Multilayer Regulation of Neisseria meningitidis NHBA at Physiologically Relevant Temperatures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Generation of Plasmids and Neisseria meningitidis Recombinant Strains
2.3. Polyacrylamide Gel Electrophoresis and Western Blotting
2.4. Quantitative Real-Time PCR (qRT-PCR) Experiments
2.5. FACS Analysis
2.6. RNA Stability Assay
2.7. Protein Stability Assay
2.8. Serum Bactericidal Assay (SBA)
3. Results
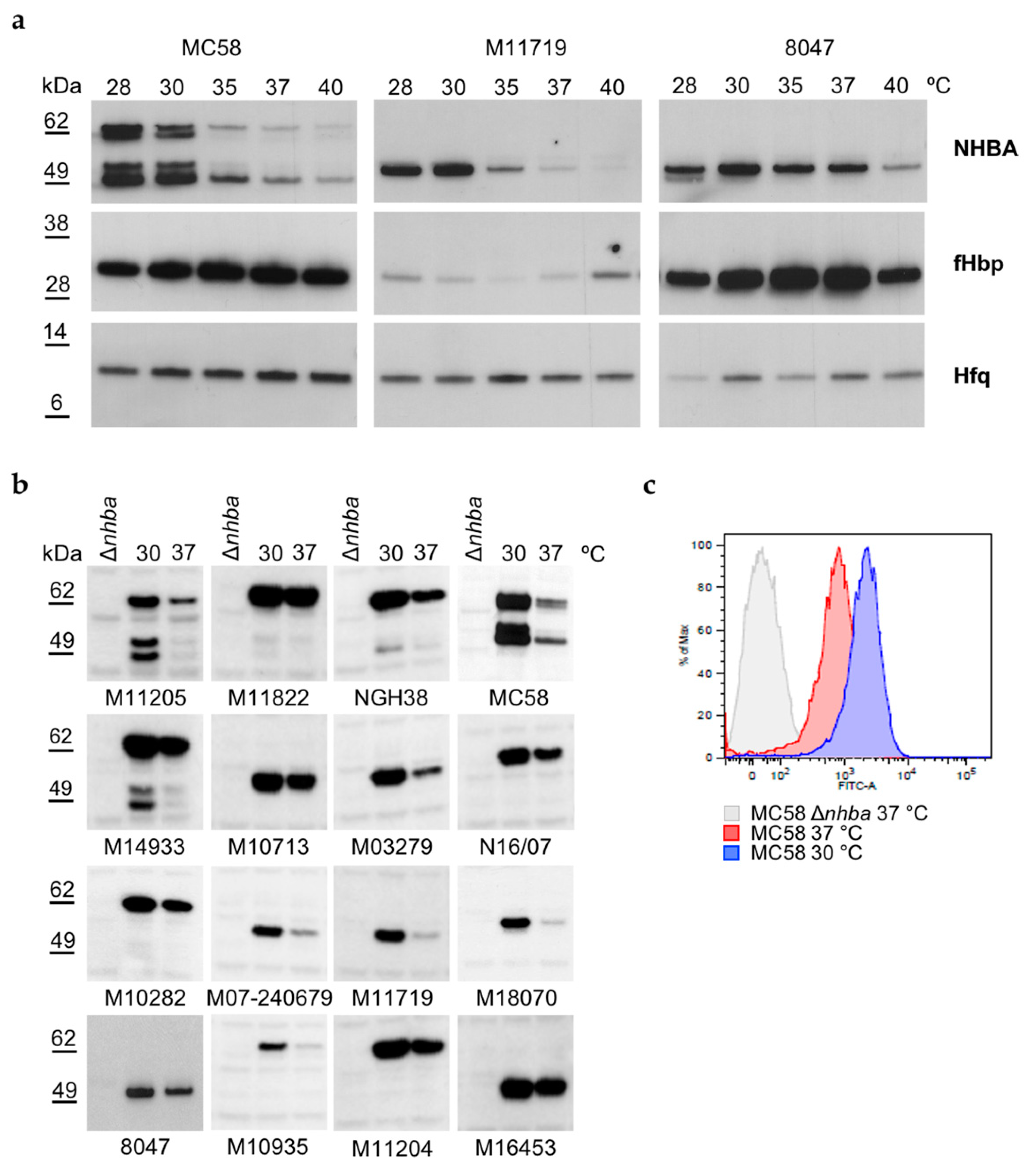
3.1. NHBA Expression and Surface Exposure Are Temperature-Dependent
3.2. NHBA Is Expressed during Active Growth
3.3. NHBA Thermoregulation Is Not Driven by the nhba Promoter
3.4. Temperature Affects nhba mRNA Half-Life
3.5. NHBA Protein Shows Higher Stability at 30 °C Respect to 37 °C
3.6. NHBA Expression Levels Correlate with Susceptibility to Complement-Mediated Killing by anti-NHBA Antibodies
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hill, D.J.; Griffiths, N.J.; Borodina, E.; Virji, M. Cellular and molecular biology of Neisseria meningitidis colonization and invasive disease. Clin. Sci. 2010, 118, 547–564. [Google Scholar] [CrossRef] [Green Version]
- Yazdankhah, S.P.; Caugant, D.A. Neisseria meningitidis: An overview of the carriage state. J. Med. Microbiol. 2004, 53 Pt 9, 821–832. [Google Scholar] [CrossRef] [Green Version]
- Stephens, D.S. Biology and pathogenesis of the evolutionarily successful, obligate human bacterium Neisseria meningitidis. Vaccine 2009, 27 (Suppl. 2), B71–B77. [Google Scholar] [CrossRef] [Green Version]
- Carbonnelle, E.; Hill, D.J.; Morand, P.; Griffiths, N.J.; Bourdoulous, S.; Murillo, I.; Nassif, X.; Virji, M. Meningococcal interactions with the host. Vaccine 2009, 27 (Suppl. 2), B78–B89. [Google Scholar] [CrossRef] [PubMed]
- Virji, M. Pathogenic neisseriae: Surface modulation, pathogenesis and infection control. Nat. Rev. Microbiol. 2009, 7, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, T.C.; Quattroni, P.; Exley, R.M.; Tang, C.M. Transcellular passage of Neisseria meningitidis across a polarized respiratory epithelium. Infect. Immun. 2010, 78, 3832–3847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrile, R.; Kasendra, M.; Rossi-Paccani, S.; Merola, M.; Pizza, M.; Baldari, C.; Soriani, M.; Aricò, B. Neisseria meningitidis subverts the polarized organization and intracellular trafficking of host cells to cross the epithelial barrier. Cell. Microbiol. 2015, 17, 1365–1375. [Google Scholar] [CrossRef]
- Deghmane, A.E.; Giorgini, D.; Larribe, M.; Alonso, J.M.; Taha, M.K. Down-regulation of pili and capsule of Neisseria meningitidis upon contact with epithelial cells is mediated by CrgA regulatory protein. Mol. Microbiol. 2002, 43, 1555–1564. [Google Scholar] [CrossRef]
- Hey, A.; Li, M.S.; Hudson, M.J.; Langford, P.R.; Kroll, J.S. Transcriptional profiling of Neisseria meningitidis interacting with human epithelial cells in a long-term in vitro colonization model. Infect. Immun. 2013, 81, 4149–4159. [Google Scholar] [CrossRef] [Green Version]
