zheng, J.H.; Zou, L.Y. Incidence of Fusarium wilt in Cucumis sativus L. is promoted by cinnamic acid, an autotoxin in root exudates. Plant Soil 2004, 2631, 143–150. [Google Scholar] [CrossRef]
- Van Eerd, L.L.; Congreves, K.A.; Hayes, A.; Verhallen, A.; Hooker, D.C. Long-term tillage and crop rotation effects on soil quality, organic carbon, and total nitrogen. Can. J. Soil Sci. 2014, 943, 303–315. [Google Scholar] [CrossRef]
- Eisenhauer, N.; Bessler, H.; Engels, C.; Gleixner, G.; Habekost, M.; Milcu, A.; Partsch, S.; Sabais, A.C.W.; Scherber, C.; Steinbeiss, S.; et al. Plant diversity effects on soil microorganisms support the singular hypothesis. Ecology 2010, 912, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, C.; Kunito, T.; Aono, T.; Liu, C.T.; Oyaizu, H. Microbial indices of soil fertility. J. Appl. Microbiol. 2005, 985, 1062–1074. [Google Scholar] [CrossRef] [PubMed]
- Venter, Z.S.; Jacobs, K.; Hawkins, H.-J. The impact of crop rotation on soil microbial diversity: A meta-analysis. Pedobiologia 2016, 594, 215–223. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 681, 1–13. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, J.; Wu, F. Soil microbial communities in cucumber monoculture and rotation systems and their feedback effects on cucumber seedling growth. Plant Soil 2017, 415, 507–520. [Google Scholar] [CrossRef]
- Xiong, W.; Li, R.; Ren, Y.; Liu, C.; Zhao, Q.; Wu, H.; Jousset, A.; Shen, Q. Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biol. Biochem. 2017, 107, 198–207. [Google Scholar] [CrossRef]
- Comeau, D.; Balthazar, C.; Novinscak, A.; Bouhamdani, N.; Joly, D.L.; Filion, M. Interactions Between Bacillus Spp., Pseudomonas Spp. and Cannabis sativa Promote Plant Growth. Front. Microbiol. 2021, 12, 715758. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yu, G.; Wu, F. Responses of soil microbial communities in the rhizosphere of cucumber (Cucumis sativus L.) to exogenously applied p-hydroxybenzoic acid. J. Chem. Ecol. 2012, 388, 975–983. [Google Scholar] [CrossRef]
- Li, X.-G.; Ding, C.-F.; Zhang, T.-L.; Wang, X.-X. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014, 72, 11–18. [Google Scholar] [CrossRef]
- Gossen, B.D.; Deora, A.; Peng, G.; Hwang, S.-F.; McDonald, M.R. Effect of environmental parameters on clubroot development and the risk of pathogen spread. Can. J. Plant Pathol. 2014, 36, 37–48. [Google Scholar] [CrossRef]
- Macfarlane, I. Factors affecting the survival of Plasmodiophora brassicae Wor. in the soil and its assessment by a host test. Ann Appl Biol. Ann. Appl. Biol. 2008, 39, 239–256. [Google Scholar] [CrossRef]
- Rashid, A.; Ahmed, H.U.; Xiao, Q.; Hwang, S.F.; Strelkov, S.E. Effects of root exudates and pH on Plasmodiophora brassicae resting spore germination and infection of canola (Brassica napus L.) root hairs. Crop Prot. 2013, 48, 16–23. [Google Scholar] [CrossRef]
- Ohi, M.; Kitamura, T.; Hata, S. Stimulation by caffeic acid, coumalic acid, and corilagin of the germination of resting spores of the clubroot pathogen Plasmodiophora brassicae. Biosci. Biotechnol. Biochem. 2003, 671, 170–173. [Google Scholar] [CrossRef]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudates. Plant Cell Environ. 2009, 326, 666–681. [Google Scholar] [CrossRef]
- Suzuki, K.; Matsumiya, E.; Ueno, Y.; Mizutani, J. Some Properties of Germination-Stimulating Factor from Plants for Resting Spores of Plasmodiophora brassicae. Jpn. J. Phytopathol. 1992, 585, 699–705. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, X.; Yu, H.; Wu, F. Root exudates of potato onion are involved in the suppression of clubroot in a Chinese cabbage-potato onion-Chinese cabbage crop rotation. Eur. J. Plant Pathol. 2017, 1503, 765–777. [Google Scholar] [CrossRef]
- Bailey, K.L.; Lazarovits, G. Suppressing soil-borne diseases with residue management and organic amendments. Soil Tillage Res. 2003, 722, 169–180. [Google Scholar] [CrossRef]
- Gill, J.S.; Sivasithamparam, K.; Smettem, K.R.J. Soil moisture affects disease severity and colonisation of wheat roots by Rhizoctonia solani AG-8. Soil Biol. Biochem. 2001, 3310, 1363–1370. [Google Scholar] [CrossRef]
- Edreva, A.; Kostoff, D. Pathogenesis-Related Proteins: Research Progress in the Last 15 Years. Gen. Appl. Plant Physiol. 2005, 31, 105–124. [Google Scholar]
- Lee, Y.S.; Park, Y.S.; Anees, M.; Kim, Y.C.; Kim, Y.H.; Kim, K.Y. Nematicidal activity ofLysobacter capsiciYS1215 and the role of gelatinolytic proteins against root-knot nematodes. Biocontrol Sci. Technol. 2013, 2312, 1427–1441. [Google Scholar] [CrossRef]
- Puopolo, G.; Cimmino, A.; Palmieri, M.C.; Giovannini, O.; Evidente, A.; Pertot, I. Lysobacter capsici AZ78 produces cyclo(L-Pro-L-Tyr), a 2,5-diketopiperazine with toxic activity against sporangia of Phytophthora infestans and Plasmopara viticola. J Appl Microbiol. 2014, 1174, 1168–1180. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Li, H.; Wei, L.; Yang, J.; Liu, Q.; Wang, Y.; Wang, X.; Ji, G. Antifungal and Biocontrol Evaluation of Four Lysobacter Strains Against Clubroot Disease. Indian J. Microbiol. 2018, 583, 353–359. [Google Scholar] [CrossRef]
- Innerebner, G.; Knief, C.; Vorholt, J.A. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 2011, 7710, 3202–3210. [Google Scholar] [CrossRef]
- Xu, L.; Ravnskov, S.; Larsen, J.; Nilsson, R.H.; Nicolaisen, M. Soil fungal community structure along a soil health gradient in pea fields examined using deep amplicon sequencing. Soil Biol. Biochem. 2012, 46, 26–32. [Google Scholar] [CrossRef]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 409, 2407–2415. [Google Scholar] [CrossRef]




| EC (mS/cm) | pH | SOM (%) | AN (mg/kg) | AP (mg/kg) | AK (mg/kg) | TN (g/kg) | ||
|---|---|---|---|---|---|---|---|---|
| 2018 | CK | 0.31 ± 0.02a | 6.32 ± 0.04c | 3.43 ± 0.23a | 116.74 ± 3.71a | 107.67 ± 18.06a | 652.50 ± 32.50a | 2.63 ± 0.16b |