- Lappann, M.; Claus, H.; van Alen, T.; Harmsen, M.; Elias, J.; Molin, S.; Vogel, U. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol. Microbiol. 2010, 75, 1355–1371. [Google Scholar] [CrossRef]
- Lappann, M.; Vogel, U. Biofilm formation by the human pathogen Neisseria meningitidis. Med. Microbiol. Immunol. 2010, 199, 173–183. [Google Scholar] [CrossRef]
- Larson, J.A.; Higashi, D.L.; Stojiljkovic, I.; So, M. Replication of Neisseria meningitidis within epithelial cells requires TonB-dependent acquisition of host cell iron. Infect. Immun. 2002, 70, 1461–1467. [Google Scholar] [CrossRef] [Green Version]
- Schryvers, A.B.; Stojiljkovic, I. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 1999, 32, 1117–1123. [Google Scholar] [CrossRef]
- Laver, J.R.; Hughes, S.E.; Read, R.C. Neisserial Molecular Adaptations to the Nasopharyngeal Niche. Adv. Microb. Physiol. 2015, 66, 323–355. [Google Scholar]
- Jamet, A.; Rousseau, C.; Monfort, J.B.; Frapy, E.; Nassif, X.; Martin, P. A two-component system is required for colonization of host cells by meningococcus. Microbiology 2009, 155 Pt 7, 2288–2295. [Google Scholar] [CrossRef] [Green Version]
- Mendum, T.A.; Newcombe, J.; Mannan, A.A.; Kierzek, A.M.; McFadden, J. Interrogation of global mutagenesis data with a genome scale model of Neisseria meningitidis to assess gene fitness in vitro and in sera. Genome Biol. 2011, 12, R127. [Google Scholar] [CrossRef] [Green Version]
- Lomholt, H.; Poulsen, K.; Caugant, D.A.; Kilian, M. Molecular polymorphism and epidemiology of Neisseria meningitidis immunoglobulin A1 proteases. Proc. Natl. Acad. Sci. USA 1992, 89, 2120–2124. [Google Scholar] [CrossRef] [Green Version]
- Keck, T.; Leiacker, R.; Riechelmann, H.; Rettinger, G. Temperature profile in the nasal cavity. Laryngoscope 2000, 110, 651–654. [Google Scholar] [CrossRef]
- McFadden, E.R.; Pichurko, B.M.; Bowman, H.F.; Ingenito, E.; Burns, S.; Dowling, N.; Solway, J. Thermal mapping of the airways in humans. J. Appl. Physiol. 1985, 58, 564–570. [Google Scholar] [CrossRef]
- Loh, E.; Kugelberg, E.; Tracy, A.; Zhang, Q.; Gollan, B.; Ewles, H.; Chalmers, R.; Pelicic, V.; Tang, C.M. Temperature triggers immune evasion by Neisseria meningitidis. Nature 2013, 502, 237–240. [Google Scholar] [CrossRef] [Green Version]
- Barnwal, R.P.; Loh, E.; Godin, K.S.; Yip, J.; Lavender, H.; Tang, C.M.; Varani, G. Structure and mechanism of a molecular rheostat, an RNA thermometer that modulates immune evasion by Neisseria meningitidis. Nucleic Acids Res. 2016, 44, 9426–9437. [Google Scholar] [PubMed] [Green Version]