| B | 0.10 ± 0.01c | 6.48 ± 0.06b | 3.42 ± 0.21a | 117.39 ± 6.13a | 115.41 ± 1.25a | 611.67 ± 30.55a | 2.84 ± 0.20b | |
| O | 0.12 ± 0.01bc | 6.73 ± 0.06a | 3.10 ± 0.47a | 110.93 ± 2.84a | 107.86 ± 6.88a | 634.17 ± 23.23a | 2.09 ± 0.14c | |
| W | 0.13 ± 0.01b | 6.76 ± 0.03a | 3.36 ± 0.39a | 132.00 ± 17.52a | 117.51 ± 8.74a | 535.83 ± 18.09b | 4.24 ± 0.15a | |
| 2019 | CK | 0.22 ± 0.02a | 6.56 ± 0.06c | 3.93 ± 0.34a | 148.59 ± 19.12a | 90.45 ± 12.75a | 569.33 ± 12.86a | 1.13 ± 0.12a |
| B | 0.13 ± 0.01b | 6.55 ± 0.06c | 4.12 ± 0.50a | 157.82 ± 3.33a | 92.48 ± 5.57a | 401.33 ± 4.62c | 1.08 ± 0.04a | |
| O | 0.14 ± 0.01b | 7.08 ± 0.02a | 3.96 ± 0.43a | 160.97 ± 5.37a | 91.08 ± 3.77a | 528.00 ± 6.93b | 1.09 ± 0.02a | |
| W | 0.12 ± 0.00b | 6.67 ± 0.01b | 4.25 ± 0.18a | 154.84 ± 2.57a | 96.26 ± 7.48a | 396.00 ± 17.44c | 1.12 ± 0.04a |
| CK | B | O | W | |
|---|---|---|---|---|
| Sphingomonas | 15.99 ± 0.44a | 12.60 ± 0.37b | 12.53 ± 0.41b | 13.02 ± 0.08b |
| Gemmatimonas | 4.42 ± 0.30a | 3.48 ± 0.18b | 3.26 ± 0.210b | 3.19 ± 0.07b |
| Candidatus_Udaeobacter | 2.52 ± 1.50a | 3.27 ± 1.50a | 3.94 ± 1.91a | 4.76 ± 0.40a |
| RB41 | 1.41 ± 0.12c | 0.45 ± 0.25bc | 1.91 ± 0.19ab | 2.17 ± 0.18a |
| Pirellula | 0.86 ± 0.31b | 1.84 ± 0.53a | 2.02 ± 0.40a | 1.69 ± 0.07ab |
| Lysobacter | 1.68 ± 0.05bc | 2.23 ± 0.19a | 1.52 ± 0.13c | 1.84 ± 0.07b |
| Gemmata | 0.78 ± 0.23a | 0.94 ± 0.31a | 1.29 ± 0.35a | 1.09 ± 0.11a |
| Streptomyces | 0.42 ± 0.09a | 0.20 ± 0.06a | 0.21 ± 0.13a | 0.19 ± 0.04a |
| Marmoricola | 1.08 ± 0.34a | 0.63 ± 0.09a | 0.83 ± 0.09a | 0.65 ± 0.21a |
| Nocardioides | 0.92 ± 0.30a | 0.50 ± 0.01ab | 0.72 ± 0.06ab | 0.61 ± 0.07b |
| Chthoniobacter | 0.31 ± 0.16b | 0.61 ± 0.29ab | 0.74 ± 0.25ab | 0.90 ± 0.06a |
| Flavisolibacter | 0.55 ± 0.10a | 0.59 ± 0.05a | 0.60 ± 0.09a | 0.61 ± 0.10a |
| Haliangium | 0.51 ± 0.02a | 0.69 ± 0.05a | 0.63 ± 0.09a | 0.50 ± 0.12a |
| Rhodanobacter | 0.93 ± 0.17a | 0.44 ± 0.03b | 0.38 ± 0.02b | 0.55 ± 0.09b |
| Bryobacter | 0.65 ± 0.10a | 0.39 ± 0.01b | 0.65 ± 0.05a | 0.59 ± 0.10ab |
| Ellin6067 | 0.51 ± 0.09a | 0.52 ± 0.12a | 0.57 ± 0.06a | 0.50 ± 0.07a |
| Subgroup_10 | 0.28 ± 0.00b | 0.48 ± 0.10a | 0.53 ± 0.05a | 0.58 ± 0.08a |
| Luteimonas | 0.63 ± 0.10a | 0.36 ± 0.03b | 0.52 ± 0.03a | 0.31 ± 0.03b |
| Arenimonas | 0.24 ± 0.03c | 0.60 ± 0.01a | 0.42 ± 0.05b | 0.50 ± 0.07ab |
| CK | B | O | W | |
|---|---|---|---|---|
| Fusarium | 30.33 ± 15.22a | 30.06 ± 1.92a | 38.96 ± 16.44a | 31.36 ± 4.80a |
| Cladorrhinum | 3.59 ± 3.54b | 18.61 ± 9.49a | 1.84 ± 0.44b | 3.68 ± 2.43b |
| Acephala | 6.09 ± 4.47a | 0.30 ± 0.50a | 1.62 ± 1.08a | 2.44 ± 1.14a |
| Penicillium | 2.01 ± 1.09a | 0.31 ± 0.230a | 1.78 ± 0.94a | 0.48 ± 0.69a |
| Gibberella | 2.01 ± 1.28a | 1.81 ± 1.53a | 2.07 ± 0.22a | 0.86 ± 0.13a |
| Talaromyces | 1.47 ± 1.20a | 0.27 ± 0.33a | 0.29 ± 0.28a | 1.71 ± 1.52a |
| Conocybe | 0.15 ± 0.04a | 0.27 ± 0.13ab | 0.14 ± 0.08b | 1.06 ± 0.62b |
| Gliocladium | 0.18 ± 0.19a | 0.90 ± 0.68a | 0.62 ± 0.52a | 0.90 ± 0.40a |