- Loh, E.; Lavender, H.; Tan, F.; Tracy, A.; Tang, C.M. Thermoregulation of Meningococcal fHbp, an Important Virulence Factor and Vaccine Antigen, Is Mediated by Anti-ribosomal Binding Site Sequences in the Open Reading Frame. PLoS Pathog. 2016, 12, e1005794. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, J.E.H.; Andersson, C.; Jacobsson, S.; Loh, E. Novel hypercapsulation RNA thermosensor variants in Neisseria meningitidis and their association with invasive meningococcal disease: A genetic and phenotypic investigation and molecular epidemiological study. Lancet Microbe 2020, 1, e319–e327. [Google Scholar] [CrossRef]
- Kortmann, J.; Narberhaus, F. Bacterial RNA thermometers: Molecular zippers and switches. Nat. Rev. Microbiol. 2012, 10, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Loh, E.; Righetti, F.; Eichner, H.; Twittenhoff, C.; Narberhaus, F. RNA Thermometers in Bacterial Pathogens. Microbiol. Spectr. 2018, 6, 55–73. [Google Scholar] [CrossRef]
- Gualerzi, C.O.; Giuliodori, A.M.; Pon, C.L. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 2003, 331, 527–539. [Google Scholar] [CrossRef]
- Goldenberg, D.; Azar, I.; Oppenheim, A.B. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol. Microbiol. 1996, 19, 241–248. [Google Scholar] [CrossRef]
- Prud’homme-Généreux, A.; Beran, R.K.; Iost, I.; Ramey, C.S.; Mackie, G.A.; Simons, R.W. Physical and functional interactions among RNase Epolynucleotide phosphorylase and the cold-shock protein, CsdA: Evidence for a ‘cold shock degradosome’. Mol. Microbiol. 2004, 54, 1409–1421. [Google Scholar] [CrossRef]
- Cruz, J.A.; Westhof, E. The dynamic landscapes of RNA architecture. Cell 2009, 136, 604–609. [Google Scholar] [CrossRef]
- Giuliodori, A.M.; Di Pietro, F.; Marzi, S.; Masquida, B.; Wagner, R.; Romby, P.; Gualerzi, C.O.; Pon, C.L. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell 2010, 37, 21–33. [Google Scholar] [CrossRef]
- Lappann, M.; Otto, A.; Brauer, M.; Becher, D.; Vogel, U.; Johswich, K. Impact of Moderate Temperature Changes on Neisseria meningitidis Adhesion Phenotypes and Proteome. Infect. Immun. 2016, 84, 3484–3495. [Google Scholar] [CrossRef] [Green Version]
- Bambini, S.; Muzzi, A.; Olcen, P.; Rappuoli, R.; Pizza, M.; Comanducci, M. Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine 2009, 27, 2794–2803. [Google Scholar] [CrossRef]
- Jacobsson, S.; Thulin, S.; Mölling, P.; Unemo, M.; Comanducci, M.; Rappuoli, R.; Olcén, P. Sequence constancies and variations in genes encoding three new meningococcal vaccine candidate antigens. Vaccine 2006, 24, 2161–2168. [Google Scholar] [CrossRef]
- Comanducci, M.; Bambini, S.; Brunelli, B.; Adu-Bobie, J.; Aricò, B.; Capecchi, B.; Giuliani, M.M.; Masignani, V.; Santini, L.; Savino, S.; et al. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 2002, 195, 1445–1454. [Google Scholar] [CrossRef] [Green Version]
- Muzzi, A.; Mora, M.; Pizza, M.; Rappuoli, R.; Donati, C. Conservation of meningococcal antigens in the genus Neisseria. mBio 2013, 4, e00163-13. [Google Scholar] [CrossRef] [Green Version]