| Alternaria | 2.23 ± 1.47a | 0.59 ± 0.43a | 3.27 ± 3.42a | 0.90 ± 0.89a |
| Aspergillus | 1.57 ± 0.96a | 0.89 ± 0.75a | 0.13 ± 0.10a | 0.63 ± 0.49a |
| Bacterial OTUs (Relative Abundance in Soil %) | Correlation Coefficient with Resting Spores | Phylum | Family | Genus |
|---|---|---|---|---|
| OTU 1 (8.591) | 0.765 ** | Proteobacteria | Sphingomonadaceae | Sphingomonas |
| OTU 3 (2.225) | 0.755 ** | Proteobacteria | Sphingomonadaceae | Sphingomonas |
| OTU 8 (1.148) | 0.687 * | Proteobacteria | Sphingomonadaceae | Sphingomonas |
| OTU 9 (0.972) | −0.684 * | Proteobacteria | Xanthomonadaceae | Lysobacter |
| OTU 11 (0.795) | 0.697 * | Gemmatimonadetes | Gemmatimonadaceae | Unclassified |
| OTU 17 (0.608) | 0.826 ** | Proteobacteria | Sphingomonadaceae | Sphingomonas |
| OTU 22 (0.433) | 0.626 * | Proteobacteria | Sphingomonadaceae | Sphingomonas |
| OTU 25 (0.428) | 0.754 ** | Acidobacteria | Unclassified | Unclassified |
| OTU 39 (0.590) | −0.717 ** | Gemmatimonadetes | Gemmatimonadaceae | Unclassified |
| OTU 63 (0.320) | 0.766 ** | Gemmatimonadetes | Gemmatimonadaceae | Gemmatimonas |
| OTU 92 (0.191) | 0.832 ** | Gemmatimonadetes | Gemmatimonadaceae | Unclassified |
| Fungal OTUs (Relative Abundance in Soil%) | Correlation Coefficient with Resting Spores | Phylum | Family | Genus |
|---|---|---|---|---|
| OTU 9 (2.609) | 0.720 ** | Ascomycota | Vibrisseaceae | Acephala |
| OTU 15 (0.571) | 0.907 ** | Ascomycota | Trichocomaceae | Penicillium |
| OTU 27 (0.919) | 0.822 ** | Basidiomycota | Agaricomycetes GS29 | Unclassified |
| OTU 31 (0.742) | 0.616 * | Ascomycota | Unclassified | Unclassified |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, W.; Lu, P.; Xu, T.; Pan, K. Three Preceding Crops Increased the Yield of and Inhibited Clubroot Disease in Continuously Monocropped Chinese Cabbage by Regulating the Soil Properties and Rhizosphere Microbial Community. Microorganisms 2022, 10, 799. https://doi.org/10.3390/microorganisms10040799
Zhang Y, Li W, Lu P, Xu T, Pan K. Three Preceding Crops Increased the Yield of and Inhibited Clubroot Disease in Continuously Monocropped Chinese Cabbage by Regulating the Soil Properties and Rhizosphere Microbial Community. Microorganisms. 2022; 10(4):799. https://doi.org/10.3390/microorganisms10040799
Chicago/Turabian StyleZhang, Yiping, Wei Li, Peng Lu, Tianyu Xu, and Kai Pan. 2022. "Three Preceding Crops Increased the Yield of and Inhibited Clubroot Disease in Continuously Monocropped Chinese Cabbage by Regulating the Soil Properties and Rhizosphere Microbial Community" Microorganisms 10, no. 4: 799. https://doi.org/10.3390/microorganisms10040799
APA StyleZhang, Y., Li, W., Lu, P., Xu, T., & Pan, K. (2022). Three Preceding Crops Increased the Yield of and Inhibited Clubroot Disease in Continuously Monocropped Chinese Cabbage by Regulating the Soil Properties and Rhizosphere Microbial Community. Microorganisms, 10(4), 799. https://doi.org/10.3390/microorganisms10040799