- Morelle, S.; Carbonnelle, E.; Nassif, X. The REP2 repeats of the genome of Neisseria meningitidis are associated with genes coordinately regulated during bacterial cell interaction. J. Bacteriol. 2003, 185, 2618–2627. [Google Scholar] [CrossRef] [Green Version]
- Vacca, I.; Del Tordello, E.; Gasperini, G.; Pezzicoli, A.; Di Fede, M.; Rossi Paccani, S.; Marchi, S.; Mubaiwa, T.D.; Hartley-Tassell, L.E.; Jennings, M.P.; et al. Neisserial Heparin Binding Antigen (NHBA) Contributes to the Adhesion of Neisseria meningitidis to Human Epithelial Cells. PLoS ONE 2016, 11, e0162878. [Google Scholar] [CrossRef] [Green Version]
- Arenas, J.; Nijland, R.; Rodriguez, F.J.; Bosma, T.N.; Tommassen, J. Involvement of three meningococcal surface-exposed proteins, the heparin-binding protein NHBA, the α-peptide of IgA protease and the autotransporter protease NalP, in initiation of biofilm formation. Mol. Microbiol. 2013, 87, 254–268. [Google Scholar] [CrossRef]
- Serruto, D.; Spadafina, T.; Ciucchi, L.; Lewis, L.A.; Ram, S.; Tontini, M.; Santini, L.; Biolchi, A.; Seib, K.L.; Giuliani, M.M.; et al. Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc. Natl. Acad. Sci. USA 2010, 107, 3770–3775. [Google Scholar] [CrossRef] [Green Version]
- Esposito, V.; Musi, V.; de Chiara, C.; Veggi, D.; Serruto, D.; Scarselli, M.; Kelly, G.; Pizza, M.; Pastore, A. Structure of the C-terminal domain of Neisseria heparin binding antigen (NHBA), one of the main antigens of a novel vaccine against Neisseria meningitidis. J. Biol. Chem. 2011, 286, 41767–41775. [Google Scholar] [CrossRef] [Green Version]
- Serruto, D.; Bottomley, M.J.; Ram, S.; Giuliani, M.M.; Rappuoli, R. The new multicomponent vaccine against meningococcal serogroup, B.; 4CMenB: Immunological, functional and structural characterization of the antigens. Vaccine 2012, 30 (Suppl. 2), B87–B97. [Google Scholar] [CrossRef] [Green Version]
- Giuliani, M.M.; Biolchi, A.; Serruto, D.; Ferlicca, F.; Vienken, K.; Oster, P.; Rappuoli, R.; Pizza, M.; Donnelly, J. Measuring antigen-specific bactericidal responses to a multicomponent vaccine against serogroup B meningococcus. Vaccine 2010, 28, 5023–5030. [Google Scholar] [CrossRef]
- Sambrook, J.F.E.M.T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Klock, H.E.; Lesley, S.A. The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol. Biol. 2009, 498, 91–103. [Google Scholar]
- Giuliani, M.M.; Santini, L.; Brunelli, B.; Biolchi, A.; Aricò, B.; Di Marcello, F.; Cartocci, E.; Comanducci, M.; Masignani, V.; Lozzi, L.; et al. The region comprising amino acids 100 to 255 of Neisseria meningitidis lipoprotein GNA 1870 elicits bactericidal antibodies. Infect. Immun. 2005, 73, 1151–1160. [Google Scholar] [CrossRef] [Green Version]
- Seib, K.L.; Oriente, F.; Adu-Bobie, J.; Montanari, P.; Ferlicca, F.; Giuliani, M.M.; Rappuoli, R.; Pizza, M.; Delany, I. Influence of serogroup B meningococcal vaccine antigens on growth and survival of the meningococcus in vitro and in ex vivo and in vivo models of infection. Vaccine 2010, 28, 2416–2427. [Google Scholar] [CrossRef] [Green Version]
- Lam, O.; Wheeler, J.; Tang, C.M. Thermal control of virulence factors in bacteria: A hot topic. Virulence 2014, 5, 852–862. [Google Scholar] [CrossRef] [Green Version]
- Hussein, H.; Fris, M.E.; Salem, A.H.; Wiemels, R.E.; Bastock, R.A.; Righetti, F.; Burke, C.A.; Narberhaus, F.; Carroll, R.K.; Hassan, N.S.; et al. An unconventional RNA-based thermosensor within the 5′ UTR of Staphylococcus aureus cidA. PLoS ONE 2019, 14, e0214521. [Google Scholar] [CrossRef]
- Catalan-Moreno, A.; Cela, M.; Menendez-Gil, P.; Irurzun, N.; Caballero, C.J.; Caldelari, I.; Toledo-Arana, A. RNA thermoswitches modulate Staphylococcus aureus adaptation to ambient temperatures. Nucleic Acids Res. 2021, 49, 3409–3426. [Google Scholar] [CrossRef]
- Carrier, M.C.; Ng Kwan Lim, E.; Jeannotte, G.; Massé, E. Acting Effectors Versus RNA. Front. Microbiol. 2020, 11, 609237. [Google Scholar] [CrossRef]
- Turner, D.P.; Wooldridge, K.G.; Ala’Aldeen, D.A. Autotransported serine protease A of Neisseria meningitidis: An immunogenic, surface-exposed outer membrane, and secreted protein. Infect. Immun. 2002, 70, 4447–4461. [Google Scholar] [CrossRef] [Green Version]
- Van Ulsen, P.; Van Alphen, L.; Ten Hove, J.; Fransen, F.; Van Der Ley, P.; Tommassen, J. A Neisserial autotransporter NalP modulating the processing of other autotransporters. Mol. Microbiol. 2003, 50, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Roussel-Jazede, V.; Jongerius, I.; Bos, M.P.; Tommassen, J.; van Ulsen, P. NalP-mediated proteolytic release of lactoferrin-binding protein B from the meningococcal cell surface. Infect. Immun. 2010, 78, 3083–3089. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borghi, S.; Antunes, A.; Haag, A.F.; Spinsanti, M.; Brignoli, T.; Ndoni, E.; Scarlato, V.; Delany, I. Multilayer Regulation of Neisseria meningitidis NHBA at Physiologically Relevant Temperatures. Microorganisms 2022, 10, 834. https://doi.org/10.3390/microorganisms10040834
Borghi S, Antunes A, Haag AF, Spinsanti M, Brignoli T, Ndoni E, Scarlato V, Delany I. Multilayer Regulation of Neisseria meningitidis NHBA at Physiologically Relevant Temperatures. Microorganisms. 2022; 10(4):834. https://doi.org/10.3390/microorganisms10040834
Chicago/Turabian StyleBorghi, Sara, Ana Antunes, Andreas F. Haag, Marco Spinsanti, Tarcisio Brignoli, Enea Ndoni, Vincenzo Scarlato, and Isabel Delany. 2022. "Multilayer Regulation of Neisseria meningitidis NHBA at Physiologically Relevant Temperatures" Microorganisms 10, no. 4: 834. https://doi.org/10.3390/microorganisms10040834
APA StyleBorghi, S., Antunes, A., Haag, A. F., Spinsanti, M., Brignoli, T., Ndoni, E., Scarlato, V., & Delany, I. (2022). Multilayer Regulation of Neisseria meningitidis NHBA at Physiologically Relevant Temperatures. Microorganisms, 10(4), 834. https://doi.org/10.3390/microorganisms10040834






